Abstract
Age at onset of alcohol dependence (AO-AD) is a defining feature of multiple drinking typologies. AO-AD is heritable and likely shares genetic liability with other aspects of alcohol consumption. We examine whether polygenic variation in AO-AD, based on a genome-wide association study (GWAS), was associated with AO-AD and other aspects of alcohol consumption in two independent samples. Genetic risk scores (GRS) were created based on AO-AD GWAS results from a discovery sample of 1788 regular drinkers from extended pedigrees from the Collaborative Study of the Genetics of Alcoholism (COGA). GRS were used to predict AO-AD, AD and Alcohol dependence symptom count (AD-SX), age at onset of intoxication (AO-I), as well as maxdrinks in regular drinking participants from two independent samples—the Study of Addictions: Genes and Environment (SAGE; n=2336) and an Australian sample (OZ-ALC; n=5816). GRS for AO-AD from COGA explained a modest but significant proportion of the variance in all alcohol-related phenotypes in SAGE. Despite including effect sizes associated with large numbers of single nucleotide polymorphisms (SNPs; >110 000), GRS explained, at most, 0.7% of the variance in these alcohol measures in this independent sample. In OZ-ALC, significant but even more modest associations were noted with variance estimates ranging from 0.03 to 0.16%. In conclusion, there is modest evidence that genetic variation in AO-AD is associated with liability to other aspects of alcohol involvement.
Similar content being viewed by others
Introduction
Multiple epidemiological and genetically informed studies have documented the importance of age at onset of alcohol dependence (AO-AD) as a key feature of sub-types of alcoholism that vary in etiology and severity.1, 2, 3, 4, 5, 6 For instance, Cloninger et al.3 identified Type I and II alcoholics who were distinguished by, among other features, age at onset of alcohol problems. Similarly, Babor et al.2 defined Type A and B alcoholics—the latter were distinguished by early onset of alcohol problems. Across these typologies, early-onset problematic use was consistently associated with a more severe form of the disorder, which was often accompanied by polysubstance use and other psychiatric comorbidity, particularly externalizing disorders.
AO-AD is also correlated with other features of drinking. For instance, earlier AO-AD is associated with more alcohol dependence symptoms,2 and this relationship may be influenced by shared genetic liability. Cloninger et al.7 also posited that Type II/early-onset alcoholism may represent a more heritable form of the disorder. While there have not been studies that examine the heritability of AO-AD, numerous studies have robustly documented the role of additive genetic influences on AD itself8, 9 and on both age at first drink10, 11, 12 and the speed of transitioning from first use to the development of alcohol problems.13, 14 In support of these findings, a recent genome-wide association study (GWAS) in extended pedigrees from the Collaborative Study of the Genetics of Alcoholism (COGA) found several single nucleotide polymorphisms (SNPs) that were significantly associated with AO-AD (P<5E−8).15 For rs2168784, the most significant SNP, 30% of those homozygous for the minor allele met criteria for alcohol dependence while only 19% of those homozygous for the major allele did. This SNP was also associated with AD diagnosis in the COGA dataset.
To our knowledge, none of the top SNP effects identified in the previous study by Kapoor et al.15 have yet been replicated. However, the significance associated with individual top SNPs might be subject to sample-specific characteristics (for example, families densely affected for alcoholism). Recently, investigators have begun to use genome-wide risk scores (GRS) that reflect the polygenic and aggregate nature of genotypic effects. Effect sizes generated for one phenotype in a given sample can be used to generate GRS in additional samples; the association between these GRS and a similar phenotype may be seen as evidence for replication while correlations between the GRS and other related phenotypes provide support for shared genetic underpinnings.16
The present study utilizes GRS generated from the analysis of AO-AD in COGA (discovery sample) conducted by Kapoor et al.15 and applies these scores to two independent samples, the Study of Addictions Genes and Environment (SAGE; using the portion independent of the COGA subjects who were in the discovery sample)17 and a sample of Australian subjects (OZ-ALC).18 In contrast to COGA, which is comprised of extended pedigrees with a high rates of alcohol dependence, SAGE consisted of alcohol-dependent cases and alcohol exposed but non-dependent controls. Even though SAGE subjects were drawn from studies that used family history of alcohol and drug dependence to ascertain cases, including COGA, all overlapping subjects were removed and all subjects were unrelated to each other. OZ-ALC, on the other hand, consisted of pedigrees that were derived from various sources, including family studies ascertained for heavy drinking and heavy smoking and a sample consisting of large sibships. Despite being similar to COGA for sibship size, the density of alcohol-related problems in the OZ-ALC pedigrees is substantially lower. The variability in SAGE and OZ-ALC allowed us to investigate the generalizability of the COGA findings.
Overall, we were interested in replicating and generalizing our prior findings and extending them to other alcohol-related phenotypes. Thus, our goals were twofold: first, we examine whether GRS created from the GWAS of AO-AD in the COGA discovery sample15 is associated with AO-AD in the independent portion of SAGE and in OZ-ALC. Second, we examine whether GRS for AO-AD is associated with other features of alcohol involvement, including age at first intoxication, lifetime maximum drinks in a 24-h period and number of symptoms and diagnosis of alcohol dependence in the SAGE and OZ-ALC datasets.
Materials and methods
Sample
Data were drawn from the three sources described below. The institutional review board at each contributing institution reviewed and approved the protocols.
Discovery sample
The discovery sample was genome-wide SNP data on 1788 regular drinkers (defined below) from 118 large European-American families densely affected with alcoholism;15 subjects from that study who were not regular drinkers were excluded. Ascertainment was based on a proband in treatment for alcohol dependence who had at least two first-degree relatives affected by alcohol dependence. Of these subjects, 685 met criteria for DSM-IV alcohol dependence (mean age of onset 22.5 years). A genome-wide Cox proportional hazards regression model was used to test the association between age at onset of AD and 4 058 415 imputed SNPs with minor allele frequency ⩾5%.15 A robust sandwich variance estimators approach was used to account for the familial correlation among observations (https://cran.r-project.org/web/packages/survival/survival.pdf).
Replication samples
SAGE consisted of 2593 unrelated European-American subjects. Of these, 2336 individuals who reported regular drinking were included in these analyses. Subjects were selected from three large, complementary studies: COGA,19 Family Study of Cocaine Dependence (FSCD)20 and Collaborative Genetic Study of Nicotine Dependence (COGEND).21 Further details of the SAGE sample are available elsewhere.17 One hundred and twenty nine individuals who were both in SAGE and the COGA discovery sample were excluded. The sample consisted of alcohol-dependent cases (N=1167, mean age at onset 24.7 years) and alcohol-exposed controls (N=1169).
The OZ-ALC sample consisted of 6169 individuals (for this study, 5816 regular drinkers were included) from 2356 families ascertained from 3 coordinated studies derived from a larger Australian twin registry: (i) the Nicotine Addiction Genetics (NAG) Study which ascertained heavy smoking index cases; (ii) the OZ-ALC-EDAC study, which ascertained index cases with a history of alcohol dependence or scoring above the 85th centile for heaviness of drinking factor score (operationalized as in the study by Grant et al.22); (iii) the OZ-ALC-BIGSIB study, which ascertained large sibships (4–14 full siblings), regardless of sibling phenotypes. Further details regarding recruitment may be found in the study by Heath et al.18 OZ-ALC was not ascertained for alcohol dependence (although some contributing studies were ascertained for heavy smoking and heavy alcohol consumption) and includes 1714 alcohol-dependent individuals (mean age of onset 26.3 years).
Phenotypic assessments
The discovery and replication samples utilized versions of the Semi-Structured Assessment for the Genetics of Alcoholism (SSAGA)23, 24 to obtain interview-based self-report data on ages of onset and other alcohol-related measures. In the discovery sample, AO-AD was defined as the age at which individuals reported first experiencing three or more of seven DSM-IV alcohol dependence criteria clustering within a 12-month period. Only individuals who were regular drinkers (that is, reported a lifetime history of drinking at least once a month for 6 months or longer) were included. As is the norm for Cox survival modeling, those who did not meet criteria for DSM-IV AD were censored at their age at interview.
For the present analyses, in addition to AO-AD, which was coded identically as in the discovery sample, the following measures were drawn from SAGE and OZ-ALC:
Alcohol-related measures
AO-I, defined as the age at which the respondent reported first getting drunk (that is, their speech was slurred or they felt unsteady on their feet).
AD diagnosis (binary) was based on DSM-IV; individuals who endorsed three or more criteria that clustered within a single 12-month period were diagnosed with AD.
Alcohol dependence symptom count (AD-SX) was defined as the sum of the seven DSM-IV dependence criteria.
Maxdrinks, defined as the maximum number of drinks consumed in a single 24-h period during their lifetime. The measure was Winsorized at the 95th percentile (>100 drinks) and log (10) transformed for analyses.
Negative control
Height, via self-report, was used as a negative control.
Genotyping in discovery sample
Genotyping was conducted using the Illumina OmniExpress array (Illumina, San Diego, CA, USA). A total of 4 058 415 SNPs that were imputed in BEAGLE (https://faculty.washington.edu/browning/beagle/beagle.html) were analyzed. Further details are available in the manuscript by Kapoor et al.;15 data are available at dbGaP phs000763.
Genotyping in replication samples
For SAGE, DNAs were genotyped on the Illumina Human 1 M beadchip (Illumina) by the Center for Inherited Diseases Research (CIDR) at the Johns Hopkins University; data are available at dbGaP phs000092. A total of 948 658 SNPs passed data-cleaning procedures and further within sample filtering for autosomal and X-chromosome markers yielded 948 142 markers. HapMap genotyping controls, duplicates, related subjects and outliers were removed from the sample set.17 The software package EIGENSTRAT25 was used to calculate principal components reflecting ancestral differences. Only genotyped SNPs were selected from SAGE, resulting in 669 984 overlapping SNPs which were further pruned for linkage disequilibrium (maximum pairwise r2=0.25 within sliding windows of 100 SNPs), resulting in 90 365 SNPs that were used for all subsequent analyses.
For OZ-ALC, most subjects (N=4601) were genotyped on the Illumina CNV370-Quadv3 (Illumina); genotyping on a small number of additional individuals was conducted on the Illumina 317 K (N=20) and 610 Quad v1 (N=517) platforms; data are available at dbGaP phs000181 (see the study by Medland et al.,26 for additional details). To account for the lower density of genotyped SNPs and variation in platform contents, imputation to HapMap (http://hapmap.ncbi.nlm.nih.gov) CEU I+II data (release 22, build 36) was conducted in MACH27 and best guess genotypes were selected based on Rsq⩾0.3 and imputation quality⩾0.9, resulting in 112 594 autosomal SNPs. Nuanced admixture was determined using EIGENSTRAT25 and outliers were removed, as outlined in the study by Heath et al.18
Association using GRS in replication datasets
Based on effect sizes for the analysis of AO-AD generated in the discovery (COGA) sample, GRS at P-value thresholds of 0.01 (GRS0.01), 0.05 (GRS0.05), 0.10 (GRS0.1) and 0.50 (GRS0.5) were created in SAGE and OZ-ALC sample using PLINK28 and SAS (SAS Institute, Cary, NC, USA). Briefly, SNPs in COGA that were significant at each P-value threshold (for example, P<0.01) were selected. For each SNP, the effect size was calculated as the natural logarithm transformation of the hazard ratio from COGA. For every individual in SAGE and OZ-ALC, this effect size was multiplied by the number of copies of reference allele, and this product was summed across all SNPs.28 For the SAGE dataset, the number of SNPs for each score was: GRSall (110 797), GRS0.5 (58 374), GRS0.1 (12 254), GRS0.05 (6147) and GRS0.01 (1441), while for OZ-ALC number of SNPs for each score was: GPSall (112 594), GPS0.5 (57 053), GPS0.1 (12 161), GPS0.05 (6268) and GPS0.01 (1402). The resulting GRS was used to predict AO-AD, as well as other measures (AO-R, AO-I, AD-SX and Maxdrinks) in those datasets. Associations between age of onset measures and GRS were conducted using Cox proportional hazards analysis. Logistic and linear regression was used for dichotomous (AD) and continuously distributed (AD-SX, Maxdrinks) measures, respectively. Similar to the analysis in the discovery sample, a robust sandwich variance estimator approach was used to account for the familial correlation among OZ-ALC families. For both studies, sex, age at last interview and study source (COGEND vs FSCD vs COGA; NAG vs EDAC vs BIGSIB) were included as covariates.
Sensitivity analysis
Based on recommendations by Dudbridge,29 that replication of polygenic scores is optimized when the size of the discovery and test samples is approximately equal; we performed 10 000 iterations in which we randomly resampled 1788 individuals from the pool of 2336 subjects in SAGE. Cox proportional hazard models were fit to each randomly drawn sample to examine whether the magnitude of the association was modified by selection of a comparatively sized test sample. Similar analyses were not conducted in OZ-ALC as random selection of subsets of individuals nested in pedigrees would not be representative of the sampling design nor would selection of subsets of whole pedigrees allow for adequate numbers of individuals with AO-AD.
Results
Sample characteristics
Characteristics of the replication samples, SAGE and OZ-ALC, are presented in Table 1. In both samples, those meeting criteria for AD (NSAGE=1167; NOZ-ALC=1714) were more likely to report an earlier AO-I. They also reported higher Maxdrinks. In general, individuals from SAGE were heavier drinkers and have a greater number of AD-SX than those from OZ-ALC. This is unsurprising given the differences in ascertainment strategies. Modestly, earlier onset of drinking to intoxication and AD was noted in SAGE relative to OZ-ALC. No differences in height were noted across individuals with and without AD or across SAGE and OZ-ALC.
Association between GRS and alcohol measures
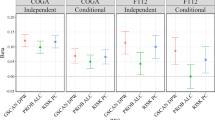
As shown in Table 2 and Figure 1, GRS for AO-AD were significantly associated with AO-AD and also with AD in SAGE for cutoffs above GRS0.05. Increasing P-value thresholds resulted in greater proportion of variance explained with the adjusted R2 ranging from 0.3% for GRS0.1 to 0.7% for GRS0.5. The variance explained was maximum when we included the SNPs with P⩽0.5. In addition to AO-AD, GRS explained a modest proportion of the variance in AO-I (≈0.3%). GRS from COGA were also related to AD-SX and Maxdrinks in SAGE, explaining 0.2 to 0.8% of the variance in these measures.
GRS generated from an analysis of AO-AD in a discovery sample, at varying P-value thresholds, predicting (a) AO-AD, (b) AO-I, (c ) AD, (d) AD-SX, (e) Maxdrinks and (f) height in SAGE dataset. The x-axis represents the GRS thresholds and y-axis represents the adjusted R2 for the trait. Each bar represents the values of adjusted R2 for SAGE. Colors of the bar represent the level of significance achieved. AD, alcohol dependence; AD-SX, total number of DSM4 AD symptoms endorsed; AO-AD, age at onset of AD; AO-I, age at onset of intoxication; GRS, genome-wide polygenic scores; SAGE, Study of Addictions: Genes and Environment.
In contrast, AO-AD GRS from COGA explained only a very modest (but significant) proportion of variation in AO-AD (0.06–0.07%) and AD (0.03–-0.06%) in OZ-ALC (Table 2 and Figure 2). The GRS were modestly associated with AO-I (0.2–0.3%), as well as with AD-SX (0.09–0.13%) and Maxdrinks (0.004–0.05%) in OZ-ALC. The associations were far less significant than those noted in SAGE, explaining ⩽0.16% of the variance in alcohol-related phenotypes. Height was included as a negative control and was not associated (P>0.05) with any GRS across SAGE and OZ-ALC.
GRS generated from an analysis of AO-AD in a discovery sample, at varying P-value thresholds, predicting (a) AO-AD, (b) AO-I, (c ) AD, (d) AD-SX, (e) Maxdrinks and (f) height in OZ-ALC dataset. The x-axis represents the GRS thresholds and y-axis represents the adjusted R2 for the trait. Each bar represents the values of adjusted R2 for OZ-ALC. Colors of the bar represent the significance level achieved. AD, alcohol dependence; AD-SX, total number of DSM4 AD symptoms endorsed; AO-AD, age at onset of AD; AO-I, age at onset of intoxication; GRS, genome-wide polygenic scores.
Resampling 10 000 subsets of SAGE individuals to create a sample size that was equivalent to COGA resulted in similar results. GRS showed statistically significant association with the AO-AD (P<0.05) in the reduced SAGE dataset in all 10 000 permutations. However, the reduction in sample size influenced the magnitude of P-values and only about 12% of the time, the association P-values were equal to or more significant than the original P=6.70 × 10−06 observed with the full SAGE sample.
Discussion
Using effect sizes generated via a prior GWAS of AO-AD in the COGA family sample,15 we created GRS at varying P-value thresholds and examined their association with AO-AD, AD, AO-I, as well as liability to problematic drinking (AD-SX and Maxdrinks) in two independent and differently structured and ascertained datasets, SAGE and OZ-ALC. GRS, especially when including SNPs associated with AO-AD at more liberal P-value thresholds, were significantly associated with a range of these alcohol-related measures in those two independent and distinctly ascertained samples. In contrast, there was no evidence for replication of the top 10 most significantly associated variants from COGA in either SAGE (6.6 × 10−1–8.1 × 10−1) or OZ-ALC (8.7 × 10−1–8.9 × 10−1). This strongly underscores the idea that multiple common genetic effects contribute to the etiology of complex disorders like addictions.
GRS comprised of >110 000 SNPs only captured very modest proportions of the variance in any alcohol-related measure (<1%). This observation is consistent with other studies.30, 31, 32 For instance, Vink et al.32 used GRS constructed from a large meta-analysis of GWAS of tobacco smoking measures to predict variance in alcohol, tobacco and cannabis-related outcomes. In that study, polygenic scores that were associated with tobacco-related measures at P<10−70 explained, at most, 1.5% of the variance in any substance-related outcome. Similarly, Power et al.30 examined the relationship between cannabis involvement and GRS generated from a meta-analysis of schizophrenia (N=13 833 cases, 18 310 controls) which included 13 genome-wide significant loci. Even though schizophrenia GRS were significantly associated with cannabis use, the scores, even at P<0.05 explained <1% of the variance in cannabis-related phenotypes. For alcohol-related measures, Salvatore et al.31 found that GRS generated for alcohol problems (N=4304) only predicted 0.6% of the variance in a similar measure in an independent sample. Despite relying on a smaller discovery sample (COGA, N=1788), our findings are consistent with these estimates. Nonetheless, the small sample size of the discovery set limits the accuracy of predicted SNP effect sizes and likely influenced our ability to generate GRS that might be reliable predictors of alcohol involvement in independent samples.
An additional consideration when viewing these results is the difference in ascertainment method across COGA, SAGE and OZ-ALC. The discovery sample (COGA) consisted of extended pedigrees ascertained for a dense family history of alcoholism and it was not expected that all variants associated with alcohol-related measures in such densely affected pedigrees would generalize to other cohorts. SAGE cases were selected for DSM-IV alcohol dependence from among several studies focused on alcohol, tobacco and cocaine and thus, as expected, we note a stronger degree of replication in this sample. In contrast, OZ-ALC comprises of samples ascertained for heavy smoking, discordance of heavy alcohol consumption measures and also for large sibship size (without any oversampling for substance-related phenotypes). Therefore, it is not surprising that replication in OZ-ALC, a less severely affected sample, is weaker for AO-AD, but occurs for ages of onset for earlier drinking milestones (for example, AO-I) and for measures that are quantitative indices of problem drinking (for example, AD-SX). In OZ-ALC, AO-I and AO-R, as well as AD-SX and Maxdrinks may serve as proxies for genetic liability to problematic drinking, while in COGA and SAGE this liability may be appropriately captured by AO-AD itself. Even so, the statistical significance of the associations and the proportions of variance explained in OZ-ALC are markedly lower than those in SAGE. Nonetheless, as OZ-ALC is so markedly distinct from COGA and SAGE, any level of association between COGA GRS and alcohol-related measures in OZ-ALC may be considered as support for the generalizability of the COGA results.
Dudbridge et al.29 has noted that there are two purposes for GRS: association testing (that is, replication, reliant on significance/P-value) and prediction of phenotypic variance (for example, reliant on R2 estimates). Based on numerous simulations, he concluded that while most studies with approximately equally sized training (that is, COGA) and testing (for example, SAGE) samples are well-powered for association testing, current training samples are underpowered for prediction. Consistent with this observation, we were interested in the former rather than the latter and even though we report R2 values, the emphasis of our analysis was association testing. We confirm that selecting a testing sample of approximately the same size as the training cohort, as was achieved via our bootstrapping approach, yields significant association. Dudbridge29 notes that 10-fold cross-validation might be a more efficient approach to maximizing prediction. As two of our samples are family-based (COGA and OZ-ALC), such cross-validation approaches, which may necessitate disaggregating members of pedigrees or selecting subsets of pedigrees that may or may not be informative for the etiology of genetically transmitted AD, may not be applicable. Hence, our findings should be observed in the context of association (and not prediction) testing alone.
Finally, it is worth noting that we did not attempt to characterize the SNPs that comprised each GRS nor did we create biologically informed GRS by selecting variants that related to a specific neurotransmitter pathway (for example, dopamine variant profile). Given the high false discovery rates typically associated with ascribing a functional direction to such variants of purported biological importance (except, perhaps, the alcohol dehydrogenase variants) and our currently limited understanding of the etiology of AD, we employed a more conservative and agnostic approach of utilizing genome-wide data. Future studies that contrast such genome-wide PRS with biologically informed risk scores may be valuable in the construction of the architecture underlying the polygenicity associated with complex traits such as AO-AD. Nonetheless, our approach precludes any mechanistic interpretation of the polygenicity represented by each GRS.
In conclusion, using GRS generated for AO-AD, we document that similar genetic factors might underpin the liability to alcohol dependence, drinking to intoxication and indices of problematic drinking in independent datasets. Future studies could strengthen these interpretations by conducting meta-analyses across multiple samples to produce larger discovery samples for generating GRS that could be applied to a wider range of alcohol and other substance phenotypes.
References
Babor TF . Types of alcoholics, I. Arch Gen Psychiatry 1992; 49: 599.
Babor TF, Dolinsky ZS, Meyer RE, Hesselbrock M, Hofmann M, Tennen H . Types of alcoholics: concurrent and predictive validity of some common classification schemes. Addiction 1992; 87: 1415–1431.
Cloninger C . Neurogenetic adaptive mechanisms in alcoholism. Science 1987; 236: 410–416.
Finn PR, Sharkansky EJ, Viken R, West TL et al. Heterogeneity in the families of sons of alcoholics: the impact of familial vulnerability type on offspring characteristics. J Abnorm Psychol 1997; 106: 26–36.
Morey LC, Skinner HA Empirically derived classifications of alcohol-related problems. In: Recent Developments in Alcoholism. Springer USA, 1986, pp 145–168.
Zucker RA, Ellis DA, Fitzgerald HE . Developmental evidence for at least two alcoholisms. Ann NY Acad Sci 1994; 708, (1 Types of Alco) 134–146.
Cloninger CR, Sigvardsson S, Gilligan SB, von Knorring A-L, Reich T, Bohman M . Genetic heterogeneity and the classification of alcoholism. Adv Alcohol Subst Abuse 1988; 7: 3–16.
Heath AC, Bucholz KK, Madden PAF, Dinwiddie SH, Slutske WS, Bierut LJ et al. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med 1997; 27: 1381–1396.
Prescott CA, Kendler KS . Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry 1999; 156: 34–40.
Agrawal A, Sartor CE, Lynskey MT, Grant JD, Pergadia ML, Grucza R et al. Evidence for an interaction between age at first drink and genetic influences on DSM-IV alcohol dependence symptoms. Alcoholism 2009; 33: 2047–2056.
Prescott CA, Kendler KS . Age at first drink and risk for alcoholism: a noncausal association. Alcoholism 1999; 23: 101–107.
Sartor CE, Lynskey MT, Bucholz KK, Madden PAF, Martin NG, Heath AC . Timing of first alcohol use and alcohol dependence: evidence of common genetic influences. Addiction 2009; 104: 1512–1518.
Sartor CE, Agrawal A, Lynskey MT, Bucholz KK, Heath AC . Genetic and environmental influences on the rate of progression to alcohol dependence in young women. Alcoholism 2008; 32: 632–638.
Stallings MC, Hewitt JK, Beresford T, Heath AC, Eaves LJ . A twin study of drinking and smoking onset and latencies from first use to regular use. Behav Genet 1999; 29: 409–421.
Kapoor M, Wang JC, Wetherill L, Le N, Bertelsen S, Hinrichs AL et al. Genome-wide survival analysis of age at onset of alcohol dependence in extended high-risk COGA families. Drug Alcohol Depend 2014; 142: 56–62.
Cross-Disorder Group of the Psychiatric Genomics C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 2013; 381: 1371–1379.
Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA 2010; 107: 5082–5087.
Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA et al. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry 2011; 70: 513–518.
Begleiter H, Reich T, Nurnberger J, Li TK, Conneally PM, Edenberg H et al. Description of the genetic analysis workshop 11 collaborative study on the genetics of alcoholism. Genet Epidemiol 1999; 17: S25–S30.
Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB . Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend 2008; 95: 14–22.
Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 2007; 16: 24–35.
Grant JD, Agrawal A, Bucholz KK, Madden PAF, Pergadia ML, Nelson EC et al. Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry 2009; 66: 795–800.
Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI et al. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol 1994; 55: 149–158.
Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V . A validity study of the SSAGA-a comparison with the SCAN. Addiction 1999; 94: 1361–1370.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Medland SE, Nyholt DR, Painter JN, McEvoy BP, McRae AF, Zhu G et al. Common variants in the trichohyalin gene are associated with straight hair in Europeans. Am J Hum Genet 2009; 85: 750–755.
Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR . MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Dudbridge F . Power and predictive accuracy of polygenic risk scores. PLoS Genet 2013; 9: e1003348.
Power RA, Verweij KJH, Zuhair M, Montgomery GW, Henders AK, Heath AC et al. Genetic predisposition to schizophrenia associated with increased use of cannabis. Mol Psychiatry 2014; 19: 1201–1204.
Salvatore J, Aliev F, Edwards A, Evans D, Macleod J, Hickman M et al. Polygenic scores predict alcohol problems in an independent sample and show moderation by the environment. Genes 2014; 5: 330–346.
Vink JM, Hottenga JJ, de Geus EJC, Willemsen G, Neale MC, Furberg H et al. Polygenic risk scores for smoking: predictors for alcohol and cannabis use? Addiction 2014; 109: 1141–1151.
Acknowledgements
AA and MK received support from, R21AA021235; AA receives support from K02DA32573. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01CA089392), and the Family Study of Cocaine Dependence (FSCD; R01DA013423, R01DA019963). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract 'High throughput genotyping for studying the genetic contributions to human disease' (HHSN268200782096C). The Collaborative Study on the Genetics of Alcoholism (COGA): COGA, Principal Investigators B Porjesz, VH, HJE, LJB includes 10 different centers: the University of Connecticut (VH); the Indiana University (HJE, J Nurnberger Jr, TF); the University of Iowa (SK, J Kramer); SUNY Downstate (B Porjesz); the Washington University in Saint Louis (LJB, AMG, J Rice, KKB); the University of California at San Diego (M Schuckit); the Rutgers University (JT); the Southwest Foundation (L Almasy), the Howard University (R Taylor) and the Virginia Commonwealth University (D Dick). A Parsian and M Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and CoPI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). OZ-ALC: Supported by NIH grants AA07535, AA07728, AA13320, AA13321, AA14041, AA11998, AA17688, DA012854, DA019951; by grants from the Australian National Health and Medical Research Council (241944, 339462, 389927, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 552485, 552498); by grants from the Australian Research Council (A7960034, A79906588, A79801419, DP0770096, DP0212016, DP0343921); and by the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). GWAS genotyping at CIDR was supported by a grant to the late Richard Todd, former PI of grant AA13320 and a key contributor to research described in this manuscript. We acknowledge the contributions of project investigator Alexandre Todorov, at Washington University. We also thank Dixie Statham, Ann Eldridge, Marlene Grace, Kerrie McAloney (sample collection); Lisa Bowdler, Steven Crooks (DNA processing); David Smyth, Harry Beeby, and Daniel Park (IT support) at the Queensland Institute of Medical Research, Brisbane, Australia. Last, but not least, we thank the twins and their families for their participation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
LJB, AMG and JCW are listed as inventors on the patent 'Markers for Addiction' (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. In addition, AA has previously received peer-reviewed grant funding and an honorarium from ABMRF/Foundation for Alcohol Research, which receives part of its funding from brewers. The remaining authors declare no conflicts of interest.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kapoor, M., Chou, YL., Edenberg, H. et al. Genome-wide polygenic scores for age at onset of alcohol dependence and association with alcohol-related measures. Transl Psychiatry 6, e761 (2016). https://doi.org/10.1038/tp.2016.27
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/tp.2016.27
This article is cited by
-
Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism
Translational Psychiatry (2019)
-
Effects of autozygosity and schizophrenia polygenic risk on cognitive and brain developmental trajectories
European Journal of Human Genetics (2018)