Abstract
Mitochondria have a central position in innate immune response via the adaptor protein MAVS in mitochondrial outer membrane to limit viral replication by inducing interferon production. Here, we reported that C11orf83, a component of complex III of electronic transfer chain in mitochondrial inner membrane, was a potent antiviral protein independent of interferon production. C11orf83 expression significantly increased in response to viral infection, and endows cells with stronger capability of inhibiting viral replication. Deletion of C11orf83 permits viral replication easier and cells were more vulnerable to viral killing. These effects mainly were mediated by triggering OAS3-RNase L system. C11orf83 overexpression induced higher transcription of OAS3, and knockdown either OAS3 or RNase L impaired the antiviral capability of C11orf83. Interestingly, the signaling from C11orf83 to OAS3-RNase L was independent of interferon production. Thus, our findings suggested a new antiviral mechanism by bridging cell metabolic machinery component with antiviral effectors.
Similar content being viewed by others
Introduction
The innate immune system limits viral replication during early stages of infections before the development of adaptive immunity1,2. The inducement of antiviral cytokines interferon (IFN) is an essential part of the antiviral response. Extensive research has provided detailed insights into the IFN signaling, however, much remains unknown about the mechanism that cell inhibits viral replication independent of IFN pathway.
Mitochondria functions as a signaling platform that is centrally positioned in the innate immune response against viral pathogens3. Retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are one of four pattern-recognition receptors (PRRs) families of the innate immune system, which bind conserved molecular patterns that are shared by different kinds of microorganisms4. RLRs are expressed in the cytosol and are required for type I IFN and pro-inflammatory cytokine production in response to viral infection5,6,7. Mitochondrial antiviral signaling protein (MAVS), an adaptor molecule localized in the outer mitochondrial membrane (OMM), is essential for RLR signal transduction8. As downstream of RLRs, MAVS induces the production of type I IFN and pro-inflammatory cytokines during viral infection by activating NF-κB, IRF3 and IRF7 signaling cascades8,9. Mitochondrial cofactors take part in the regulation of RLR-MAVS signaling10,11,12. Moreover, mitochondrial dynamics govern antiviral signaling by enhancing mitochondria–MAMs interactions and RLR–MAVS signalosome formation around intracellular sites of viral infection13,14,15,16. Mitochondrial ROS is also shown to have a critical role in regulating RLR–MAVS signaling during viral infection17.
The 2′-5′-oligoadenylate synthetases (OASs) belongs to a nucleotidyltransferase superfamily18,19. The transcription of OAS genes is induced by both virus infection and IFN stimulation20,21. Upon binding to double-stranded RNA (dsRNA), OASs are activated through a conformational change to synthesize 2′-5′-phosphodiester-linked oligoadenylates (2-5As) and 2-5As acts as second messengers to activate RNase L22,23. Activation of RNase L leads to degradation of cellular and viral RNA and thereby suppresses viral replication24. Thus OASs-RNase L system offers protection against a wide range of RNA and DNA viruses. In humans, the OAS family consists of four members: OAS1, OAS2, OAS3, and OASL25. OAS1, OAS2, and OAS3 have 2′-5′-oligoadenylate synthetase activity, and OASL lacks this activity26,27. OAS3 is activated at a substantially lower concentration of dsRNA than OAS1, and synthesizes considerably longer 2-5As that are sufficient to activate RNase L intracellularly19,28.
Mammalian mitochondria may contain up to 1500 different proteins, and most of their functions have not been well revealed29. C11orf83, also named UQCC3, is a mitochondrial inner membrane protein. It is specifically associated with the complex III of the electron transport chain and is involved in the early stages of its assembly by stabilizing supercomplexes that contain complex III, especially the III2/IV supercomplex30. Patient cells with a homozygous c.59T > A missense mutation in C11orf83 have reduced complex III activity. The patient displays lactic acidosis, hypoglycemia, hypotonia, delayed development and severely delayed psychomotor development31. In the present study, we reported the identification of C11orf83 as a novel antiviral protein. Higher expression of C11orf83 responding to viral infection endows cells with stronger capability of inhibiting viral transcription, whereas the loss of C11orf83 expression renders viral replication easier in cells and cells were vulnerable to viral killing. Importantly, this effect mainly was mediated by triggering OAS3-RNase L system and was independent of interferons production, which is significantly different to MAVS.
Results
C11orf83 expression increases after virus infection
C11orf83 is a mitochondrial inner membrane protein30. To investigate whether C11orf83 is associated with virus infection, we used vesicular stomatitis virus (VSV) as the model virus8 and examined C11orf83 expression in human cells (HEK293, HUVEC and HepG2) at different time points after VSV infection. HEK293 is immortal human embryonic kidney cells, HUVEC is primary human endothelial cells, and HepG2 is malignant human hepatocellular carcinoma cells. The results clearly demonstrated that C11orf83 expression significantly increased after VSV infection. C11orf83 was upregulated early at 2 hours and higher at 4 and 8 hours after virus infection (Fig. 1A), suggesting C11orf83 is an early responsive protein during viral infection. Then, we detected C11orf83 expression responding to different amounts of VSV (MOI = 0, 0.1, 1.0 and 10), and found that more viral infection induced higher expression of C11orf83 (Fig. 1B). Moreover, qRT-PCR showed C11orf83 mRNA significantly increased at 2, 4, 8 hours post VSV infection (MOI = 1.0) in all three kinds of cells, and more amounts of VSV infection induced more significant increase of C11orf83 mRNA (Supplementary Fig. S1). These results suggested that the increased C11orf83 protein after VSV infection is most likely due to an increased transcription. Moreover, plaque assay and q-PCR results showed that the load of virus and viral RNA in cells was proportional to C11orf83 expression (Supplementary Fig. S2), indicating that C11orf83 is sensitive to virus load in cells.
C11orf83 Is a Potent Antiviral Protein
Since C11orf83 is upregulated during virus infection, we examined whether C11orf83 participate in the anti-viral process in cells. VSV replicates rapidly in 293 cells8. We used a recombinant VSV line expressing GFP (rVSV-GFP) to demonstrate viral replication in infected cells32. C11orf83 Overexpression (Supplementary Fig. S3A) significant inhibited rVSV-GFP replication in 293 cells (Fig. 2A). Both fluorescent microscopic observations and flow cytometry analysis demonstrated the obviously decrease of GFP-positive cells in C11orf83-overexpressed 293 cells (Fig. 2A,B). Consistently, measurement of the viral titer in cells supernatants showed that C11orf83 overexpression made the decrease of viral titer by more than 7-fold as compared to control cells (Fig. 2C). N gene codes the nucleocapsid protein that is necessary to VSV. The detection to N gene at mRNA level clearly showed the much less of VSV mRNA than controls when C11orf83 was overexpressed (Fig. 2D). Infection of HEK293 cells by rVSV-GFP led to nearly complete killing of the cells within 24 hours (MOI = 0.01). However, only about 30% cells were killed by rVSV-GFP infection in C11orf83-overexpression cells (Fig. 2E).
(A–E) Effects of C11orf83 overexpression to rVSV-GFP replication in HEK293 cells. All analysis was performed at 12 hours post rVSV-GFP (MOI = 1.0) infection. **p < 0.01. (A) Representative imaging fluorescent shows the replication of rVSV-GFP in cells. (B) Quantitative analysis of GFP positive cells by using flow cytometry. (C) Quantitative analysis of virus titer in cells supernatants. (D) Transcription detection of C11orf83 and N gene of VSV using RT-PCR. (E) Quantitative analysis of VSV-induced cells death using Trypan Blue staining. (F) Graphical representation of the Human genomic loci of C11orf83 showed the targeting sites for Cas9, sgRNA, and mutation initiation site. The sgRNA targeting sequences are labeled in magenta. The PAM sequences are labeled in red. The mutation initiation sites are indicated by the black triangle and the DNA scissors. (G) Detection of cells viability using MTT.(H–L) Effects of the loss of C11orf83 to rVSV-GFP replication in HEK293 cells. All analysis was performed at 12 hours post rVSV-GFP (MOI = 0.1) infection. Cas9-Control, HEK293 cells transfected with Cas9 and control sgRNA. Cas9-KO#1 and Cas9-KO#2, HEK293 cells transfected with Cas9 and sgRNA#1 or sgRNA#2 as shown in F. **p < 0.01. (H) Representative imaging fluorescent shows the replication of rVSV-GFP in cells. (I,K) Quantitative analysis of GFP positive cells and mean fluorescence intensity in cells by using flow cytometry. (J) Quantitative RT-PCR results shows the relative change of N gene transcription. (L) Quantitative analysis of VSV-induced cells death using Trypan Blue staining.
Next, we deleted endogenous C11orf83 expression by using CRISPR-Cas9 system (Supplementary Fig. S3B,C). We used two sgRNAs that effectively cleaved C11orf83 gene and produced mutations in C11orf83 exon 1 (Fig. 2F). The C11orf83 deletion did not caused any obvious effects to cell growth (Fig. 2G), but fluorescence and N gene detection and plaque assay to rVSV-GFP demonstrated that C11orf83 deletion leaded to an increase of viral replication by about 5-fold as compared to control cells (Fig. 2H–K and Supplementary Fig. S4). Consequently, C11orf83 deficiency greatly sensitized cell killing by rVSV-GFP at a much lower MOI (0.01) (Fig. 2L). Similarly, viral replication in C11orf83-deleted human HepG2 cell also markedly increased (Supplementary Fig. S5). The plaque assay to VSV growth kinetic in control and C11orf83-KO cells demonstrated that C11orf83 significantly inhibited VSV replication within 24 hours post viral infection (Supplementary Fig. S6). Importantly, the effects caused by Crispr/Cas9 could be partial rescued by C11orf83 reconstitution (Supplementary Fig. S7). Together, these results show that C11orf83 is a pivotal cellular antiviral protein.
C11orf83 inhibits virus replication through OAS3-RNase L system
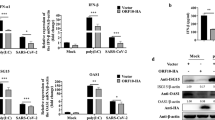
OASs-RNase L system is one of the most important cellular antiviral mechanism, providing protection against a wide range of RNA and DNA viruses. The 42-kDa OAS1 consists of one OAS domain, while the 69-kDa OAS2 consists of two OAS domains and the 120-kDa OAS3 consists of three OAS domains25,33. To investigate whether the antiviral effect of C11orf83 is through OASs-RNase L system, we examined the expression of OAS family four members (OAS1, OAS2, OAS3, and OASL) at the mRNA level in HEK293 cells that only overexpressed C11orf83 but no viral infection. qPCR results revealed that C11orf83 resulted in the upregulation of OAS1, OAS3 and OASL expression, whereas OAS3 (p100) had the most significantly alternation by about 4 folds of increase (Fig. 3A). Also, OAS3 transcription dramatically increased after VSV infection, and is significantly impaired when c11orf83 is absent (Supplementary Fig. S8). These suggested that OAS3 might play an important role during antiviral process triggered by C11orf83.
(A) qPCR analysis of OAS variants in C11orf83 overexpressed HEK293 cells. **p < 0.01. (B–F) Effects of the loss of OAS3 to rVSV-GFP replication in C11orf83 overexpressed HEK293 cells. HEK293 cells were transfected with pcDNA3.1 (vector) or pcDNA3.1-C11orf83, and 48 hours later cells were transfected with siControl or siOAS3. In the next day, cells were infected with rVSV-GFP (MOI = 1.0) and 12 hours later assays were performed. **p < 0.01. (B,D) Quantitative analysis of GFP positive cells and mean fluorescence intensity in cells by using flow cytometry. (C) Quantitative RT-PCR results shows the relative change of N gene transcription. (E) Quantitative analysis of VSV-induced cells death using Trypan Blue staining. (F) RNA integrity analysis using Agilent Bioanalyzer 2100. (G–J) Effects of the loss of RNase L to rVSV-GFP replication in C11orf83 overexpressed HEK293 cells. HEK293 cells were transfected with pcDNA3.1 (vector) or pcDNA3.1-C11orf83, and 48 hours later cells were transfected with siControl or siRNase L. In the next day, cells were infected with rVSV-GFP (MOI = 1.0) and 12 hours later assays were performed. **p < 0.01. (G,I) Quantitative analysis of GFP positive cells and mean fluorescence intensity in cells by using flow cytometry. (H) Quantitative RT-PCR results shows the relative change of N gene transcription. (J) Quantitative analysis of VSV-induced cells death using Trypan Blue staining.
OAS family is the activator of endoribonuclease RNase L and RNase L is known to terminate virus replication by cleaving viral and cellular RNAs. We found that when endogenous OAS3 or RNase L was silenced in C11orf83-overexperssed HEK293 cells by using small interfering RNA (siRNA) (Supplementary Fig. S9)34, C11orf83 lost the anti-viral capability evidenced by the analysis to viral green fluorescence, viral N gene, viral titer (plaque analysis), cell death and mRNA degradation detection (Fig. 3B–J and Supplementary Fig. S10). All siRNAs had no obvious toxic affects to cells (Supplementary Fig. S11). OAS3 knockdown did not further affect viral titer in c11orf83KO cells, suggesting OAS3 is the downstream of C11orf83 (Supplementary Fig. S12). These results together indicated that c11orf83 inhibits virus replication through the OAS3-RNase L system, and were confirmed in HepG2 cells (Supplementary Fig. S13A–I).
The antiviral effect of C11orf83 is independent of IFN production
Interferons (IFNs) play important roles during anti-viral process by inducing expression of interferon stimulated genes (ISGs). To reveal whether C11orf83 is an ISG, we detected the kinetics of C11orf83 expression upon type I, type II and type III IFNs at early and late time points post stimulation. The results clearly showed that c11orf83 expressions were not significantly affected by type I, type II and type III IFNs (Supplementary Fig. S14). Then, we detected c11orf83 expression in IFNR1 or IFNR2 knock-out cells with or without viral infection, and examined transcriptional alternation of c11orf83 in the presence of type I IFN (IFNα and IFNβ) neutralizing antibodies (Supplementary Fig. S15 and Fig. 4). We found that neither IFN receptors deficiency nor IFN blocking impaired viral infection-induced c11orf83 expression (Fig. 4A–D). The results indicated that C11orf83 is induced in an interferon independent manner, suggesting c11orf83 is not an ISG itself. Moreover, C11orf83 overexpression also did not trigger expression of IFNα, IFNβ and IFNγ (Fig. 4E).
(A,B) VSV infection upregulating c11orf83 expression in IFNR1- and IFNR2-deficient cells. (C,D) VSV infection upregulating c11orf83 expression in the presence of type I IFN (IFNα and IFNβ) neutralizing antibodies. (E) RT-PCR analysis to the transcription of C11orf83 and IFNs (IFN-λ2/3, IFN-λ1, IFN-α and IFN-β) in HEK293 cells at 0, 16, 24 and 48 hours after the transfection of C11orf83 gene.
OASs are a family of interferon-stimulated genes (ISGs). The transcription of OAS family genes can be induced by TypeI IFN stimulation. To further reveal whether C11orf83 upregulating OAS3 depends on TypeI IFN or not, we examined OAS3 expression in Vero and Vero E6 cells that are intrinsically deficient in type I IFNs production because lack the gene cluster for type I IFN35. We found that overexpression of C1H11orf83 (the Chlorocebus sabaeus homology of human c11orf83) still induced the upregulation of OAS3 (Fig. 5A,B) and markedly decreased viral replication in Vero and Vero E6 cells (Fig. 5C). When endogenous OAS3 was knocked-down using siRNA, C11orf83 lost the capability of inhibiting rVSV-GFP replication in Vero cells (MOI = 0.1) (Fig. 5D–F).
(A,B) qPCR analysis to the expression of OAS3 in the mRNA level in IFN-deficient Vero (B) and Vero E6 (C) cells. Vero and Vero E6 cells were transfected with pcDNA3.1 (vector) or pcDNA3.1-C11orf83-monkey. **p < 0.01. (C–F) Effects of the loss of OAS3 to rVSV-GFP replication in C11orf83 overexpressed Vero cells. Vero cells were transfected with pcDNA3.1 (vector) or pcDNA3.1-C11orf83-monkey, and 48 hours later cells were transfected with siControl or siOAS3. In the next day, cells were infected with rVSV-GFP (MOI = 0.1) and 12 hours later assays were performed. **p < 0.01. (C,D) Quantitative analysis of GFP positive cells and mean fluorescence intensity in cells by using flow cytometry. (E) Quantitative RT-PCR results shows the relative change of N gene transcription. (F) Quantitative analysis of VSV-induced cells death using Trypan Blue staining.
In addition, we deleted endogenous C11orf83 expression in Vero cells by using CRISPR-Cas9 system (Fig. 6). We used two sgRNAs that effectively cleaved C11orf83 gene in exon 1 and terminated C11orf83 protein expression in Vero cells (Supplementary Fig. S16 and Fig. 6A). Viral fluorescence, N gene detection, plaque assay and cell death analysis demonstrated that C11orf83 deletion leaded to the significant increase of viral replication in Vero cells (Fig. 6B–G). Together, these results indicated that C11orf83 functions as an antiviral protein independent of IFNs signaling.
(A) The absence of C11orf83 protein in Vero cells by Crispr-Cas9 system. (B–G) Effects of c11orf83 deletion to rVSV-GFP replication in Vero cells. (B) Representative imaging fluorescent shows the replication of rVSV-GFP in cells. (C) Quantitative RT-PCR results showing the relative change of N gene transcription. (D–F) Quantitative analysis of GFP positive cells and mean fluorescence intensity in cells by using flow cytometry. (G) Quantitative analysis of VSV-induced cells death using Trypan Blue staining.
Discussion
Mitochondria are dynamic double-membrane-bound organelles in eukaryotic cells. Mitochondria are involved in a broad range of cellular processes, such as ATP generation, apoptosis, calcium homeostasis, and the biosynthesis of amino acids, lipids and nucleotides. Recent research has unveiled that mitochondria participate in innate immune responses, such as antiviral signaling, anti-bacterial immunity and sterile inflammation. MAVS, a mitochondrial antiviral signaling protein (also named IFNβ promoter stimulator 1 (IPS1), CARD adaptor inducing IFNβ (CARDIF) or virus-induced signalling adaptor (VISA)), localized in the outer mitochondrial membrane and signaling downstream of RLRs during viral infection8,9,36,37. Viral dsRNA recruits the RIG-I and MDA-5 to interact with MAVS directly or indirectly. The signal of viral replication thus is transmitted to the mitochondria, and further triggers antiviral responses through activation of NF-κB, IRF3 and IRF7 signaling8,9,36,37. In addition, many common innate immune signaling molecules have been shown to be downstream of MAVS6. Therefore, mitochondria are centrally positioned in the innate immune response functioning as signaling platforms against viral pathogens.
Mitochondrial own genome encodes 13 proteins of the oxidative phosphorylation machinery. However, mitochondrial proteome comprises about 1500 proteins, most of which are encoded by nuclear genome. C11orf83 is a nuclear genome coded small protein, targeted and anchored in the mitochondrial inner membrane by its N-terminal section. C11orf83 is currently known as an assembly factor of Complex III of the electron transport chain and is required for proper mitochondrial morphology and function. C11orf83-deficient cells displayed complex III assembly deficiency and abnormal mitochondria with an impaired oxidative phosphorylation30,31. In the present study, we found that C11orf83 also is sensitive to viral infection, which was rapidly upregulated when cells were infected by VSV. Higher expression of C11orf83 endows cells with stronger immunity to viral infection, whereas the loss of C11orf83 expression renders VSV replication easier in cells and cells were vulnerable to viral killing. Thus, we propose that C11orf83 is novel antiviral protein in mitochondria. At least to our known, our findings also firstly bridge mitochondrial energy metabolism (oxidative phosphorylation) machinery to antiviral mechanism in innate immune response.
Detection of virus-derived nucleic acids plays a central role in initiating antiviral immunity. Nucleic acid recognition by PRRs results in the secretion of type I interferon (IFN) cytokines and IFN-stimulated genes (ISGs), which function to impede viral replication. We have known two kinds of cellular sensors to viral infection: first is cGMP-AMP synthase (cGAS) as a DNA sensor that recognizes cytosolic DNA and subsequently produces cGMP-AMP (cGAMP) to trigger interferon production38,39. Second are RNA sensors including cytoplasmic RNA helicase RIG-I as the sensor of 5′-triphosphorylated RNA and MDA5 as the sensor of long double-stranded RNA40. C11orf83 localizes in mitochondrial inner membrane as an important component of complex III30, it does not seem to be reasonable that C11orf83 functions as an adapter molecule liking MAVS. So, we would like to believe that C11orf83 might represent a novel kind of cellular sensor, a metabolic sensor, to virus infection.
Type I interferon responses are considered the primary means by which viral infections are controlled in mammal cells. In response to virus infection, RIG-I senses viral RNA and activates mitochondrial MAVS, the adaptor protein that stimulates downstream signaling pathways leading to type I interferon production. Then interferon induces the expression of antiviral effectors, Interferon-stimulated gene (ISG) such as OASs and Mx, to restrict virus propagation in cells. Mitochondrial dynamics and mitochondrial ROS also participate in governing antiviral process by modulating RLR–MAVS signaling. C11orf83 is a potent antiviral protein in mitochondrial inner membrane. Different to above known mechanisms, C11orf83 induces higher expression of interferon effectors OAS3 and results in the cleavage of cellular and viral RNA. Although we do not know how C11orf83 induces the upregulation of OAS3, we show that this process is independent of interferons production. To the best of our knowledge, this the first report that a mitochondrial protein triggers the activation of antiviral effector system of OAS3-RNase L. Together, our studies have uncovered a new mechanism of mitochondria in innate immunity against viral infection, but much more work is required in the future to elucidate the biochemical mechanism of C11orf83 signaling, at least including the relationship of C11orf83 to MAVS, the signaling that mediates C11orf83 activating OAS3-RNase L system, and whether C11orf83 can efficiently prevent the production of viral proteins by inhibiting translational machinery.
Materials and Methods
Cells and Virus
HEK293, HepG2, HUVEC-CS, Vero and VeroE6 cells were grown in DMEM (Gibco) supplemented with 10% of heat-inactivated FBS and 1% penicillin and streptomycin at 5% CO2 and 37 °C.
The Vesicular Stomatitis Virus (Indiana strain) and the VSV-GFP recombinant (rVSV-GFP) virus were kindly provided by Dr. Chunsheng Dong in Soochow University. GFP was fused to the coding sequence of the G protein in VSV genome. VSV and rVSV–GFP replication in infected cells was indicated by green fluorescence.
Plasmid Constructs
The coding sequence (CDS) of human and monkey (Chlorocebus sabaeus) C11orf83 and its mutant of human C11orf83 cDNA was inserted into the pcDNA3.1 expression vector between the restriction sites BamH I and XhoI. The recombinant constructs were named pcDNA3.1-C11orf83, pcDNA3.1-C11orf83-Mutant, and pcDNA3.1- C11orf83-monkey. The empty pcDNA3.1 was used as a control.
To construct a recombinant eukaryotic vector bearing the fusion gene of human C11orf83 gene and EGFP (enhanced green fluorescent protein), and to transfect it into HEK293T cells for its subcellular localization, the pEGFP-N1 was used to construct pEGFP-N1-C11orf83 and pEGFP-N1-C11orf83-Mutant between the restriction sites HindIII.
CRISPR/Cas9 system was used to disrupt the endogenous C11orf83, IFNAR1 and IFNAR2 in human HEK293T or HepG2 cells, and monkey (Chlorocebus sabaeus) Vero cells. The annealed double-stranded single-guide RNAs (sgRNAs) downstream of the C11orf83 start codon were cloned into LentiCRISPRv2 (Addgene) by BsmB I (NEB). Two sgRNAs targeting human C11orf83 (#1 agtcgcaatgctgggcgca and #2 cccggggtcacgataacg), two sgRNAs targeting human IFNAR1(#1 gacaggagcgatgagtctgt and #2 gcaggagcgatgagtctgtc), two sgRNAs targeting human IFNAR2(#1 gttccggtccatcttatcat and #2 gtttccggtccatcttatca) and two sgRNAs targetting monkey (Chlorocebus sabaeus) C11orf83 (#1 gctgatcacagtcgcagtgc and #2 gcagtcgcagtgctgggcgc) were effective to produce mutation of respective gene in genomic DNA.
Transfection
For transient transfection, cells (HEK293, HepG2, Vero or Vero E6) were seeded in 6-well plates and were grown to 80–90% confluence. An aliquot of plasmid DNA (5 μg) was mixed with Lipofectamine 3000 reagent (7.5 μL) (Life Technology) and added to cells as suggested by the manufacturer. Twelve hours after incubation, the cells were placed in serum-containing medium and further incubated at 37 °C.
Viral infection and plaque assays
VSV and rVSV-GFP were propagated and amplified by infecting the monolayer of HEK293 cells. The supernatant was harvested 24 hours later and clarified by centrifugation.
Viral titers were determined by evaluating plaque assays in HEK293 cells. The HEK293 cells (1 × 105) were transfected with the indicated plasmids for 48 h and seeded into 24-well plate. The viral sample were diluted 1:101~6 and infect confluent HEK293 cells seeded before. At 1 h after infection, uninfected viruses were washed three times. Then, 3% methylcellulose was overlaid. At 24 hours after infection, overlay was removed and cells were fixed with 4% formaldehyde for 20 min and stained with 0.25% crystal violet. Plaques were counted and multiplied by the dilution factor to determine viral titer as LOG10 (pfu/ml).
Western blotting
Cells were harvested and lysed with RIPA (20 mM Tris PH7.5, 150 mM NaCl, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM EDTA, 1% Na3VO4, 0.5 μg/ml leupeptin, 1 mM PMSF). For immunoblotting, proteins from whole-cell lysates were resolved by 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to PVDF membranes. C11orf83 antibodies (Abmart, China) were used at 1:1000 and secondary antibodies conjugated with horseradish peroxidase (HRP) were used at 1:1000 dilutions in 5% non-fat dry milk. After the final washing, PVDF membranes were exposed for an enhanced chemiluminescence assay using the Gel imaging instrument (BIO-RAD ChemiDoc MP). The gray value of all the bands is measured by IMAGEJ.
qPCR
Total RNA was isolated with the TRIZOL reagent (Life technology) and reverse transcribed with PrimeScript RT reagent Kit with gDNA Eraser Kit (Takara RR047a). Quantitative PCR analysis was performed using a BIO-RAD CFX96 Real-Time PCR System (BIO-RAD) with SYBR Green qPCR Master Mix for all the target genes (Takara, 638320). Primers for ACTB or GAPDH were used as the internal control. All of the primer sequences are listed in Table 1.
siRNA
Cells were trypsinized and incubated overnight to achieve 60–70% confluence before siRNA transfection. The siRNA sequences were respectively shown below (Bréhin, A. C. et al.34): OAS3 siRNA (Ribobio Corporation; 200 nM; sense, 5′-UUUGGU GCCAGAACUGAGC(dTdT)-3′; antisense, 5′-GCUCAGUUCUGGCACCAAATT (dTdT)-3′), RNase L siRNA (Ribobio Corporation; 200 nM; sense, 5′-AGUGGA CGACUAAGAUUAATT(dTdT)-3′; antisense, 5′-UUAAUCUUAGUCGUCCACU TG(dTdT)-3′) and negative control siRNA (Ribobio Corporation) were mixed with Lipofectamine 3000 (Invitrogen). The cells were incubated with the transfection mixture for 48 h and then rinsed with DMEM containing 10% fetal bovine serum. The cells were then infected with rVSV-GFP.
Flow Cytometry
Single-colour FCM was used to determine the percentage of GFP positive cells. Briefly, cells were collected and suspended in PBS and analyzed by flow cytometer (BD FACSCalibur) with excitation and emission wavelengths at 488 nm and 530 nm, respectively. The data were analyzed using FlowJo 10.0.7 software.
Confocal microscopy
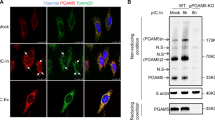
For experiments confirming proper localization of the C11orf83 and mitochondria, HEK293T expressing pEGFP-N1-C11orf83 or pEGFP-N1-C11orf83-Mutant were seeded into six-well plates (Corning). After VSV infection, cells were incubated with 25 nM MitoTracker Red FM (Thermo Fisher Scientific M22425) for 30 minutes and were fixed in 2% paraformaldehyde for 20 min. Cells were then washed three times with PBS-T (PBS containing 0.05% Tween-20) and were then stained with DAPI at 25 °C for 10 min. Images were acquired under a LEICA TCS SP5II confocal microscope.
RNA fragment analysis
HEK293 Cells first were transfected by pcDNA3.1, pcDNA3.1-C11orf83, OAS3-siRNA or control siRNA, respectively. 24 hours later cells were infected with VSV (MOI = 1.0) for twelve hours, and then cell total RNA was isolated with Trizol (Life Technologies) and rRNA degradation was monitored in RNA chips with an Agilent 2100 bioanalyzer as the standard protocol.
Statistical analysis
Student’s t-test was used in all cellular experiments and the results from three independent experiments are presented as mean SEM. In addition, all the mRNA expression data represent the mean values with standard error of the mean (SEM). The differences in mRNA expression data were compared using the ratio t-test. All statistical analyses were performed using the GraphPad Prism software. Asterisks denote statistical significance as follows: ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01.
Additional Information
How to cite this article: Yang, Y. et al. Mitochondrial C11orf83 is a potent Antiviral Protein Independent of interferon production. Sci. Rep. 7, 44303; doi: 10.1038/srep44303 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Alexopoulou, L., Holt, A. C., Medzhitov, R. & Flavell, R. A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–738 (2001).
Yoneyama, M. & Fujita, T. Recognition of viral nucleic acids in innate immunity. Rev Med Virol. 20, 4–22 (2010).
Koshiba, T., Bashiruddin, N. & Kawabata, S. Mitochondria and antiviral innate immunity. Int J Biochem Mol Biol. 2, 257–262 (2011).
Yoneyama, M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 5, 730–737 (2004).
Hayden, M. S., West, A. P. & Ghosh, S. NF-kappaB and the immune response. Oncogene. 25, 6758–6780 (2006).
Seth, R. B., Sun, L. & Chen, Z. J. Antiviral innate immunity pathways. Cell Res. 16, 141–147 (2006).
West, A. P., Koblansky, A. A. & Ghosh, S. Recognition and signaling by toll-like receptors. Annu Rev Cell Dev Biol. 22, 409–437 (2006).
Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 122, 669–682 (2005).
Xu, L. G. et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol cell. 19, 727–740 (2005).
Jia, Y. et al. Negative regulation of MAVS-mediated innate immune response by PSMA7. J Immunol. 183, 4241–4248 (2009).
Liu, X. Y., Wei, B., Shi, H. X., Shan, Y. F. & Wang, C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 20, 994–1011 (2010).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 455, 674–678 (2008).
Yasukawa, K. et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2, ra47 (2009).
Onoguchi, K. et al. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS pathog. 6, e1001012 (2010).
Koshiba, T., Yasukawa, K., Yanagi, Y. & Kawabata, S. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci signal. 4, ra7 (2011).
Castanier, C., Garcin, D., Vazquez, A. & Arnoult, D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 11, 133–138 (2010).
Tal, M. C. et al. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc Natl Acad Sci USA 106, 2770–2775 (2009).
Holm, L. & Sander, C. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem Sci. 20, 345–347 (1995).
Ibsen, M. S. et al. The 2′-5′-oligoadenylate synthetase 3 enzyme potently synthesizes the 2′-5′-oligoadenylates required for RNase L activation. J Virol. 88, 14222–14231 (2014).
Williams, B. R., Golgher, R. R., Brown, R. E., Gilbert, C. S. & Kerr, I. M. Natural occurrence of 2-5A in interferon-treated EMC virus-infected L cells. Nature. 282, 582–586 (1979).
Silverman, R. H. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 81, 12720–12729 (2007).
Han, Y., Whitney, G., Donovan, J. & Korennykh, A. Innate immune messenger 2-5A tethers human RNase L into active high-order complexes. Cell Rep. 2, 902–913 (2012).
Huang, H. et al. Dimeric structure of pseudokinase RNase L bound to 2-5A reveals a basis for interferon-induced antiviral activity. Mol Cell 53, 221–234 (2014).
Clemens, M. J. & Williams, B. R. Inhibition of cell-free protein synthesis by pppA2′p5′A2′p5′A: a novel oligonucleotide synthesized by interferon-treated L cell extracts. Cell. 13, 565–572 (1978).
Chebath, J., Benech, P., Hovanessian, A., Galabru, J. & Revel, M. Four different forms of interferon-induced 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J Biol Chem. 262, 3852–3857 (1987).
Kristiansen, H., Gad, H. H., Eskildsen-Larsen, S., Despres, P. & Hartmann, R. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res. 31, 41–47 (2011).
Hartmann, R., Olsen, H. S., Widder, S., Jorgensen, R. & Justesen, J. p59OASL, a 2′-5′ oligoadenylate synthetase like protein: a novel human gene related to the 2′-5′ oligoadenylate synthetase family. Nucleic Acids Res. 26, 4121–4128 (1998).
Li, Y. et al. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc Natl Acad Sci USA 113, 2241–2246 (2016).
Bonawitz, N. D., Clayton, D. A. & Shadel, G. S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 24, 813–825 (2006).
Desmurs, M. et al. C11orf83, a mitochondrial cardiolipin-binding protein involved in bc1 complex assembly and supercomplex stabilization. Mol Cell Biol. 35, 1139–1156 (2015).
Wanschers, B. F. et al. A mutation in the human CBP4 ortholog UQCC3 impairs complex III assembly, activity and cytochrome b stability. Hum Mol Genet. 23, 6356–6365 (2014).
Wu, F., Fan, X., Yue, Y., Xiong, S. & Dong, C. A vesicular stomatitis virus-based mucosal vaccine promotes dendritic cell maturation and elicits preferable immune response against coxsackievirus B3 induced viral myocarditis. Vaccine. 32, 3917–3926 (2014).
Melchjorsen, J. et al. Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res. 29, 199–207 (2009).
Brehin, A. C. et al. The large form of human 2′,5′-Oligoadenylate Synthetase (OAS3) exerts antiviral effect against Chikungunya virus. Virology. 384, 216–222 (2009).
Osada, N. et al. The genome landscape of the african green monkey kidney-derived vero cell line. DNA Res. 21, 673–683 (2014).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 6, 981–988 (2005).
Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Hornung, V., Hartmann, R., Ablasser, A. & Hopfner, K. P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat Rev Immunol. 14, 521–528 (2014).
Goubau, D., Deddouche, S. & Reis e Sousa, C. Cytosolic sensing of viruses. Immunity. 38, 855–869 (2013).
Acknowledgements
This project was supported by the grant from Natural Science Funding of China (30700699 and 81572402). We would like to thank Dr. Bisen Ding and Dr. Zhongwei Cao for informative comments and suggestions for this study.
Author information
Authors and Affiliations
Contributions
H.Y. conceived the initial concept and designed the study. Y.Y., S.X., B.C., H.L., E. D., Q.L., G.J. and C.Z. carried out the experiments. Y.W. and Y.W. offered critical opinion to improve the design and study. Y.Y. and H.Y. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yang, Y., Xiong, S., Cai, B. et al. Mitochondrial C11orf83 is a potent Antiviral Protein Independent of interferon production. Sci Rep 7, 44303 (2017). https://doi.org/10.1038/srep44303
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep44303
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.