Abstract
Santalum album L. (Indian sandalwood) is an economically important plant species because of its ability to produce highly valued perfume oils. Little is known about the mechanisms by which S. album adapts to low temperatures. In this study, we obtained 100,445,724 raw reads by paired-end sequencing from S. album leaves. Physiological and transcriptomic changes in sandalwood seedlings exposed to 4 °C for 0–48 h were characterized. Cold stress induced the accumulation of malondialdehyde, proline and soluble carbohydrates, and increased the levels of antioxidants. A total of 4,424 differentially expressed genes were responsive to cold, including 3,075 cold-induced and 1,349 cold-repressed genes. When cold stress was prolonged, there was an increase in the expression of cold-responsive genes coding for transporters, responses to stimuli and stress, regulation of defense response, as well as genes related to signal transduction of all phytohormones. Candidate genes in the terpenoid biosynthetic pathway were identified, eight of which were significantly involved in the cold stress response. Gene expression analyses using qRT-PCR showed a peak in the accumulation of SaCBF2 to 4, 50-fold more than control leaves and roots following 12 h and 24 h of cold stress, respectively. The CBF-dependent pathway may play a crucial role in increasing cold tolerance.
Similar content being viewed by others
Introduction
Low temperature is one of the major abiotic factors that impedes plant growth and limits crop yield and geographical distribution1. Plants have evolved several adaptive mechanisms to cope with cold stress to protect them against damage2. A diverse range of plant species can withstand cold stress to varying degrees, but this depends on reprogramming gene expression to adjust their metabolism and developmental programs3. Not all plants are able to tolerate low nonfreezing temperatures while temperate plants can adapt to a cold environment and increase their cold tolerance in response to low temperatures, such as Arabidopsis, winter wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.), among others4. By contrast, plants of tropical and subtropical origin generally have little tolerance to low temperatures and exhibit various symptoms of chilling injury, such as maize (Zea mays L.)5, cassava (Manihot esculenta Crantz)6, and soybean (Glycine max Merr.)7.
The mechanism of cold stress in plants has been extensively studied over the last 20 years but the complete mechanisms by which plants perceive low temperatures still remain unknown. Research has shown that cold stress tolerance involves the remodeling of cell structures and reprogramming gene expression by triggering a series of protective mechanisms against cold damage, including an increase in the level of intracellular solutes, the accumulation of cryoprotectants and antioxidants, as well as the induction of antifreeze proteins and cold-regulated (COR) proteins8,9. During cold stress, these metabolites and proteins help to protect plant membranes and prevent cell disruption by stabilizing membrane lipids, maintaining ion homeostasis and scavenging reactive oxygen species (ROS)3.
The molecular mechanisms underlying cold stress tolerance have been investigated in the model plant Arabidopsis, as well as in other crop species such as maize, wheat and barley1,10,11. Plants have developed C-repeat (CRT)-binding factor/dehydration-responsive element binding factor (CBF/DREB)-dependent and CBF/DREB-independent pathways, allowing them to adapt to cold stress. The most extensively studied mechanism of the response to cold has focused on the CBF/DREB-mediated transcriptional regulatory cascade4. Recently, it was revealed that the number of genes (about 100) induced by CBF genes only accounted for a small percentage of COR genes in freezing tolerance while other cold-induced genes showed expression patterns similar to those of CBFs in Arabidopsis, such as ZAT12 (C2H2-type zinc finger), RAV1 (related to ABI3/VP1), and MYB7312. Besides Arabidopsis, the CBF/DREB1 transcription factors (TFs) are present in a wide array of plants, including crops, such as wild tomato (Solanum habrochaites S. Knapp & D.M. Spooner) and rice (Oryza sativa L.)13,14, as well as woody plants, such as grey poplar (Populus tremula × alba)15, frost grape (Vitis riparia Michx.)16, silver birch (Betula pendula Roth.)17, and cider gum (Eucalyptus gunnii Hook.f.)18. In addition, signaling molecules such as Ca2+, protein kinase, ROS, and hormones (auxin, abscisic acid (ABA), and gibberellin (GA)) play important roles in cold-induced gene expression19,20.
Santalum album L., commonly known as Indian sandalwood, is the best known economically important member of the Santalaceae, and is highly valued for its aromatic heartwood that contains sandal oil, which has various applications in cosmetics, religion and medicine21,22. Four sesquiterpenols (α-, β-, and epi-β-santalol, and α-exo-bergamotol) make up approximately 90% of S. album sandalwood oil21. Relevant terpenoid synthase genes producing these major constituents were cloned and characterized23,24,25,26,27,28. Külheim et al. discovered that several key genes in the terpenoid pathway were induced by spraying a 0.1% methyl jasmonate solution over the foliage, but valuable heartwood essential oils were not be detected in the leaves and stems of seedlings following this treatment29. Whether environmental stresses, including cold stress, affect the expression of terpene synthase genes and the accumulation of essential oil in sandalwood is unclear.
S. album is a tropical and subtropical species, and cold damage is the main factor limiting the introduction and cultivation of this species. It occurs in coastal dry forests and also grows at 700 m above sea level in India and displays a wide temperature tolerance ranging from 4.5 to 38 °C30. As the price for the heartwood of S. album has increased at an average rate of more than 50% per year over the past two decades31, natural populations have been heavily exploited and thus wild populations are vulnerable to extinction32. S. album is now cultivated in South China, Sri Lanka, Indonesia, Malaysia, the Philippines and Northern Australia33. This species was first introduced to South China Botanical Garden (SCBG) in 1970 and its cultivation was then limited to Guangdong, Fujian, and Guangxi provinces, which have a subtropical climate. Every year, these regions experience extremely low temperatures (<−2 °C) in January. A statistical survey showed that these trees grow safely above −1 °C throughout winter34. However, little is known about the physiological and molecular mechanisms that underlie the cold stress response in S. album.
In this study, we completed the in-depth transcriptome sequencing of leaf tissues of S. album seedlings. Comprehensive and dynamic changes in the leaves in response to cold stress were monitored using physiological and RNA-seq analyses. Genes involved in terpenoid biosynthesis were identified from the transcriptome data. Eight genes were differentially expressed by cold stress. We further investigated the TFs involved in regulating the response of sandalwood to cold stress using transcriptome data combined with RACE and quantitative real time PCR (qRT-PCR). This study presents the first global transcriptomic overview of the genes involved in the cold stress response in sandalwood. Our findings lay a solid foundation for further investigation of the molecular mechanisms responsible for this tree’s response to cold stress.
Results
Morphological and physiochemical changes of S. album in response to cold stress
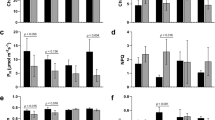
Sandalwood seedlings displayed no obvious morphological alterations prior to 24 h of cold stress (Fig. 1a,b), but young leaves began to wilt as cold treatment was prolonged from 24 to 48 h (Fig. 1c–f). The terminal shoots of seedlings recovered within 48 h after being returned to normal conditions (Fig. 1g). Malondialdehyde (MDA) and electrolyte leakage (EL) are generally used as direct stress markers to reflect membrane damage by cold stress35. In this study, MDA content showed no significant change at 12 h (relative to the control) and a slight increase at 24 h after cold treatment, but peaked at 48 h with a 1.77-fold higher change than the control (Fig. 2a). EL decreased slightly at 12 h, increased dramatically at 24 h, then decreased after 48 h of cold stress. The content of proline, a well-known osmoprotectant, showed no significant difference at 12 h compared to the control, but increased rapidly and peaked at 24 h, then decreased slightly after 48 h of cold stress. These results strongly suggest that the cold-stressed sandalwood seedlings actively mounted a stress response after they were exposed to 4 °C. Moreover, the activities of superoxide dismutase (SOD) and peroxidase (POD) showed continuously increasing trends, with approximately two-fold higher levels than the control at 48 h, implying that these ROS-scavenging enzymes were involved in the detoxification of ROS induced by cold. The total soluble carbohydrate content increased at 12 h, but did not change significantly after 24 and 48 h of exposure to cold.
(a) Control plant. (b–d) Cold-stressed plants for 12, 24, and 48 h, respectively. (e,f) The terminal shoots of seedlings at 24 h (e) and 48 h (f) after cold treatment. Arrows show leaves harmed by cold. (g) Seedlings recovered within 48 h after cold treatment. Potted host plant in (a,b,c,d) is Kuhnia rosmarnifolia. Scale bar = 2 cm.
(a) Changes in malondialdehyde (MDA), electrolyte leakage, proline content, superoxide dismutase (SOD) activity, peroxidase (POD) activity, and soluble sugar content in response to cold stress; (b) Photosynthetic indices under cold stress. Three measurements were averaged and statistically treated using a t-test. *P ≤ 0.05 > 0.005; **P ≤ 0.005 < 0.001; ***P < 0.001. FW, fresh weight.
The dynamic net photosynthetic rate (A), H2O stomatal conductance (Gs), transpiration rate (E), intercellular CO2 concentration (Ci), and respiration rate (R) of leaves were further monitored during cold stress (Fig. 2b). The level of A decreased sharply at 12 h after exposure to cold and declined gradually from 12 to 24 h, but reached a minus value at 48 h. Both Gs and E showed a similar trend as A. A stable level of Ci was sustained from 0 to 24 h, but increased sharply by 60–80% at 48 h. It is likely that the rapid decline in A caused much CO2 to fill the intercellular spaces. Water-use efficiency (WUE) was higher at 12 and 24 h compared to the control, implying that an increase in WUE occurred in response to moderate water deficits caused by cold. However, WUE became sharply minus after 48 h of cold stress, indicating that plants suffered from severe cold stress. R decreased gradually from 0 to 48 h.
Development of de novo transcriptome reference sequence and mapping of stress-induced transcriptome sequences from sandalwood leaves
In total, 97,349,766 clean reads out of 100,445,724 raw reads (GenBank Accession: SRR3731808, SRR3731809) were obtained from S. album leaves after adapter trimming and low quality trimming. De novo sequences were assembled to yield the S. album Gene Index Version 2 (SaGI02) with a total of 135,938 non-redundant unigenes, having an average size of 1203 bp and an N50 value of 2253 (Table S1). The length distributions of unigenes revealed that more than 60,507 unigenes (44.51%) were greater than 1000 bp (Fig. S1). A total of 80,426 sequences (59.16%) were annotated as homologs against the NCBI non-redundant protein (Nr) database, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), eukaryotic orthologous groups (COG) and GO databases with a cut-off E-value of <10−5. For the E-value distribution, more than half of the homologous sequences (50.5%) had a threshold E-value less than 1.0E−60 that showed strong homology (Fig. S2a). The similarity distributions showed that 68.9% of sequences had more than 60% similarity with sequences within the Nr database, while 31.1% of matches had lower similarity that ranged from 17% to 60% (Fig. S2b). The species that provided the best BLASTx match was V. vinifera, which showed 52.7% homology (Fig. S2c).
To gain a dynamic view of the gene profiles regulated by cold stress, we further performed RNA-seq analysis of non-cold-treated leaves (0 h) and cold-treated leaves at 12, 24, and 48 h using three biological replicates. A total of 12 libraries that were constructed were subjected to the Illumina sequencing platform. The sequenced raw reads have been submitted to the SRA at NCBI with the following accession numbers: SRR3490434, SRR3490436, SRR3490438 through to SRR3490442, SRR3490480, SRR3490561, SRR3490688, SRR3490711, and SRR3490712. After filtering out low-quality data (tags containing unknown base N and only adaptor tags), the clean reads of all 12 libraries accounted for more than 99.9% of raw reads (Fig. S3). Approximately 89.87–91.45% of reads in the 12 libraries were mapped to SaGI02, suggesting that the transcriptome was a reliable reference, in which approximately 72.24–74.14% of reads were perfectly matched (Table 1).
Correlation analysis of samples and expression pattern of differentially expressed genes under cold treatment
The global abundance of gene expression showed that most transcripts were activated by cold, in which the number of genes with FPKM, i.e. Fragments Per Kilobase of transcript per Million mapped reads, ranging from 1 to 10 was most abundant, as assessed by box plot analysis (Fig. 3a). The square of Pearson’s correlation coefficient (R2) was greater than 0.92 among the three biological replicates at each time point, indicating both operational stability and the reliability of the experimental results (Table S2). Principal component analysis showed that the expression levels between all biological replicates at each time point were highly correlated (Fig. 3b). These data suggest that alterations in quantitative expression could be estimated by statistical analysis of this dataset. A total of 4,424 differentially expressed genes (DEGs) were identified as being cold responsive over 48 h of cold stress, including 3,075 cold-inducible and 1,349 cold-repressed genes (Fig. 4a,b). A heat map representing the transcript levels for these transcripts at 0, 12, 24 and 48 h showed that these transcripts could be divided into eight clusters based on the modulation of their expression (Fig. 4c,d; Table S3). Most genes in cluster 2 were rapidly up-regulated at 12 h, but were down-regulated 24 and 48 h post cold treatment, indicating that these transcripts were transiently expressed by cold. Clusters 4, 6 and 8 showed similar patterns of changes in transcript, with an increase in expression at 12, 24, and 48 h compared to the control.
(a) Venn diagrams represent the overlaps of induced (left) and repressed (right) genes across three comparisons. (b) Induced and repressed genes for each pairwise comparison. (c) Heat map of the expression profiles of 4424 DEGs after cold stress. (d) Hierarchical clustering of DEGs in clusters in Fig. 3(c). The X axis represents time points of cold treatments. The Y axis represents the value of the relative expression level (log2 (FPKM + 1))(Fragments Per Kilobase of transcript per Million mapped reads). Hierarchical clustering was performed for DEGs with a ≥2-fold change and deviation probability ≥0.8 in at least one pairwise comparison of 0 vs 12 h/24 h/48 h using the Manhattan distance matrix of the heatmap.2 function in the gplots package of the R environment.
Gene ontology and pathway enrichment analyses of differentially expressed genes
In order to identify the biological functions associated with the cold stress response in sandalwood leaves, pairwise gene set enrichment analyses using a hypergeometric test were performed to search for over- and under-represented GO terms (0 vs 12 h/24 h/48 h). The datasets of three time points in comparison with the control yielded many significant over-represented GO terms (Fig. 5; Table S4). As shown in Fig. 5a, there were seven over-represented GO terms in all three pair-wise comparisons. Some overlapping and unique GO terms were observed at different time points, suggesting that a dynamic response occurred over the time course of cold stress. Prior to 24 h of cold stress, GO terms enriched in up-regulated transcripts were associated with plasma membrane transporters, cell wall reconstruction, etc. (Fig. 5a). GO-terms for down-regulated transcripts included water transport, malate metabolism, and regulation of ROS metabolic processes (Fig. 5b). Many underlying genes involved in membrane transporters, calcium signaling and protein kinases that were induced by cold stress are shown in Tables S5 and S6. These results suggest that the cell membrane, cell homeostasis, and calcium signaling play important roles in the early response to cold stress. GO terms related to flavonoid biosynthetic, ketone biosynthetic, and phenylpropanoid metabolic processes indicate that these secondary metabolites might be an integral part of sandalwood’s adaptation to cold stress. Interestingly, 36 over-represented GO terms were significantly enriched at 48 h/0 h (Fig. 5a). Among these, GO terms related to the response to endogenous stimuli, osmotic stress, hormones, and regulation of the defense response, were most dominant. The most notable are the enrichment of GO-terms related to the response to indole butyric acid or brassinosteroid (BR) stimulus, auxin polar transport, GA catabolism and the GA- or BR-mediated signaling pathway, as well as jasmonic acid (JA) biosynthesis. A number of DEGs related to hormones were found in the list of DEGs (Table S7). These suggest that trends in the expression of genes regulating cold response-related processes are enhanced as cold treatment proceeds, allowing sandalwood plants to adapt to environmental variations in which phytohormones play an important role. Based on this assessment, the transcriptomic data was divided into two groups, representing early (12, 24 h) and late (48 h) responses in S. album.
(a) Processes enriched for cold-induced transcripts. (b) Processes enriched for cold-repressed transcripts. The induced or repressed DEGs were annotated to the GO database using a hypergeometric test. The calculated P value underwent a Bonferroni correction and the threshold of the corrected P value was less than 0.05.
According to the KEGG function annotations, a total of 12, 22, and 10 pathways that were significantly enriched (Qvalue ≤ 0.05) were identified at 12 h/0 h, 24 h/0 h, and 48 h/0 h, respectively (Table 2). The “pyruvate metabolism”, “starch and sucrose metabolism”, and “carbon fixation in photosynthetic organisms” pathways that are involved in plastid photosynthesis were predominant in all three pairwise comparisons, suggesting that these pathways play important roles in the cold stress response. DEGs associated with membrane damage and cell construction were significantly enriched, including those involved in linoleic acid and fatty acid metabolism, the biosynthesis of unsaturated fatty acids, and other processes. As shown in Table 2 and Fig. S4, a number of genes in the “plant hormone signal transduction” pathway changed significantly. In addition, we noticed that monoterpenoid, diterpenoid or terpenoid backbone biosynthesis were significantly enriched in early and late responses.
Photosynthesis under cold treatment
To further explore the effect of cold stress on photosynthesis, a total of 98 DEGs associated with the KEGG photosynthesis pathway in the KEGG database were detected (Table S8). The expression of these genes was classified into eight groups (Fig. S5). Genes in clusters 2, 3, and 8 were persistently up-regulated, the majority of which were induced by cold, whereas the expression of genes in cluster 4 was down-regulated. Interestingly, genes in clusters 1 and 6 were down-regulated at 12 h, but were once again induced at 24 h. These results indicate that diverse activation and repression of gene expression is associated with the photosynthesis of sandalwood plants exposed to cold treatment.
Among these DEGs, 23 out of the 32 induced transcripts were annotated as malate dehydrogenases (NADP-MDHs), implying that carbon dioxide fixation might be enhanced under cold treatment. Genes coding for electron transporters of photosystem I (PSI) and photosystem II (PSII), PSI P700 chlorophyll a apoprotein A2 (PsaB) and PSII CP47 chlorophyll apoprotein (PabB), and oxygen-evolving enhancer protein 2 (OEE2), a key protein of the photosynthetic light reaction in PSII, were induced by cold. Several genes encoding phytochrome-interacting factor 3 (PIF3), which is involved in the light-responsive transcriptional network, accumulated persistently. In contrast, two light-harvesting complex II chlorophyll a/b binding (LHCB) protein-coding genes declined sharply at 12 h, but accumulated significantly at 24 h and 48 h. Taken together, these results suggest that photoprotection, to some extent, increased in response to cold damage. In contrast, the expression of genes annotated as phosphoenolpyruvate carboxylase (PEPC), fructose-bisphosphate aldolase (FBPA), triosephosphate isomerase (TPI), and ribulose-phosphate 3-epimerase (RPE), was persistently down-regulated, suggesting that carbohydrate metabolism had weakened during cold stress.
Response of genes involved in terpene biosynthesis under cold treatment
All terpenoids are synthesized through two independent pathways: the cytosolic mevalonic acid (MVA) pathway and the plastidial methyerythriol phosphate (MEP) pathway27. In this study, 51 unigenes were found to be potentially related to terpenoid backbone biosynthesis. Among these, at least one unigene from the transcriptome database of S. album leaves corresponded to the enzyme participating in core biosynthetic steps of the MVA and MEP pathways (Table S9). These unigenes were expressed to varying degrees, but there were no significant differences among them at 12, 24 or 48 h of cold stress compared to the control except for several homologs of the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase gene, a first rate-limiting enzyme in the MVA pathway. This gene had log2Ratio >10 values in treated leaves than in the control (CL5305.Contig8). More importantly, of the 41 transcripts encoding terpenoid synthases, seven genes were differentially expressed, including five up-regulated and two down-regulated by cold stress (Fig. 6a).
(a) Heat map of changes of transcript abundance for eight genes. (b) qRT-PCR determination of transcript levels of these genes in leaves and roots following cold treatment (4 °C). Three measurements were averaged from the results of three replicated experiments and statistically treated using a t-test. *P ≤ 0.05 > 0.005; **P ≤ 0.005 < 0.001; ***P < 0.001. L, leaves. R, roots.
The full lengths of these seven genes besides one HMG-CoA were obtained by RACE. The sequences of two genes (unigene64433 and CL737.Contig6) were identical to SaMonoTPS1 and S. album bergamotene oxidase (SaCYP76F38) submitted to NCBI (accession No. EU798692 and KC533715, respectively). The remaining six genes are being functionally characterized. We further measured the accumulation of these DEGs in leaves and roots under cold treatment using qRT-PCR. The expression patterns obtained by qPCR in leaves were in agreement with those inferred by the RNA-seq data (Fig. 6b). One gene encoding (E)-β-ocimene/(E,E)-α-farnesene synthase (BOS/AFS) was persistently induced in leaves, but increased slowly in roots in response to cold stress. Strikingly, three genes annotated as santalene/bergamotene synthase 1 (SS/BS), SaMonoTPS1, and SaCYP76F38 were repressed in leaves, but significantly up-regulated in roots in response to cold stress, suggesting that organ-specific terpenes might be involved in the response of S. album to cold stress. The level of expression of putative (3 S,6E)-nerolidol synthase 1-like and myrcene synthase peaked significantly at 12 h of cold stress in leaves. These results confirmed that terpenoid synthase genes play a role in the cold stress response of S. album.
Identification, isolation and characterization of cold-induced transcription factors
A total of 15 TF families consisting of 63 DEGs were identified (Table S10). Following cluster analysis, these TFs were clustered into four groups, i.e., G1, G2, G3, and G4 (Fig. 7). Strikingly, most of these TFs were up-regulated in the three pairwise comparisons, as shown in G2, G3, and G4, revealing that transcriptional activation, but not repression, was significantly involved in the cold stress response. DEGs in the AP2/ERF family were the most abundant. Of these, transcripts of three genes coding for an AP2 domain class TF, one for CBF4, and two for ethylene-responsive TFs, increased dramatically at 12 h and decreased gradually until 48 h of cold stress, suggesting that these TFs might play crucial roles in the early cold response. One zinc finger CCCH domain protein-coding transcript accumulated significantly, with a similar expression pattern as the AP2 domain class of TFs described above. Members of MYB, bHLH (basic helix-loop-helix), WRKY, NAC (NAM, ATAF1/2, CUC2), and bZIP families of TFs exhibited cold-inducible expression. Furthermore, a decline in the transcripts of developmental regulators was observed, including GRAS and NFY.
The full lengths of nine genes were obtained by RACE. Homology searches showed that these nine proteins are members of the AP/ERF domain and ANK superfamilies, and the bHLH family. Based on sequence similarities via BLAST in NCBI, these genes were designated as SaCBF1, SaCBF2, SaCBF3, SaCBF4, SaERF017, SaERF109-like, SaRAP2.4, SaC3H29, and SaICE1 (Table S11). The full-length sequences of SaCBF1 encode small proteins 215 amino acids long while SaCBF2 to 4 contain 220 amino acids. Alignment of the deduced amino acid sequences of SaCBF1 to 4 with those of grape CBF1 to 4, poplar CBF1 to 4, and Arabidopsis CBF1 to 4 demonstrated significant similarities in the AP2/ERF binding domain and the CBF signature sequences between species (Fig. 8a). This result suggests that sandalwood CBFs share remarkable similarities at the amino acid sequence level with known CBF/DREB proteins in other plants and carry critical amino acids that are needed for binding to the CRT elements in the target genes. A phylogenetic analysis of the deduced protein sequences of the four sandalwood CBF genes with Arabidopsis CBF1 to 4 proteins, poplar CBF1 to 4 proteins, and grape CBF1 to 4 proteins showed that sandalwood CBF2 and 4 were closely grouped, whereas SaCBF1 was more closely related to poplar CBF1 and 3 (Fig. 8b).
(a) Alignment of AP2 and flanking region of CBF factors. The AP2/ERF domain, putative nuclear localization signal (NLS), and DSAWR motif are marked with a solid line and a dotted line, respectively. (b) A phylogenetic analysis of sandalwood, Arabidopsis, poplar, and grape proteins, based on minimum evolution, was performed with the full-length protein sequences using OsCBF1 as an out-group. Sequences were aligned using four sandalwood CBF proteins: SaCBF1 (KX009414), SaCBF2 (KX009415), SaCBF3 (KX009416), and SaCBF4 (KX009417), four poplar CBF proteins, PtCBF1 (JGI666968), PtCBF2 (JGI346104), PtCBF3 (JGI548519), and PtCBF4 (JGI63608), four Arabidopsis CBF proteins, AtCBF1 (ABV27072.1), AtCBF2 (ABV27106.1), AtCBF3 (ABV27138.1), and AtCBF4 (ABV27155.1), and four grape CBF proteins, VvCBF1 (AIL00526.1), VvCBF2 (AIL00632.1), VvCBF3 (AIL00730.1), and VvCBF4 (AIL00786.1). CBF proteins were initially aligned using CLUSTALX (v1.83) and used to conduct phylogenetic analyses using MEGA version 4. A phylogenetic tree was constructed using the neighbor-joining method on 1,000 bootstrap replications. Bootstrap percentages are shown on the dendrogram branch points.
The expression patterns of genes in leaves obtained by qPCR were in agreement with those inferred by the RNA-seq data, which further confirmed the reliability of the RNA-seq dataset. SaCBF1 in leaves and roots was slightly up-regulated in response to cold stress (Fig. 9). However, transcripts of SaCBF2 to 4 accumulated dramatically at 12 h in leaves, with a more than 50-fold increase compared to the control. Similarly, the expression levels of SaCBF 2 to 4 peaked in roots at 24 h of cold treatment, suggesting that leaves were more susceptible to cold stress than roots. It is likely that leaves were directly exposed to cold air and responded quickly to cold stress more than roots, which were immersed in, and thus somewhat protected by, soil. The expression of SaERF017, SaERF109-like, SaRAP2.4, SaC3H29, and SaICE1 exhibited similar expression patterns as SaCBFs over the 48-h time series of cold stress, but the relative expression ratios of these TFs were less than 10.
Novel genes induced by cold stress in S. album leaves
As shown in Table S3, approximately 51% genes of all DEGs were found to not have any annotation (ca. 2000 genes) or to only have an Nr annotation (250 genes). These were considered to be novel genes in response to cold stress. Of these, about 80% genes were activated by cold. A Venn diagram showed that 894, 778, and 883 novel genes were induced at 12, 24, and 48 h of cold treatment, respectively (Fig. S6a). Moreover, 184 genes were commonly expressed at all three time points compared with 0 h. Notably, there were 442, 284, and 485 genes that were uniquely induced at 12 h/0 h, 24 h/0 h, and 48 h/0 h, implying that these genes might play important roles at different time points of cold stress. These novel genes could be divided into 27 clusters, and clusters 1 to 8 were significantly different, showing P values less than 0.05 (Fig. S6b). The expression profile of genes in these significant clusters showed that some genes accumulated persistently throughout the 48-h period of cold stress (Fig. S6ci,ii). In contrast, genes were activated at 12 h, but were then completely repressed as the cold stress period was lengthened to 48 h (Fig. S6ciii–v). Interestingly, genes were induced at 12 h, repressed at 24 h, but further activated at 48 h of cold stress (Fig. S6cvi,vii). Moreover, the expression level of many induced genes remained relatively stable from 12 to 48 h of cold stress (Fig. S6cviii). Thus, the potential response mechanism of cold stress is complex in S. album.
Discussion
S. album is an economically important plant species because of its highly valued essential oils used in the perfumery industry. Most research has focused on the identification of sandalwood oil components and on the functional characterization of genes coding for enzymes that generate major constituents22,23,24. In recent years, RNA-seq has emerged as a powerful and cost-efficient tool for global analyses of transcriptomic changes when genomic information is not available. The molecular characterization of the biosynthesis of oil production in S. album has been conducted based on transcriptome data from heartwood tissues being particularly rich in sequiterpenoids26,27,28. In this study, we provided a transcriptome database of approximately 10.0 GB of data from S. album leaves and assembled into 135,938 unigenes, more than half of which were annotated against the NCBI NR database. This work will provide an important public genomic resource and facilitate further functional genomic studies of this essential oil-bearing plant.
Although plants are exposed to fluctuating external temperatures, they can enhance their cold tolerance by modulating gene induction and transcription in response to cold stress4. A total of 3075 cold-inducible and 1349 cold-repressed genes were identified in the present study. This predominance of gene induction during cold stress indicates a continuous adaptation over an extended period of time. More than 50% of the S. album DEGs had no annotated homologs in the Nr database and these may represent cold-responsive genes with homologs that have not been identified in previous studies with other plants or that are specific to S. album. Further research of these genes may reveal new mechanisms of cold-stress responses for S. album plants. In addition, many metabolites that contribute to the induction of stress tolerance have long been linked to stress responses36. An increase in the contents of MDA, proline and soluble sugars, as well as EL have been demonstrated in cold-tolerant plants such as Vitis amurensis Rupr., wheat, and wild tomato9,37. Consistent with those reports, an increase in MDA, EL, proline and solute sugar content were observed over the 48-h time course of cold stress, indicating that osmolytes might protect plant cell membranes and increase membrane stabilization during cold-induced dehydration in S. album. Taken together, these results suggest that S. album have the ability to withstand the stress of short-period when exposed to cold.
All aspects of photosynthesis including the light reactions, the Calvin cycle, and photorespiration can be affected by low temperature stress38,39. The cessation of growth resulting from cold stress reduces the capacity for energy utilization and causes feedback inhibition of photosynthesis40. In the present study, significant GO terms for “malate metabolic process” and “carbon utilization” appeared in the early response but were not detected in the late response to cold stress. Although photo-protection was activated during cold stress, cold-induced photo-inhibition finally occurred as the period of cold treatment was prolonged, as confirmed by our physiological analysis. This supports the fact that photosynthetic efficiency is negatively affected by cold stress. ROS accumulation is connected to decreased photosynthetic efficiency, and is caused by a failure in the electron transfer reaction of PSI41. The enzymatic antioxidant system is one of the protective mechanisms employed to eliminate or reduce ROS and increase a plant’s capacity for stress tolerance under abiotic stress42,43. Increased SOD and POD activity was detected in our study. The significant enrichment of GO terms related to increased expression of cold-responsive genes, including a response to oxygen-containing compounds or alcohol, and regulation of a plant-type hypersensitive response, were found in the early and late cold responses. This indicates that a dynamic balance in ROS formation and removal from cells to protect them against oxidative damage occurred during cold stress. Moreover, evidence in Arabidopsis showed that flavonoids as antioxidants in photo-protection played important roles in freezing tolerance44. Our identification of robust GO terms related to flavonoid biosynthesis implied that flavonoids might be involved in reducing the oxidative damage caused by cold stress. Importantly, the GO terms of up-regulated transcripts related to “response to wounding”, “regulation of defense”, and “regulation of immune response” were detected in the late cold response (Fig. 5a). Therefore, it can be hypothesized that S. album plants recruit a defense mechanism to increase cold tolerance when exposed to a prolonged period of cold.
Terpenes are a very large and structurally diverse group of secondary metabolites which are widely distributed in the plant kingdom45. Terpene biosynthesis in plants is of great interest because terpenoids play important roles in enhancing biotic or abiotic stress tolerance46. Research suggested that plants might enhance their tolerance to environmental stress (drought, high light intensity or high temperature) by increasing the emission of isoprenes or monoterpenes47. Evidence showed that sesquiterpenes, (E)-β-farnesene and (E)-α-bergamotene quench ozone but could not protect wild Nicotiana attenuata Torr. ex S. Watson from ozone, UVB light, and drought stresses48. Previous studies and our study indicate that transcripts generating terpenoids were expressed in multiple tissues of sandalwood, including leaves, heartwood, and roots. However, the molecular regulation of santalol biosynthesis in sandalwood is largely unknown. A foliar application of methyl jasmonate caused a higher expression level of HMGR and the farnesyl diphosphate synthase gene in leaves and stems of treated S. album seedlings compared to untreated plants at 4 h, but expression of cytochrome P450 showed no change in both leaves and stems29. In the present study, several homologs of HMGR were dramatically up-regulated in leaves by cold stress, implying that this key rate-limiting enzyme in MVA is likely to play an important role in the response of S. album to external stress. To date, all terpenes that have shown enhanced abiotic stress tolerance were synthesized in plastids45. Under stress, photosynthetic efficiency is closely associated with the accumulation of ROS which constrains the electron transfer reaction of PSI41. As evidenced in our study, cold stress led to a decrease in photosynthetic efficiency in S. album. We speculate that these putative terpene synthase genes may play a role in protection against localized oxidative stress in plastids. A sesquiterpene-rich essential oil is very abundant in the heartwood and roots of sandalwood21,23. High-level accumulation of SS/BS, SaMonoTPS1, and SaCYP76F38 in sandalwood roots implies that these genes play a role in protecting sandalwood from cold stress damage. In contrast, high expression of BOS/AFS in leaves was detected, suggesting that there is likely a different protection mechanism in different organs of S. album under cold stress. Functional roles for terpene synthase genes need to be further investigated in S. album.
Research in recent years has indicated that hormones play important roles in regulating plant freezing tolerance by either CBF-dependent or -independent pathways19. Our robust GO term annotation and KEGG pathway analysis showed the involvement of auxin, cytokinin, GA, ET, JA, BRs, ABA and salicylic acid in the response of S. album to cold stress (Fig. 5; Fig. S4), indicating that a complex regulatory network of hormones exists. What most deserves to be mentioned is the role of GA in response to cold stress. Exposure of Arabidopsis seedlings to cold stress stimulated the accumulation of DELLA proteins, suggesting that a link between CBFs and GA in cold stress-induced growth retardation exists49,50. Similarly, the overexpression of GhDREB1 and CbCBF in Gossypium hirsutum L. and N. tabacum L., respectively, caused a reduction of bioactive GA51,52. Other studies provide evidence that GA signaling components can affect the low temperature stress responses of plants53. In our study, up-regulated GA2 oxidases (GA2ox) and the GO term “GA mediated signaling pathway” were detected, highlighting the fact that GA metabolism and signaling are involved in the cold-stress response of S. album plants. Conversely, one GA20 oxidase (GA20ox, CL2514.Contig1) showed increased expression, with a log2 ratio of more than 8 compared to the control at 24 and 48 h (Table S7). It is worth exploring the significance of genes in the biosynthesis of GA in response to cold stress in the future.
Transcriptome profiling indicates that multiple regulatory pathways other than the CBF cold response pathway are activated during cold acclimation54. Park et al.12 found that there are at least 17 cold-induced TFs with similar expression patterns as those of CBFs in Arabidopsis, including ZF, CZF1, RAV1, MYB73, ZAT12, HSFC1, MYB44, ERF5, etc.12. Transcriptomic analysis showed that several RAP and ERF homologs were up-regulated under cold (4 °C) and freezing (−4 °C) treatment in desert poplar (Populus euphratica Oliv.)55. Our study indicates that SaCBF1 to 4, besides SaERF017, SaERF109-like, SaRAP2.4, and SaC3H29, play important roles in the regulation of the cold stress response in sandalwood. This is in agreement with the known fact that a complex regulatory network exists besides the CBF pathway in response to cold stress4. However, Park et al.12 thought that the increase in freezing tolerance that occurs with cold acclimation is only partially dependent on the CBF-CRT/DRE regulatory module in Arabidopsis. Since cold stress leads to cellular dehydration and an elevation of salt concentration, a dynamic crosstalk exists between the cold stress-signaling pathway and other signaling systems associated with dehydration and salt stress56,57. This study demonstrated that SaCBF2 to 4 was strongly responsive to cold stress in the leaves and roots of sandalwood, although the expression patterns of other TFs, SaERF017, SaERF109-like, SaRAP2.4, and SaC3H29 were similar to those of SaCBFs (Fig. 8, Table S9). This suggests that SaCBFs are critical contributors to the cold regulatory mechanism in S. album and that other cold-inducible TFs might be involved in the regulation of COR gene expression.
The present study generated a robust and well-annotated transcriptome for the cold stress response in sandalwood leaves. Based on the comprehensive physiological and transcriptomic analyses, when combined with RACE and qRT-PCR, our study provides fundamental knowledge related to the physiological changes and gene regulatory networks that operate when a sandalwood seedling is exposed to cold treatment. After a seedling perceives low temperature through cell membrane receptors, cold signal transduction induces changes to cellular metabolism, including calcium influx, the activation of Ca2+-dependent protein kinase cascades, the accumulation of cryoprotectants, proline, soluble sugar, and the antioxidants SOD and POD. Photosynthetic efficiency is negatively affected by cold stress. Terpenoid synthase genes were also involved in the cold stress response. Phytohormones play important roles in the response to cold stress, although further validation is required. The CBF-dependent signaling pathway might play a crucial role in the regulation of cold tolerance in sandalwood. Thus, a hypothetical model for sandalwood cold response networks has been proposed (Fig. S7). The functions of candidate genes involved in the regulation of cold signaling and cold-resistance genes will be verified in the future. This study contributes to uncovering the molecular background of S. album’s defense response to cold stress that will allow for the planting area to be expanded and for improving the cold tolerance of sandalwood by genetic engineering. Moreover, the finding that terpene synthase genes are induced by cold stress provides a clue as to how to stimulate santalol biosynthesis and accumulate essential oil in this species.
Methods
Plant material
Fully ripe S. album seeds collected from 20-year-old trees growing in SCBG were germinated according to our previously described method58. Seedlings were transplanted to 2-L pots filled with a mixture of sand, peat, and soil (3:1:1, v/v/v) and fertilized with 10 mL of 5% fertilizer whose optimum N: P: K ratio was 2.1: 1: 0.4 for each pot per week. Plants were cultivated in a greenhouse under a natural photoperiod (Feb-May, 2015, Guangzhou, China) with a day/night temperature of about 20–28 °C, 70–80% relative humidity, and a photosynthetic photon flux density (PPFD) of 140–260 μmol m−2 s−1. Seedlings were obtained after three months of culture. The host plant, Kuhnia rosmarnifolia Vent., which is a potential pot host for S. album in China34, was assembled to encourage host-parasite infection following our previous method59. Plants were watered twice daily with tap water. Pots were randomly moved every week to minimize position effects.
Cold treatment and sample collection
Five and a half month old seedlings with uniform growth were transferred to a phytotron under standardized growth conditions (16-h photoperiod, 28/23 °C (day/night), PPFD = 80 μmol m−2 s−1) and acclimated for two weeks prior to cold treatment. Seedlings were subjected to 4 °C for 0, 12, 24, and 48 h, serving as the cold stress treatments, under the same light conditions in the phytotron. The first and second fully expanded leaves were harvested immediately after the completion of each period of cold exposure, quickly frozen in liquid nitrogen, and stored at −80 °C until use. Physiological and biochemical indexes were measured using leaves at the same position as those used for RNA sample collection. The samples from five individuals were pooled for each time point and the collection of leaf material was repeated three times from different seedlings, serving as three independent biological replicates.
Physiological analyses of cold-treated leaves from S. album
To analyze the physiological changes in S. album leaves under cold stress prior to the RNA-seq study, three indicators of the cold response were estimated: MDA and proline content, and EL. MDA content was determined by the thiobarbituric acid reaction60. Proline concentration was measured by the sulfosalicylic acid-acid ninhydrin method61. EL was estimated as previously described62. Electrical conductivity of the bathing solution (Lt) was determined by a digital conductometer DDS-11A (Leici Instrument Factory, Shanghai, China) and a last conductivity reading (L0) was obtained upon equilibration at 25 °C after the bathing solution with leaf tissues was autoclaved at 120 °C for 20 min. In addition, the level of total soluble carbohydrates was determined using the anthrone method with glucose as the standard63. All spectrophotometric measurements were performed using a Beckmann DU640 (MI, USA) spectrophotometer.
Assay for antioxidative enzyme activities
To measure enzyme activities, 2 g of leaves were homogenized in 10 mL of 50 mM potassium phosphate buffer (pH 7.8) containing 4% polyvinylpyrrolidone (MW: 40,000). The homogenate was centrifuged at 12,000 rpm for 20 min and the supernatant was used for enzyme analyses at room temperature using a Beckmann DU640 spectrophotometer. All steps in the preparation of the enzyme extract were conducted at 0–4 °C. SOD activity was expressed on the basis of the inhibition of the photochemical reduction of nitro blue tetrazolium (NBT) at 560 nm64. The quantity of enzyme that reduced NBT by 50% in the dark was defined as one unit of SOD activity. POD activity was assayed following a method originally applied to Glycyrrhiza uralensis65. Absorbance was measured at 20-sec intervals at 460 nm for a total of 3 min. One unit of POD activity was defined as the increase by one unit per minute of the absorbance at 460 nm.
Measurement of photosynthetic parameters
Indexes of photosynthesis including A, Gs, E, Ci, and R of the third and fourth fully expanded leaves of six-month-old seedlings exposed to cold for 0, 12, 24, and 48 h were measured under standardized conditions with a portable photosynthesis system (LI-6400, Li-COR Inc., Lincoln, NE, USA). The measurement conditions were: 150 μmol m−2 s−1 PPFD, 70% relative air humidity, and 400 μmol mol−1 ambient CO2 concentration. WUE was calculated as the ratio of A to E66. Measurements were made on five plants for each of the three biological replicates of each treatment.
RNA isolation, library construction, and sequencing
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) and treated with DNase I to remove genomic DNA contamination using the Turbo DNA-free Kit (Ambion, Austin, TX, USA). RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, NC, USA). The quality of total RNA was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). One cDNA library was constructed from total RNA of non-treated and treated leaf samples for de novo assembly of the sandalwood transcriptome using an Illumina TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Also, a total of 12 libraries were separately generated from the control sample and from 12, 24, and 48 h cold-treated samples using the same protocol. Reads were generated in a 100 bp paired-end for de novo assembly using an Illumina Hiseq 2000 platform at the Beijing Genomics Institute (BGI). The libraries were sequenced with 50 bp single end reads for RNA-seq analysis (mRNA quantitative analysis) on the Illumina Hiseq 2000 platform.
Data processing and analysis
Transcriptome de novo assembly was performed using Trinity67. Firstly, shorts reads were combined with certain a length of overlap to form longer fragments without N, known as contigs. These reads were then mapped back to contigs. Using paired-end reads, we detected contigs from the same transcript as well as the distances between these contigs. Next, we used Trinity to connect the contigs and obtained sequences that cannot be extended on either end, which were defined as unigenes. Finally, we assembled all the unigenes to form a single set of non-redundant unigenes using TGICL68. After clustering, the unigenes were divided into two classes: clusters and singletons. After assembly, BLASTx alignment was performed between unigenes and the protein databases with a cut-off E-value of 10−5, following this order of priority: Nr, Swiss-Prot, KEGG, COG and GO. The best alignment results were used to decide the sequence direction of unigenes.
Raw reads of FASTQ format from RNA-seq data of control and treated samples were first processed through in-house Perl scripts. In this step, clean reads were obtained by removing reads containing an adapter, reads in which unknown bases accounted for more than 10%, and low quality reads (i.e., where more than 50% of bases in a read had a quality value Q ≤ 5) from the raw data. Clean reads were aligned to the reference unigenes using Bowtie69. Gene expression levels were quantified by RSEM (RNASeq by Expectation Maximization)70, which computes maximum likelihood abundance estimates using the Expectation-Maximization (EM) algorithm for its statistical model, including the modeling of paired-end, variable-length reads, fragment length distributions, and quality scores, to determine which transcripts are isoforms of the same gene. Gene expression level was calculated with the FPKM method71. The number of transcripts after filtering among all samples was analyzed by a bar graph. A correlation between samples was performed based on FPKM results by calculating Pearson’s correlation for pairs of samples. Principal component analysis was performed to visualize the distance of the relationship between each sample using R statistical software (version 3.2.3)72.
DEGs under cold stress were screened using the Noiseq package with the following criteria: fold change ≥2 and deviation probability ≥0.8 at 12 h/0 h, 24 h/0 h, and 48 h/0 h, respectively73. Hierarchical clustering was performed for DEGs with a ≥8-fold change and deviation probability ≥0.8 in at least one of the three pairwise comparisons using the Manhattan distance matrix of the heatmap.2 function in the gplots package of the R environment. We then separately mapped induced or repressed DEGs to each term of the GO database (http://www.geneontology.org/) and found significantly enriched GO terms in cold-inducible and cold-repressed genes using a hypergeometric test55. The calculated P value underwent a Bonferroni correction until the threshold of the corrected P value was less than 0.05. GO terms fulfilling this condition were defined as significantly enriched GO terms to detect the main biological functions that DEGs exercise during cold stress.
Screening and cloning of cold-induced transcription factors
In order to identify exactly which TF genes were highly expressed during cold stress, the predicted coding sequences of seven genes encoding AP2 domain-containing TFs, CBF4, RAP2-4-like, ethylene-responsive TF, and zinc finger CCCH domain-containing protein 29-like, in addition to one inducer of CBF expression 1 and one CBF/DREB-like gene, were obtained from assembled unigenes. The SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories Inc., Takara Bio, Mountain View, CA, USA) was used to generate both 5′ and 3′ cDNA ends. The 5′ and 3′ RACE PCR products were purified by a Gel Extraction Kit (Dongsheng Biotech, Guangzhou, China), cloned into the pMD18-T vector (Takara Bio Inc., Dalian, China) and sequenced at the BGI. Full-length cDNAs were amplified with primers designed from 5′ and 3′ untranslated regions (UTR) by PCR using the LA PCR Amplification Kit (Takara Bio Inc.). All primers used to amplify 3′ and 5′ UTR regions and full-length cDNAs are listed in Table S12. The multiple alignments were used by DNAMAN8.0 (Lynnon Biosoft, San Ramon, CA, USA) and a phylogenetic tree was constructed by the neighbor-joining (NJ) method using MEGA 4.074,75.
qRT-PCR analyses
Total RNA extracted from S. album leaves and roots after 0, 12, 24, 48 h of cold stress were treated with DNase I to remove genomic DNA contamination using the TurboDNA-free Kit (Ambion, Austin, TX, USA). First-strand cDNA synthesis and purification followed our published method58. Gene-specific primers are listed in Table S13. Melting curve analysis was performed for each primer pair prior to further analyses. qRT-PCR reactions were performed with the SYBR Green Real Master Mix (Bio-RAD, Foster, CA, USA) in the ABI 7500 fast Real-Time PCR system (Applied Biosystems, Foster, CA, USA). The expression level was calculated using our protocol58. qRT-PCR results were obtained from three biological replicates and three technical repeats for each gene and sample.
Additional Information
How to cite this article: Zhang, X. et al. Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. leaves. Sci. Rep. 7, 42165; doi: 10.1038/srep42165 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Jeon, J. & Kim, J. Cold stress signaling networks in Arabidopsis. J. Plant Biol. 56, 69–76, (2013).
Chinnusamy, V., Zhu, J. & Zhu, J. K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451 (2007).
Yadav, S. K. Cold stress tolerance mechanisms in plants. A review. Agronomy Sustain. Dev. 30, 515–527 (2010).
Zhao, C. Z., Lang, Z. B. & Zhu, J. K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 20, 466–468 (2015).
Rodríguez, V. M., Butrón, A., Rady, M. O. A., Soengas, P. & Revilla, P. Identification of quantitative trait loci involved in the response to cold stress in maize (Zea mays L.). Mol. Breed. 33, 363–371 (2014).
An, D., Yang, J. & Zhang, P. Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to cold stress. BMC Genomics 13, 64 (2012).
Tian, X. et al. Comparative proteomic analysis of seedling leaves of cold-tolerant and -sensitive spring soybean cultivars. Mol. Biol. Rep. 42, 581–601 (2015).
Uemura, M., Warren, G. & Steponkus, P. L. Freezing sensitivity in the sfr4 mutant of Arabidopsis is due to low sugar content and is manifested by loss of osmotic responsiveness. Plant Physiol. 131 (2003).
Chen, H. Y. et al. A comparison of the low temperature transcriptomes of two tomato genotypes that differ in freezing tolerance: Solanum lycopersicum and Solanum habrochaites . BMC Plant Biol. 15, 132 (2015).
Sanghera, G. S., Wani, S. H., Hussain, W. & Singh, N. B. Engineering cold stress tolerance in crop plants. Curr. Genomics 12, 30–43 (2011).
Morran, S. et al. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249 (2011).
Park, S. et al. Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207 (2015).
Zhang, X. et al. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis . Plant J. 39, 905–919 (2004).
Ito, Y. et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47, 141–153 (2006).
Benedict, C. et al. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29, 1259–1272 (2006).
Xiao, H., Siddiqua, M., Braybrook, S. & Nassuth, A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 29, 1410–1421 (2006).
Welling, A. & Palva, E. T. Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 147, 1199–1211 (2008).
Navarro, M. et al. Complementary regulation of four Eucalyptus CBF genes under various cold conditions. J. Exp. Bot. 60, 2713–2724 (2009).
Shi, Y. T., Ding, Y. L. & Yang, S. H. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 56, 7–15 (2015).
Miura, K. & Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 14, 5312–5337 (2013).
Baldovini, N., Delasalle, C. & Joulain, D. Phytochemistry of the heartwood from fragrant Santalum species: a review. Flavour Frag. J. 26, 7–26 (2011).
Misra, B. B., Das, S. S. & Dey, S. Volatile profiling from heartwood of East Indian sandalwood tree. J. Pharm. Res. 7, 299–303 (2013).
Jones, C. G. et al. Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Arch. Biochem. Biophys. 477, 121–130 (2008).
Jones, C. G. et al. Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J. Biol. Chem. 286 (2011).
Rani, A., Ravikumar, P., Reddy, M. D. & Kush, A. Molecular regulation of santalol biosynthesis in Santalum album L. Gene 527, 642–658 (2013).
Diaz-Chavez, M. L. et al. Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS One 8, e75053 (2013).
Srivastava, P. L. et al. Functional characterization of novel sesquiterpene synthases from Indian sandalwood, Santalum album . Sci. Rep. 5, 10095 (2015).
Celedon, J. M. et al. Heartwood-specific transcriptome and metabolite signatures of tropical sandalwood (Santalum album) reveal the final step of (Z)-santalol fragrance biosynthesis. Plant J. 86, 289–299 (2016).
Kulheim, C. et al. Foliar application of methyl jasmonate does not increase terpenoid accumulation, but weakly elicits terpenoid pathway genes in sandalwood (Santalum album L.) seedlings. Plant Biotechnol. 31, 585–591 (2014).
Jain, S. H. et al. Identification of provenances of sandal in India for genetic conservation. In Sandal and its Products (eds Radomiljac, A. M., Ananthapadmanabho, H. S., Welbourn, R. M. & Satyanarayana, K. ) Australian Centre for International Agricultural Research (ACIAR) Proceedings (84), Canberra, pp. 117–120 (1998).
TFS. TFS (Tropical Forestry Services) Sandalwood Project 2015, Indian Sandalwood. Product Disclosure Statement. Tropical Forestry Services Ltd. 169 Broadway, Nedlands WA 6009, Australian (2015).
Rashkow, E. D. Perfumed the axe that laid it low: The endangerment of sandalwood in southern India. Indian Econ. Soc. Hist. Rev. 51, 41–70 (2014).
Teixeira da Silva, J. A. et al. Sandalwood: basic biology, tissue culture, and genetic transformation. Planta 243, 847–887 (2016).
Li, Y. L. Study on the introduction of sandalwood. Science Press, Beijing, China, pp 97–98 (in Chinese) (2003).
Campos, P. S., Quartin, V. n., Ramalho, J. c. & Nunes, M. A. Electrolyte leakage and lipid degradation account for cold sensitivity in leaves of Coffea sp. plants. J. Plant Physiol. 160, 283–292 (2003).
Guy, C. L. Cold acclimation and freezing stress tolerance: role of protein metabolism. Ann. Rev. Plant Physiol. Plant Mol. Biol. 41, 187–223 (1990).
Xu, W. R. et al. Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol. Biol. 86, 527–541 (2014).
Goulas, E. et al. The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J. 47, 720–734 (2006).
Friesen, P. C. & Sage, R. F. Photosynthetic responses to chilling in a chilling-tolerant and chilling-sensitive Miscanthus hybrid. Plant Cell Environ., doi: 10.1111/pce.12699 (2016).
Ruelland, E. & Zachowski, A. How plants sense temperature. Environ. Exp. Bot. 69, 225–232 (2010).
Liu, Y. F., Qi, M. F. & Li, T. L. Photosynthesis, photoinhibition, and antioxidant system in tomato leaves stressed by low night temperature and their subsequent recovery. Plant Sci. 196, 8–17 (2012).
Jithesh, M. N., Prashanth, S. R., Sivaprakash, K. R. & Parida, A. K. Antioxidative response mechanisms in halophytes: their role in stress defence. J. Genetics 85, 237–254 (2006).
Mittler, R., Vanderauwera, S., Gollery, M. & Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 (2004).
Schulz, E., Tohge, T., Zuther, E., Fernie, A. R. & Hincha, D. K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana . Sci. Rep. 6, 34027 (2016).
Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 9, 297–304 (2006).
Tholl, D., Sohrabi, R., Huh, J. H. & Lee, S. The biochemistry of homoterpenes–common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 72, 1635–1646 (2011).
Holopainen, J. K. & Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 15, 176–184 (2010).
Palmer-Young, E. C., Daniel, V., Gershenzon, J. & Schuman, M. C. The sesquiterpenes(E)-β-farnesene and (E)-α-bergamotene quench ozone but fail to protect the wild tobacco Nicotiana attenuata from ozone, UVB, and drought stresses. PLoS One 10, doi: 10.1371/journal. pone.0127296 (2015).
Achard, P. et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006).
Achard, P. et al. The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129 (2008).
Shan, D. P. et al. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 176, 70–81 (2007).
Zhou, M. Q. et al. CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol. Biol. 85, 259–275 (2014).
Eremina, M., Rozhon, W. & Poppenberger, B. Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 73, 797–810 (2016).
Fowler, S. & Thomashow, M. F. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690 (2002).
Chen, J. H. et al. Deep-sequencing transcriptome analysis of low temperature perception in a desert tree, Populus . BMC Genomics 15, 326 (2014).
Xiong, L. M., Schumaker, K. S. & Zhu, J. K. Cell signaling under salt, water and cold stresses. Plant Cell S165–183 (2002).
Huang, G. T. et al. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987 (2012).
Zhang, X. H. et al. RNA-Seq analysis identifies key genes associated with haustorial development in the root hemiparasite Santalum album . Front. Plant Sci. 6, 661 (2015).
Zhang, X. H. et al. Endogenous hormone levels and anatomical characters of haustoria in Santalum album L. seedlings before and after attachment to the host. J. Plant Physiol. 169, 859–866 (2012).
Dhindsa, R. S., Plumb-Dhindsa, P. & Thorpe, T. A. Leaf senescence correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J. Exp. Bot. 32, 93–101 (1981).
Bates, L., Waldren, R. & Teare, I. Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207 (1973).
Lutts, S., Kinet, J. M. & Bouharmont, J. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 78, 389–398 (1996).
Yemm, E. W. & Willis, A. J. The estimation of carbohydrates in plant extracts by anthrone. Biochemistry 57, 508–514 (1954).
Beauchamp, C. & Fridovich, I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287 (1971).
Ranieri, A., D’Urso, G., Nali, C., Lorenzini, G. & Soldatini, G. F. Ozone stimulates apoplastic antioxidant systems in pumpkin leaves. Physiol. Plant. 97, 381–387 (1996).
Li, W. D. et al. Effect of water deficit on biomass production and accumulation of secondary metabolites in roots of Glycyrrhiza uralensis . Russ. J. Plant Physiol. 58, 538–542 (2011).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Pertea, G. et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652 (2003).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Li, B. & Dewey, C. N. RSEM accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Mortazavi, A., B., W., McCue, K. & Schaeffer, L. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S, 4th ed. Springer-Verlag: New York, (2002).
Tarazona, S., Garcíaalcalde, F., Dopazo, J., Ferrer, A. & Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 21, 2213–2223 (2011).
Saitou, N. & Nei, M. The neighbor-joining method a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (Grant numbers 31470685, 31100498, and 31270720), the Natural Science Foundation of Guangdong Province (S2012010009025), and the Guangdong Province Science and Technology Program (2015B020231008).
Author information
Authors and Affiliations
Contributions
X.H.Z. and J.A.T.D.S. conceived the project; isolation of genes and cloning was done by X.H.Z. and M.Y.N.; X.H.Z., J.A.T.D.S., S.J.Z. and G.H.M. contributed to transcriptome analysis; physiological and physiochemical measurements were performed by M.Z.L., H.M.H. and J.H.Z. All authors analyzed the data, wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, X., Teixeira da Silva, J., Niu, M. et al. Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. leaves. Sci Rep 7, 42165 (2017). https://doi.org/10.1038/srep42165
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep42165
This article is cited by
-
Integrated metabolomics and transcriptomics analysis during seed germination of waxy corn under low temperature stress
BMC Plant Biology (2023)
-
Characterization of sandalwood (E,E)-α-farnesene synthase whose overexpression enhances cold tolerance through jasmonic acid biosynthesis and signaling in Arabidopsis
Planta (2023)
-
Transcriptome-based gene regulatory network analyses of differential cold tolerance of two tobacco cultivars
BMC Plant Biology (2022)
-
Cloning, characterization, and functional analysis of acetyl-CoA C-acetyltransferase and 3-hydroxy-3-methylglutaryl-CoA synthase genes in Santalum album
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.