Abstract
We have used a computational approach to identify anti-fibrotic therapies by querying a transcriptome. A transcriptome signature of activated hepatic stellate cells (HSCs), the primary collagen-secreting cell in liver, and queried against a transcriptomic database that quantifies changes in gene expression in response to 1,309 FDA-approved drugs and bioactives (CMap). The flavonoid apigenin was among 9 top-ranked compounds predicted to have anti-fibrotic activity; indeed, apigenin dose-dependently reduced collagen I in the human HSC line, TWNT-4. To identify proteins mediating apigenin’s effect, we next overlapped a 122-gene signature unique to HSCs with a list of 160 genes encoding proteins that are known to interact with apigenin, which identified C1QTNF2, encoding for Complement C1q tumor necrosis factor-related protein 2, a secreted adipocytokine with metabolic effects in liver. To validate its disease relevance, C1QTNF2 expression is reduced during hepatic stellate cell activation in culture and in a mouse model of alcoholic liver injury in vivo, and its expression correlates with better clinical outcomes in patients with hepatitis C cirrhosis (n = 216), suggesting it may have a protective role in cirrhosis progression.These findings reinforce the value of computational approaches to drug discovery for hepatic fibrosis, and identify C1QTNF2 as a potential mediator of apigenin’s anti-fibrotic activity.
Similar content being viewed by others
Introduction
The hepatic stellate cell (HSC) represents the major focus for developing anti-fibrotic therapies, whereas other cell types, e.g, portal fibroblasts, also contribute to fibrogenesis to a minor extent depending on the site, duration and nature of liver injury1,2. HSC-specific genes and proteins likely serve as clues to candidate therapeutic targets and will accelerate preclinical testing and clinical development of anti-fibrotic therapies.
Bioinformatic interrogation of public databases is a comprehensive and efficient strategy to identify disease molecular signatures, drug targets, and even candidate drugs. There are a growing number of transcriptome datasets available for the identification of genes and pathways unique to a variety of biological and clinical contexts across multiple assay formats and tissue types, including liver3. Liver disease-specific molecular signatures such as regulators of collagen deposition and hepatocellular carcinoma subtype/prognosis classifiers have been identified to date4,5,6,7. Our previous unbiased interrogation of the liver cell transcriptome compendium has identified a 122-gene HSC-specific molecular signature uniquely expressed in quiescent and/or activated HSCs compared to other cell types in the liver, which was associated with poorer clinical outcome in patients with hepatitis C virus (HCV)-related cirrhosis and hepatocellular carcinoma8.
In parallel, molecular signature-based in silico drug screening and repurposing has been successfully utilized for quick hypothesis-free identification of novel therapeutics for a variety of cancers and inflammatory diseases, among many others diseases3. In the current study, we used an experimentally-defined HSC activation gene signature in an unbiased manner as a basis for applying a computational drug discovery approach to identify candidate anti-fibrotic drugs that antagonize the HSC gene signature.
Material and Methods
Computational compound screen for candidate anti-fibrotic agents
An HSC activation signature was defined in transcriptome profiles of freshly isolated HSCs from cirrhotic rat liver treated with repeated low-dose diethylnitrosamine (low-dose DEN rat)9 (NCBI Gene Expression Omnibus [GEO] accession number, GSE63726). Differentially expressed genes between the cells isolated from cirrhotic and healthy control livers were defined after making to human orthologous genes (NCBI HomoloGene database, release 68) by random permutation t-test based on significance threshold of false discovery rate (FDR < 0.05) (Supplementary Table 1). The gene signature was used to query a database of transcriptome profiles of 1,309 unique FDA-approved drugs and bioactive compounds, the Connectivity Map (CMap) database (https://portals.broadinstitute.org/cmap/)10. Compounds with significant negative association (enrichment p ≤ 0.05) were selected as candidates for subsequent experimental evaluation.
In vitro assessment of candidate anti-fibrotic agents
TWNT-4 (human HSC line) cells11 were seeded onto a 96-well plate at 5,000 cells per well in 100 μl of assay medium (DMEM, 10% FBS, 1% penicillin/streptomycin), and cultured at 37 °C and 5% CO2. After 24 hours, the cells were treated with apigenin (Sigma-Aldrich) (≥97% purity) at final concentrations of 2.5 μM, 10 μM, 20 μM, 40 μM, 60 μM, 80 μM, 100 μM, and 200 μM dissolved in 0.5% DMSO or DMSO control in triplicate for 24 hours. Cell viability was measured by MTS assay using CellTiter 96® Aqueous One Solution Reagent (Promega) following manufacturer’s instruction. Percentage of mean absorbance of each drug-treated condition over the control was calculated.
Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR)
RNA was extracted from adherent cells using RNeasy Mini kit (Qiagen). Equimolar concentrations of RNA were converted to cDNA (Clontech), and quantitative real-time PCR was performed using SYBR green reagent (Roche) on the Lightcycler 480 system (Roche). Gene expression level in each sample was internally normalized to GAPDH expression. The following PCR primers were used (5′ to 3′): GGCTTCCCTGGTCTTCCTGG (forward) and CCAGGGGGTCCAGCCAAT (reverse) for human COL1A1; GAGGCTCCTCCCAGTCATCA (forward) and GGGATCATGGTGGTTACCCAGA (reverse) for human C1QTNF2; CCAGAAGCCATCAGCAGCAAG (forward) and AGGCCCTGAGAGATCTGTGG (reverse) for human PDGFRB: AGGCACCCCTGAACCCCAA (forward) and CAGCACCGCCTGGATAGCC (reverse) for human ASMA; CAAGGGCTACCATGCCAACT (forward) and AGGGCCAGGACCTTGCTG (reverse) for human TGFB1; CGAGTGCCAAATGAAGAGGACC (forward) and AAACCTGAGCCAGAACCTGACG (reverse) for human TGFRB1; CAATGACCCCTTCATTGACC (forward) and GATCTCGCTCCTGGAAGATG (reverse) for human GAPDH.
Western blotting
TWNT-4 cells were lysed in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 1% IGEPAL, 0.5% Sodium deoxycholate, 1% SDS) with proteinase inhibitors (Roche) and pelleted. Inguinal adipose tissue was dissected from two C57BL/6 mice (Charles River). Whole liver tissue was isolated from 5 control mice fed a normal diet for 6 weeks and 5 mice fed a Lieber DeCarli ethanol-containing diet for 6 weeks. In both cases, the tissue was lysed mechanically using steel beads in a TissueLyser LT (Qiagen) and with RIPA lysis buffer with the proteinase inhibitor. The lysate was sonicated and pelleted and the aqueous supernatant was isolated. Twenty μg of protein from each sample was suspended in NuPAGE LDS sample buffer and heated for 10 min at 70 °C. Samples were electrophoresed on 10% BisTris NuPAGE gels (Invitrogen) and then transferred to nitrocellulose membranes (Invitrogen). Membrane blotting was performed using the following primary antibodies, rabbit polyclonal anti-COL1A1 antibody (Rockland, Limerick, PA, catalog #600-401-103) (1:5000), rabbit polyclonal anti-C1QTNF2 antibody (ProSci, catalog #3561) (1:1000), mouse monoclonal anti-GAPDH antibody (Millipore, catalog #CB1001) (1:2500), mouse monoclonal anti-β-tubulin (Sigma-Aldrich, catalog #T4026) (1:2500), mouse anti-calnexin (Abcam, catalog #75801) (1:2500) and appropriate HRP-conjugated secondary antibody. Bands were visualized with chemiluminescent HRP antibody detection reagent (HyGlo e2400, Denville), captured with Amersham Imager 600 (GE Healthcare Life Sciences), and quantified using ImageJ software (https://imagej.nih.gov/ij/).
Immunofluorescence staining
A total of 50,000 TWNT-4 cells were plated onto glass coverslips and cultured until 90% confluent, and then fixed with 100% acetone for 10 minutes at −20 °C. The cells were subsequently permeabilized in Tween-20 detergent in PBS for 20 minutes and then incubated in the rabbit polyclonal anti-C1QTNF2 antibody (1:1000) with negative and positive controls, chicken polyclonal anti-GFAP (Abcam, catalog #4674) (1:200), and anti-Desmin (AbCam, catalog #15200) (1:200). Appropriate green fluorescent tagged secondary antibodies (Life Technologies) were used and DAPI was used for nuclear staining. Cells were imaged under Eclipse TS100 fluorescent microscope (Nikon).
Culture activated mouse HSCs
DNA microarray-based transcriptome profiles of mouse primary HSCs before (day 0) and after in vitro culture activation (day 7) were obtained from GEO database (NCBI Gene Expression Omnibus [GEO] accession number, GSE34949)12,13.
Clinical HCV cirrhosis cohort
DNA microarray-based transcriptome profiles of 216 patients with HCV-related compensated cirrhosis we previously reported were used to evaluate prognostic association of C1QTNF2 expression level (GSE15654)6. C1QTNF2-high.group was defined as samples with C1QTNF2 expression higher than one standard deviation above mean. Prognostic association was assessed by Kaplan-Meier curve and log-rank test.
Statistical analysis
Continuous values are presented by mean and standard error of mean (SEM). Differences were assessed by either t-test or one-way ANOVA followed by Dunnett’s multiple comparisons test. Two-tailed p-value less than 0.05 was regarded as statistically significant. All statistical analyses were performed using Graph Pad Prism version 7.0a (GraphPad Software).
Results
Computational screen to identify candidate anti-fibrotic agents
A 673-gene in vivo HSC activation signature was defined in the isolated HSC fraction from the low-dose DEN rat (Supplementary Table 1). The gene signature was used to query the compound perturbation transcriptome database (CMap) for candidate anti-fibrotic agents that potentially antagonize the HSC activation signature. Eighteen compounds with significant negative association (p ≤ 0.05) were identified (Table 1). Of note, the majority of the compounds (n = 14, 78%) are not recognized for their possible anti-fibrotic effect, highlighting the potential advantage of this unbiased in silico screen to efficiently identify candidate drugs. Among them, 9 top hit compounds commercially available and without clinically known severe toxicity were chosen for subsequent experimental evaluation.
In vitro validation of anti-fibrotic effect of apigenin in a HSC cell line
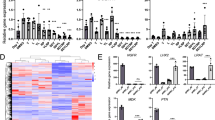
The 9 computationally prioritized candidate compounds were tested for their anti-fibrotic effect in a human HSC cell line, TWNT-4, together with a multi-kinase inhibitor, sorafenib, and an mTOR inhibitor, rapamycin, as positive controls14,15. Apigenin, a flavonoid with a known anti-fibrotic activity in a mouse model of chronic pancreatitis16,17, was the only compound that reduced COL1A1 expression at comparable level to sorafenib, a known anti-fibrotic drug, with statistical significance (Fig. 1A). The COL1A1 suppressive effect was dose-dependent (Fig. 1B), which was also confirmed at the protein level (Fig. 1C). Expression of PDGFRB, encoding platelet-derived growth factor receptor-β, was similarly reduced by 10 μM of apigenin (Fig. 1D). Cell viability assessment showed that the compound is not toxic at concentrations below 20 μM (Fig. 1E). Other known liver fibrosis-related genes, ASMA, TGFB1, and TGFBR1, were not suppressed at the non-toxic concentration (Fig. 1F–H), suggesting that apigenin’s effect is directed to a specific subset of fibrogenesis-related pathways.
(A) Modulation of COL1A1 expression by nine computationally selected candidate compounds together with sorafenib and rapamycin (positive controls) compared to DMSO-treated controls (qRT-PCR, n = 3). *p < 0.05, t-test. (B) Dose-dependent suppression of COL1A1 expression by apigenin (qRT-PCR, n = 3). *p < 0.001, Dunnett’s test. (C) Suppression of collagen 1 protein by sorafenib and apigenin (Western blotting, n = 1). Relative intensity to GAPDH was calculated. (D) Modulation of PDGFRB expression by apigenin and sorafenib (qRT-PCR, n = 3). *p < 0.001, Dunnett’s test. (E) Cell viability in association of apigenin dose (MTS assay, n = 3). *p < 0.01, **p < 0.001, Dunnett’s test. (F) Modulation of ASMA expression by apigenin and sorafenib (qRT-PCR, n = 3). (G) Modulation of TGFB1 expression by apigenin and sorafenib (qRT-PCR, n = 3). (H) Modulation of TGFRB1 expression by apigenin and sorafenib (qRT-PCR, n = 3). Bar graphs show mean and standard error of mean (SEM) (error bars) of replicated experiments.
C1QTNF2 as a potential intracellular target of apigenin
Next we sought to identify targets of apigenin in hepatic stellate cells. A 122-gene signature uniquely expressed in HSC8 was overlaid on a list of 160 genes encoding intracellular proteins that physically interact with apigenin based on phase display18. C1QTNF2 was identified as the only gene common to the 2 gene lists (Fig. 2A). With 20,354 protein-coding genes in human genome according to NCBI CCDS database (release 21) (www.ncbi.nlm.nih.gov/projects/CCDS), the number of apigenin target genes (160 genes) that could be found within the hepatic stellate cell signature (122 genes) by chance is less than 1 (0.959), although it does not reach statistical significance (p = 0.37, hypergeometric test). To date, C1q and tumor necrosis factor related protein 2 encoded (C1QTNF2) protein is an adipokine that has been identified previously in adipose tissue, where it has effects on lipid metabolism and insulin activity19, but upregulation of C1QTNF2 mRNA expression unique to HSC was confirmed in a panel of various cell types presenting in fibrotic liver, which were assembled in our previous study8 (Fig. 2B). Furthermore, immunofluorescence staining showed strong cytoplasmic expression of C1QTNF2 in TWNT-4 cells (Fig. 2C). Apigenin treatment did not alter C1QTNF2 mRNA and protein abundance (Fig. 2D,E). These results suggest that apigenin elicits its COL1A1-suppressive effect in HSC without modulating C1QTNF2 expression levels.. However, apigenin could interfere with the function of C1QTNF2 protein via a physical interaction. Interestingly, baseline C1QTNF2 mRNA expression was reduced during the process of in vivo HSC activation by carbon tetrachloride (CCl4) treatment or bile duct ligation (BDL) (Fig. 2B) and in vitro culture activation13 (Fig. 3A). C1QTNF2 protein expression was similarly reduced in the livers from mouse model of alcoholic injury by Lieber DeCarli ethanol-containing diet for 6 weeks20 (Fig. 3B). Furthermore, in a clinical cohort of 216 patients with HCV-related compensated cirrhosis, patients with high C1QTNF2 expression showed a better clinical outcome of cirrhosis, as measured by Child-Pugh classification21 (Fig. 4). These animal model- and clinical cohort-based findings suggest that C1QTNF2 plays a protective role in cirrhosis progression. If a physical interaction with C1QTNF2 protein is needed for apigenin to elicit its COL1A1-suppresive effect as we computationally predict, it may be possible that the status of C1QTNF2 expression can serve as a predictive marker of apigenin responses, which could be clarified in future studies.
(A) Overlap between HSC gene signature8 and potential intracellular targets of apigenin18. (B) C1QTNF2 expression in a panel of various cell types isolated from mouse livers (expression DNA microarray, n ≥ 3 in each cell type)8. BDL: bile duct ligation; CCl4: carbon tetrachloride. *p < 0.001, Tukey’s test. (C) Subcellular localization of C1QTNF2 protein in TWNT-4 cells (immunofluorescence staining). (D) Modulation of C1QTNF2 expression by apigenin and sorafenib in TWNT-4 cells (qRT-PCR, n = 2). (E) Modulation of C1QTNF2 protein by apigenin and sorafenib in TWNT-4 cells and adipose tissues (Western blotting, n = 1). Relative intensity to tubulin was calculated.
(A) C1QTNF2 expression in freshly isolated (“quiescent”) and 7-day-cultured (“activated”) mouse primary HSCs13 (expression DNA microarray, n = 3). *p < 0.001, t-test. (B) C1QTNF2 protein expression in livers from mice fed with Lieber DeCarli ethanol-containing diet normalized to calnexin (Western blotting, n = 5). *p < 0.05, t-test. Bar graphs show mean and standard error of mean (SEM) (error bars) of replicated experiments.
(A) clinical cohort of 216 patients with compensated HCV-related cirrhosis6 was classified into C1QTNF2-high (n = 25) and low (n = 101) groups, and evaluated for association with cirrhosis progression, i.e., progression of Child-Pugh class21 from (A) to (B) or (C). P-value was calculated by log-rank test.
Discussion
Molecular signature-based unbiased and hypothesis-free computational drug discovery has been successfully utilized primarily in cancer and inflammatory diseases3. Our study has demonstrated that this strategy can be similarly applied to anti-fibrotic drug discovery. Generation of the query gene signature in transcriptomic profiles of activated HSCs enabled the discovery of candidate compounds specific to the biological context, circumventing the need for costly large compound library screen. The identification of apigenin, already known to be anti-fibrotic in other tissue types such as pancreas22,23, clearly indicates that our approach is a viable option to discover biologically relevant anti-fibrotic agents in a cost-effective manner.
Apigenin is a flavonoid, abundant in parsley and celery, that has gained interest as a health-promoting agent because of its low intrinsic toxicity24. Despite the promising in vitro anti-fibrogenic activity comparable to sorafenib14, its poor solubility limits optimal in vivo biodistribution and needs further biochemical modifications to improve solubility. In addition, apigenin is known to modulate the immune response18, although our in vitro experimental system did not uncover an immune-related activity, which could be assessed in future studies. C1QTNF2, which we have associated with liver disease severity and prognosis, may be a factor that potentially influences the outcome of apigenin-based therapy in particular, and the biology of hepatic fibrosis in general. C1QTNF2 is a member of C1q/TNF-related proteins that represent an adipokine family less characterized than other well-studied adipocytokines implicated in liver fibrosis, such as adiponectin19,25. The only known source of C1QTNF2 production is stromal vascular cells in adipose tissue, and the highly restricted sites of production suggests its function is distinct from adiponectin19. Indeed, C1QTNF2 does not substitute for adiponectin in caloric restriction26. C1QTNF2 is known to be involved in several metabolic processes such as AMP kinase phosphorylation to stimulate glucose uptake in muscle cells27 and improvement of insulin and lipid tolerance in diet-induced obese mice28. C1QTNF2 may form heteromers with C1QTNF7 and adiponectin19. Our study suggets it has a novel role in HSC biology that could maintain the cell’s quiescent state. These findings reinforce the value of computational approaches to drug discovery for hepatic fibrosis, and identify C1QTNF2 as a potential mediator of apigenin’s anti-fibrotic activity.
Additional Information
How to cite this article: Hicks, D. F. et al. Transcriptome-based repurposing of apigenin as a potential anti-fibrotic agent targeting hepatic stellate cells. Sci. Rep. 7, 42563; doi: 10.1038/srep42563 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Mederacke, I. et al. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun 4, 2823 (2013).
Trautwein, C., Friedman, S. L., Schuppan, D. & Pinzani, M. Hepatic fibrosis: Concept to treatment. J Hepatol 62, S15–24 (2015).
Wooden, B., Goossens, N., Hoshida, Y. & Friedman, S. L. Using Big Data to Discover Diagnostics and Therapeutics for Gastrointestinal and Liver Diseases. Gastroenterology 152, 53–67 e53 (2017).
Blaauboer, M. E. et al. Novel combination of collagen dynamics analysis and transcriptional profiling reveals fibrosis-relevant genes and pathways. Matrix Biol 32, 424–431 (2013).
Hoshida, Y. et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res 69, 7385–7392 (2009).
Hoshida, Y. et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology 144, 1024–1030 (2013).
Hoshida, Y. et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 359, 1995–2004 (2008).
Zhang, D. Y. et al. A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut 65, 1754–1764 (2016).
Fuchs, B. C. et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology 59, 1577–1590 (2014).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Shibata, N. et al. Establishment of an immortalized human hepatic stellate cell line to develop antifibrotic therapies. Cell transplantation 12, 499–507 (2003).
Friedman, S. L. & Roll, F. J. Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Analytical biochemistry 161, 207–218 (1987).
Chen, Y. et al. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology 143, 1319–1329 e1311–1311 (2012).
Thabut, D. et al. Complementary vascular and matrix regulatory pathways underlie the beneficial mechanism of action of sorafenib in liver fibrosis. Hepatology 54, 573–585 (2011).
Bridle, K. R. et al. Rapamycin inhibits hepatic fibrosis in rats by attenuating multiple profibrogenic pathways. Liver Transplantation 15, 1315–1324 (2009).
Chen, H. et al. Design, synthesis, and characterization of novel apigenin analogues that suppress pancreatic stellate cell proliferation in vitro and associated pancreatic fibrosis in vivo . Bioorganic & medicinal chemistry 22, 3393–3404 (2014).
Mrazek, A. A. et al. Apigenin inhibits pancreatic stellate cell activity in pancreatitis. Journal of Surgical Research 196, 8–16 (2015).
Arango, D. et al. Molecular basis for the action of a dietary flavonoid revealed by the comprehensive identification of apigenin human targets. Proc Natl Acad Sci USA 110, E2153–2162 (2013).
Wong, G. W. et al. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J 416, 161–177 (2008).
Lieber, C. S. & DeCarli, L. M. The feeding of alcohol in liquid diets: two decades of applications and 1982 update. Alcohol Clin Exp Res 6, 523–531 (1982).
Pugh, R. N., Murray-Lyon, I. M., Dawson, J. L., Pietroni, M. C. & Williams, R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg 60, 646–649 (1973).
Chen, H. et al. Design, synthesis, and characterization of novel apigenin analogues that suppress pancreatic stellate cell proliferation in vitro and associated pancreatic fibrosis in vivo . Bioorg Med Chem 22, 3393–3404 (2014).
Mrazek, A. A. et al. Apigenin inhibits pancreatic stellate cell activity in pancreatitis. J Surg Res 196, 8–16 (2015).
Shukla, S. & Gupta, S. Apigenin: a promising molecule for cancer prevention. Pharmaceutical research 27, 962–978 (2010).
Schaffler, A. & Buechler, C. CTRP family: linking immunity to metabolism. Trends Endocrinol Metab 23, 194–204 (2012).
Rohrbach, S., Aurich, A. C., Li, L. & Niemann, B. Age-associated loss in adiponectin-activation by caloric restriction: lack of compensation by enhanced inducibility of adiponectin paralogs CTRP2 and CTRP7. Mol Cell Endocrinol 277, 26–34 (2007).
Wong, G. W., Wang, J., Hug, C., Tsao, T.-S. & Lodish, H. F. A family of Acrp30/adiponectin structural and functional paralogs. Proceedings of the National Academy of Sciences of the United States of America 101, 10302–10307 (2004).
Peterson, J. M., Seldin, M. M., Tan, S. Y. & Wong, G. W. CTRP2 overexpression improves insulin and lipid tolerance in diet-induced obese mice. PLoS One 9, e88535 (2014).
Smolen, J. S. et al. Efficacy and safety of leflunomide compared with placebo and sulphasalazine in active rheumatoid arthritis: a double-blind, randomised, multicentre trial. The Lancet 353, 259–266 (1999).
Lane, I., Lumley, P., Michael, M., Peters, A. & McCollum, C. A specific thromboxane receptor blocking drug, AH23848, reduces platelet deposition on vascular grafts in man. Thrombosis and haemostasis 64, 369–373 (1990).
De Bono, D. P. et al. Effect of the specific thromboxane receptor blocking drug AH23848 in patients with angina pectoris. British heart journal 56, 509–517 (1986).
German & Group, A.X.S. Double-blind placebo-controlled comparison of digoxin and xamoterol in chronic heart failure. The Lancet 331, 489–493 (1988).
Rougier, P. et al. Phase II study of irinotecan in the treatment of advanced colorectal cancer in chemotherapy-naive patients and patients pretreated with fluorouracil-based chemotherapy. Journal of Clinical Oncology 15, 251–260 (1997).
Pareek, A., Chandanwale, A., Oak, J., Jain, U. & Kapoor, S. Efficacy and safety of aceclofenac in the treatment of osteoarthritis: a randomized double-blind comparative clinical trial versus diclofenac–an Indian experience. Current medical research and opinion 22, 977–988 (2006).
Sandoval, M. A. & Hernandez-Vaquero, D. Preventing peridural fibrosis with nonsteroidal anti-inflammatory drugs. European Spine Journal 17, 451–455 (2008).
Singh, G., Radhakrishnan, G. & Guleria, K. Comparison of sublingual misoprostol, intravenous oxytocin, and intravenous methylergometrine in active management of the third stage of labor. International Journal of Gynecology & Obstetrics 107, 130–134 (2009).
Bateman, B. et al. Methylergonovine Maleate and the Risk of Myocardial Ischemia and Infarction. Obstetric Anesthesia Digest 34, 214–215 (2014).
Chung, F.-H. et al. Gene-Set Local Hierarchical Clustering (GSLHC)—A Gene Set-Based Approach for Characterizing Bioactive Compounds in Terms of Biological Functional Groups. PloS one 10, e0139889 (2015).
Investigators, W.G.f.t.W.s.H.I. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women’s Health Initiative randomized controlled trial. Jama 288, 321–333 (2002).
Janssen, K. et al. Effects of the flavonoids quercetin and apigenin on hemostasis in healthy volunteers: results from an in vitro and a dietary supplement study. The American journal of clinical nutrition 67, 255–262 (1998).
Ma, G., Zhang, R., Ying, K. & Wang, D. Effect evaluation of cisplatin-gemcitabine combination chemotherapy for advanced non-small cell lung cancer patients using microarray data. Eur Rev Med Pharmacol Sci 19, 578–585 (2015).
Li, J. et al. Gene expression profiling of CD133-positive cells in coronary artery disease. Molecular medicine reports 12, 7512–7516 (2015).
Balakumar, P. & Singh, M. Effect of 3-aminobenzamide, an inhibitor of poly (ADP-ribose) polymerase in experimental cardiac hypertrophy. Int J Pharmacol 2, 543–548 (2006).
Huttunen, M. O. et al. Risperidone versus zuclopenthixol in the treatment of acute schizophrenic episodes: a double‐blind parallel‐group trial. Acta Psychiatrica Scandinavica 91, 271–277 (1995).
Yoshitake, T., Asano, K., Yamamura, H., Yoshitake, J. & Arai, T. Experimental and clinical studies on the hemodynamic and metabolic effects of dibutyryl cyclic AMP. Progress in clinical and biological research 111, 211 (1983).
Miyagi, Y., Sasayama, S., Nakajima, H., Fujita, M. & Asanoi, H. Comparative hemodynamic effects of intravenous dobutamine and dibutyryl cyclic AMP, a new inotropic agent, in severe congestive heart failure. Journal of cardiovascular pharmacology 15, 138–143 (1990).
Kollros, P., Bates, S., Mathews, M., Horwitz, A. & Glagov, S. Cyclic AMP inhibits increased collagen production by cyclically stretched smooth muscle cells. Laboratory investigation; a journal of technical methods and pathology 56, 410–417 (1987).
Acknowledgements
Funding from Icahn School of Medicine Medical Student Research Office to D.F.H. Juan de la Cierva contract (JCI-2012-15124, Ministerio de Economía y Competitividad) and Fundación Juan Esplugues and FISABIO (UGP-14-153) to A.B.G. FLAGS foundation, the Nuovo-Soldati Cancer Research Foundation, and an advanced training grant from Geneva University Hospital to N.G. NIH/NIDDK DK099558, European Union ERC-2014-AdG-671231HEPCIR, Irma T. Hirschl Trust, and US Department of Defense (W81XWH-16-1-0363) to Y.H., and NIH DK56621 and AA020709 to S.L.F.
Author information
Authors and Affiliations
Contributions
Study concept and design: D.F.H., N.G., A.B.G., B.W., M.C.W., B.R., J.T.D., Y.A.L., Y.H., S.L.F. Acquisition of data: D.F.H., N.G., A.B.G., T.T., M.C.W., A.L., B.R. Analysis and interpretation of data: D.F.H., N.G., A.B.G., M.C.W., A.L., Y.A.L., Y.H., S.L.F. Drafting of the manuscript: D.F.H., Y.H., S.L.F. Critical revision of the manuscript for important intellectual content: all authors. Obtained funding: D.F.H., N.G., T.T., Y.H., S.L.F.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Hicks, D., Goossens, N., Blas-García, A. et al. Transcriptome-based repurposing of apigenin as a potential anti-fibrotic agent targeting hepatic stellate cells. Sci Rep 7, 42563 (2017). https://doi.org/10.1038/srep42563
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep42563
This article is cited by
-
Repositioning of a novel GABA-B receptor agonist, AZD3355 (Lesogaberan), for the treatment of non-alcoholic steatohepatitis
Scientific Reports (2021)
-
Weighted correlation network bioinformatics uncovers a key molecular biosignature driving the left-sided heart failure
BMC Medical Genomics (2020)
-
Apigenin inhibits fibroblast proliferation and reduces epidural fibrosis by regulating Wnt3a/β-catenin signaling pathway
Journal of Orthopaedic Surgery and Research (2019)
-
Postgenomic Properties of Natural Micronutrients
Bulletin of Experimental Biology and Medicine (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.