Abstract
The mechanisms involved in the chronic hepatitis C progression are incompletely understood. The aim was to analyze the association between 2′5′oligoadenylate synthetase 1,2 and 3 (OAS1-3) and myxovirus resistance proteins 1 (Mx1) polymorphisms and severity of liver disease in human immunodeficiency virus (HIV)/hepatitis C virus (HCV) coinfected patients. We performed a cross-sectional study in 219 patients that underwent a liver biopsy. DNA genotyping for Mx1 (rs469390), OAS1 (rs2285934), OAS2 (rs1293762) and OAS3 (rs2010604) was performed by using GoldenGate assay. The outcome variables ion liver biopsy were: (i) significant fibrosis (F ≥ 2); (ii) moderate activity grade (A ≥ 2). Additive model of inheritance for genetic association test was used. The likelihood of having significant fibrosis (F ≥ 2) was lower in patients carrying OAS2 rs1293762 A allele [adjusted odds ratio (aOR) = 0.51; p = 0.040]. Besides, the likelihood of having moderate activity grade (A ≥ 2) was higher in patients carrying Mx1 rs464397 C allele (aOR = 1.63; p = 0.028) and Mx1 rs469390 G allele (aOR = 1.97; p = 0.005), while it was lower in patients carrying OAS1 rs2285934 A allele (aOR = 0.64; p = 0.039) and OAS2 rs1293762 A allele (aOR = 0.41; p = 0.009). In conclusion, Mx1 and OAS1-2 polymorphisms were associated with the severity of liver disease in HIV/HCV-coinfected patients, suggesting a significant role in the progression of hepatic fibrosis.
Similar content being viewed by others
Introduction
Only a minority of patients may spontaneously clear the hepatitis C virus (HCV) during the acute phase of infection, which correlates with a rapid induction of innate response, especially interferon-stimulated genes (ISGs)1. However, the majority of patients is unable to clear the HCV and develops chronic hepatitis C (CHC), which may cause irreversible fibrosis, leading to cirrhosis and hepatocellular carcinoma1,2. Moreover, CHC is accelerated in human immunodeficiency virus (HIV)/HCV coinfected patients, resulting in higher rates of disease progression than HCV mono-infected patients3.
The pathogenic mechanisms involved in this progress are incompletely understood, but it is known that the alternations of liver inflammatory profiles and the impaired immune responses are thought to be an important determinant of the rate of fibrosis progression2. Among others, chronic HCV infection has a pronounced effect on gene expression of a subset of ISGs such as 2′5′oligoadenylate synthetase (OAS) family and myxovirus resistance proteins (Mx)4. The Mx1 protein is a GTPase that acts at an early step of the virus life cycle, prior to the genome replication, by trapping viral components and preventing them from reaching their cellular destination4. OAS proteins are able to synthesize 2′,5′-oligomers, which specifically activate the latent form of RNaseL, which may then mediate RNA degradation, contributing to decay of viral RNA4. Single nucleotide polymorphisms (SNPs) in the OAS gene (chromosome 12) and Mx gene (chromosome 21) have been related to severity and/or outcome of viral infections such as tick-borne encephalitis5, West Nile virus6, dengue virus7, coronavirus8, and hepatitis B9. Regarding to HCV infection, Mx1 rs2071430 polymorphism has been related to HCV clearance during acute HCV infection and after interferon (IFN) therapy, and higher severity of liver disease in HCV-monoinfected patients10,11,12,13. However, to our knowledge, there is no report related to OAS/Mx SNPs and severity of liver disease in HIV/HCV coinfected patients.
The aim of this study was to analyze the association between Mx1 and OAS1-3 polymorphisms and severity of liver disease in HIV/HCV coinfected patients.
Results
Characteristics of the study population
Table 1 shows the epidemiological and clinical characteristics of 219 HIV/HCV-coinfected patients at the time of liver biopsy. Regarding liver biopsy, 49.8% had significant fibrosis (F ≥ 2), 52.6% had moderate necroinflammatory activity (A ≥ 2), and 15.3% had alanine aminotransferase (ALT) ≥ 150 IU/L.
Mx1 and OAS1-3 polymorphisms
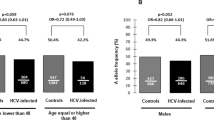
Table 2 shows the characteristics of SNPs, which displayed missing values < 5% and were in HWE (p > 0.05). When SNPs frequencies in HIV/HCV coinfected patients were compared to the SNPs frequencies in healthy subjects from the IBS, we only found differences between groups for Mx1 rs469390 AA (p = 0.011) and Mx1 rs469390 AG (p = 0.036) genotypes.
Association between Mx1 and OAS1-3 polymorphisms and liver disease
Table 3 shows the relationship of Mx1 and OAS1-3 polymorphisms with severity of liver disease under a model of additive inheritance, which was the genetic model that best fitted our data. The trend of having significant fibrosis (F ≥ 2) was lower in patients carrying of OAS2 rs1293762 A allele (p = 0.023). The trend of having moderate activity grade (A ≥ 2) was higher in patients carrying Mx1 rs464397 C allele (p = 0.015) and Mx1 rs469390 G allele (p = 0.003); while it was lower in patients carrying OAS1 rs2285934 A allele (p = 0.026), OAS2 rs1293762 A allele (p = 0.002), and it was nearly significant for OAS3 rs2010604 C allele (p = 0.095). Moreover, the likelihood of having significant fibrosis (F ≥ 2) in liver biopsy was lower in patients carrying OAS2 rs1293762 A allele [adjusted odds ratio (aOR) = 0.51; p = 0.040]. Besides, the likelihood of having moderate activity grade (A ≥ 2) was higher in patients carrying Mx1 rs464397 C allele (aOR = 1.63; p = 0.028) and Mx1 rs469390 G allele (aOR = 1.97; p = 0.005), while it was lower in patients carrying OAS1 rs2285934 A allele (aOR = 0.64; p = 0.039) and OAS2 rs1293762 A allele (aOR = 0.41; p = 0.009).
Figure 1 shows the relation between MX1 and OAS1-3 genetic polymorphisms and serum ALT. We found that patients carrying Mx1 rs464397 CC genotype and Mx1 rs469390 GG genotype had higher values of serum ALT and percentage of patients with ALT ≥ 150 IU/L. However, we did not find any significant association in multivariate regression analysis (data not shown). Moreover, we also analyzed the relation between MX1 and OAS1-3 polymorphisms and serum aspartate aminotransferase (AST), but we did not find any association (data not shown).
Table 4 shows the relationship among haplotype of Mx1 gene (rs464397, rs458582, rs469390) and OAS1-3 cluster (rs2285934, rs2285933, rs2010604, rs739903, rs1293762) and severity of liver disease. The likelihood of having moderate activity grade (A ≥ 2) in liver biopsy was higher in patients with Mx1 CTG haplotype (aOR = 1.44; p < 0.001) and lower in patients with Mx1 CTA haplotype (aOR = 0.62; p = 0.007). Moreover, the likelihood of having moderate activity grade (A ≥ 2) in liver biopsy was lower in patients with OAS1-3 AGCTA haplotype (aOR = 0.78; p = 0.008).
Discussion
To our knowledge, this study is the first description of the relationship between Mx1 and OAS1-3 polymorphisms and severity of liver disease in HIV/HCV-coinfected patients. The major findings of the present study were that Mx1 rs464397 C allele, Mx1 rs469390 G allele, and Mx1 CTG haplotype were associated with higher odds of having moderate activity grade (A ≥ 2) in liver biopsy, while OAS1 rs2285934 A allele, OAS2 rs1293762 A allele, and OAS1-3 AGCTA haplotype were related to lower likelihood of having moderate activity grade (A ≥ 2). Besides, OAS2 rs1293762 A allele was related to lower likelihood of having significant fibrosis (F ≥ 2) in liver biopsy. Thus, Mx1 and OAS1-2 polymorphisms might play a role in developing of liver disease in HIV/HCV-coinfected patients.
In our study, we included a control group to show that our study cohort had allelic and genotypic frequencies similar to the general population. However, we found significant differences in allelic and genotypic frequencies of Mx1 rs469390 between HIV/HCV coinfected patients and Iberian populations in Spain from HapMap data. This observation might suggest that Mx1 rs469390 could be associated to innate resistance to HCV infection. However, our study design is not adequate to draw such conclusions due to a number of factors that we do not control. For example, our cohort may have a survival bias because patients were more than 20 years of median with HIV and HCV infections. In addition, patients were selected due to they were candidates for antiviral treatment against HCV because progression of liver disease was suspected.
In the setting of CHC, the immune system initially attempts to eradicate the HCV, but besides, probably promotes hepatocyte damage and fibrosis through direct cellular toxicity and the release of inflammatory cytokines2. The innate immune response against viruses produce type I IFN (alpha, beta), which induces the expression of Mx1 protein4,14. Although its effect against HCV is not well documented, high levels of Mx1 protein have been detected in IFN-treated HCV patients with sustained virological response15. In fact, Mx1 rs2071430 polymorphism, in the promoter region, has been related to Mx1 gene expression, spontaneous viral clearance during acute HCV infection, higher hepatic fibrosis score and hepatitis activity grade, and response to IFN therapy in HCV-monoinfected patients10,11,12,13. In the in silico analysis performed, we found that Mx1 rs464397 seems to be implicated in both proximal (open chromatin region, transcription factor binding sites and histone markers) and distal regulation. Furthermore, rs464397 seems also be involved in RNA binding protein-mediated post-transcriptional regulation as well as to be a SNP with experimental expression quantitative trait loci (eQTL) evidence. On the other hand, Mx1 rs469390 polymorphism is a missense variant (V379I) that might not affect protein structure; but it seems influence an exonic splicing enhancer motif and it might be involved in protein domain abolishing. In this way, MX1 rs469390 polymorphism could decrease expression of MX1, getting worse the control of HCV, and might therefore be related to the increase of sustained inflammation and progression of liver disease.
The OAS proteins play an important role in the innate immune response and genetic variants in the OAS genes are known to affect OAS activity and are associated with outcome of viral infections16. The OAS1 rs2660, rs10774671, and rs3741981 polymorphisms have been related to persistent HCV infection11,17 and progression of HCV disease18; and OAS1 polymorphism at exon 7 splice acceptor site with response to IFN therapy in HCV infected patients19. The OAS2 polymorphism rs1293762 has been associated with predisposition to CHC20. In our study, OAS1-3 polymorphisms were related to lower likelihood of having significant liver disease (A ≥ 2 and/or F ≥ 2), indicating that the A-allele conferred a protective effect with respect to liver disease progression. OAS1 rs2285934 and OAS2 rs1293762 polymorphisms are located in intron 3 and intron 2, respectively. In the in silico analysis, both SNPs seem to be involved in RNA binding protein-mediated post-transcriptional gene regulation. In this way, OAS1 rs2285934 and OAS2 rs1293762 polymorphisms could increase expression of OAS1 and OAS2, improving the control of HCV, and might therefore be related to the decrease of sustained inflammation and progression of liver disease.
As discussed above, the presence of Mx1 and OAS1-3 polymorphisms may be related to have moderate or significant liver disease. This association that we observed might be due to the proximity to other SNPs that may modulate Mx1 gene expression, or a direct effect of Mx1 and OAS1-3 polymorphisms analyzed in this study. On the one hand, haplotype analysis showed a stronger association with liver disease than that found with each polymorphism alone. On the other hand, the in silico analysis showed that polymorphisms alone could have some influence on the expression of the proteins involved. However, it would be necessary to demonstrate if these Mx1 and OAS1-3 polymorphisms may modify the expression of Mx1 and OAS1-3 genes.
The impact of OAS/MX1 SNPs on fibrosis progression in CHC has not been picked up in the GWAS previously carried out21,22. However, we found a relatively strong association between OAS/MX1 SNPs and the severity of liver disease in our HIV/HCV coinfected patients. This could be because: (a) GWAS usually does not have the power to detect all, only the biggest effects. Besides, note that strong statistical adjustments on the p-value are carried out for correcting multiple testing; (b) we studied a SNP candidate with MAF greater than 20%. In these cases, an individual effect is more easily detected when the size of sample is small23; (c) we should not discard the effect of HIV infection on the immune system, which could show the influence of OAS/MX1 polymorphisms on CHC progression.
Finally, several aspects have to be taken into account for the correct interpretation of the results. Firstly, this is a cross-sectional study with a limited number of patients, which could limit achieving statistically significant values. Besides, the cross-sectional design nature of the analyses may be a confounding factor. Secondly, data were collected retrospectively, which entails a lack of uniformity. Thirdly, all selected patients met a set of criteria for starting HCV treatment (e.g., no alcohol abuse, high CD4 cell counts, controlled HIV replication, and good treatment adherence), and this may have introduced a selection bias. Fourthly, regarding the statistical significance, there is a considerable controversy about adjusting the “p-value” after multiple tests on clinical-orientated studies24,25. In our study, there was a hypothesis supported by theory and previous reports that related Mx1 and OAS1-3 polymorphisms to severity and/or outcome of viral infections5,6,7,8,9, including HCV infection10,11,12,13. Therefore, we were not literally doing a random search of a meaningful result, and our results should not be affected by the fact of carrying out a high number of statistical tests. Fifthly, this study was performed on patients with European ancestry, and it would be interesting to perform these analyses on different ethnic groups. Finally, our study only included HIV/HCV-coinfected patients and it would be interesting to know the role of studied polymorphisms in HCV monoinfected patients, but we did not have access to a cohort of HCV monoinfected patients.
In conclusion, Mx1 and OAS1-2 polymorphisms were associated with the severity of liver disease in patients coinfected with HIV and HCV, suggesting that these polymorphisms might play a significant role in the progression of hepatic fibrosis. Further analysis involving large numbers of patients are needed to corroborate these associations that we have found.
Materials and Methods
Study population
We performed a cross-sectional study in 219 HIV/HCV-coinfected patients that underwent a liver biopsy between September 2000 and November 2008. The study was conducted in accordance with the Declaration of Helsinki and patients gave their written consent for the study. The Institutional Review Board and the Research Ethic Committee of the Instituto de Salud Carlos III approved the study. Each participating patient signed an informed consent form.
Liver biopsies were performed on patients who were potential candidates for anti-HCV therapy and had not received previous IFN therapy (naïve for HCV-treatment). Selection criteria were: no clinical evidence of hepatic decompensation, detectable HCV RNA by polymerase chain reaction (PCR), negative hepatitis B surface antigen, availability of DNA sample, CD4 + lymphocyte count higher than 200 cells/μL, and stable combination antiretroviral therapy (cART) for at least 6 months before study entry or no need for cART according to treatment guidelines used in the study period26,27. Patients with active opportunistic infections, active drug addiction, and other concomitant severe diseases were excluded. All subjects included in our study were European white.
Epidemiological and clinical data
Clinical and epidemiological data were obtained from medical records. The duration of HCV infection for patients with a history of intravenous drug use was estimated starting from the first year they shared needles and other injection paraphernalia, which are the most relevant risk practices for HCV transmission28. The duration of HCV infection was not calculated when the date of initiation of their HCV infection could not be determined with certainty. Consumption of more than 50 g of alcohol per day for at least 12 months was considered as a high intake. Body mass index (BMI) was calculated as the weight in kilograms divided by the square of the height in meters.
Biochemistry panel was measured using an autoanalyzer Hitachi 912 (Boehringer Mannheim, Germany), while patients were fasting. The degree of insulin resistance was estimated for each patient using the homeostatic model assessment (HOMA)29: fasting plasma glucose (mmol/L) times fasting serum insulin (mU/L) divided by 22.5.
HCV assays
HCV infection was documented in all patients by enzyme-linked immunosorbent assay and PCR test. HCV genotype was determined by hybridization of biotin-labeled PCR products to oligonucleotide probes bound to nitrocellulose membrane strips (INNO-LiPA HCV II, Innogenetics, Ghent, Belgium). Plasma HCV-RNA viral load was measured by PCR (Cobas Amplicor HCV Monitor Test, Branchburg, NJ, USA) and real-time PCR (COBAS AmpliPrep/COBAS TaqMan HCV test); and results were reported in terms of international units per milliliter (IU/mL). The limit of detection varied between 10 IU/mL and 600 IU/mL depending on the HCV diagnostic test used at the time of biopsy. There was no patient with HCV viral loads below the limit of detection. The HCV RNA level of 500,000 UI/ml was chosen as cutoff for low vs. high viral load (circulating HCV RNA)30,31.
Liver biopsy
Liver biopsies were performed as we described previously32. The samples were always evaluated by the same pathologist, who was unaware of the patients’ clinical or laboratory data. Liver fibrosis and necroinflammatory activity were estimated according to Metavir score as follows33: F0, no fibrosis; F1, mild fibrosis; F2, significant fibrosis; F3, advanced fibrosis; and F4, definite cirrhosis. The degree of necroinflammation (activity grade) was scored as follows: A0, no activity; A1, mild activity; A2, moderate activity; A3, severe activity.
Genotyping of DNA polymorphisms
The most common SNPs in Mx1 gene (chromosome 21) and OAS1-3 genes (chromosome 12) were selected using the databases of HapMap Project (http://snp.cshl.org/cgi-perl/gbrowse/hapmap_B35/) and NCBI (dbSNP) (http://www.ncbi.nlm.nih.gov/entrez/). The selection criteria were: (i) SNPs located in putative regulatory regions, or splicing control elements (SCE), or missense variants; (ii) minor allelic frequency (MAF) greater than 20% in European people in order to have guarantees to obtain significant associations when working with small sample sizes34. With this setting, we found three SNPs in Mx1 gene (rs464397 in intron 3, rs458582 in intron 5, and rs469390 in exon 13 (missense)) and other five SNPs at OAS genes (OAS1 rs2285934 in intron 3, OAS2 rs1293762 in intron 2, OAS3 rs739903 in 3′UTR, OAS3 rs2285933 in exon 6 (missense), and OAS3 rs2010604 in 3′UTR). Finally, five polymorphisms were selected as tagSNPs according to linkage disequilibrium (LD) (Fig. 2): two SNPs at Mx1 (rs464397 and rs469390) and three SNPs at OAS (OAS1 rs2285934, OAS2 rs1293762, and OAS3 rs739903). Three SNPs (Mx1 rs458582, OAS3 rs2285933, and OAS3 rs2010604) were discarded for the analysis of individual association because these SNPs were at high linkage disequilibrium (LD) (Fig. 2).
Genomic DNA was extracted from peripheral blood with Qiagen kit (QIAamp DNA Blood Midi/Maxi; Qiagen, Hilden, Germany). DNA samples were sent at the Spanish National Genotyping Center (CeGen; http://www.cegen.org/) for DNA genotyping by using GoldenGate assay with VeraCode Technology (Illumina Inc., San Diego, California, USA).
Moreover, for healthy subjects, the frequencies of alleles and genotypes for studied polymorphisms were obtained using the 1000 Genomes Project website (http://www.1000genomes.org/home), which provide a broad representation of common human genetic variation by applying whole-genome sequencing to a diverse set of individuals from multiple populations35. We select the IBS (Iberian populations in Spain) population that included 107 individuals.
Outcome variables
The primary outcome variables were related to severity of liver disease: significant fibrosis (F ≥ 2) and moderate activity grade (A ≥ 2). These outcomes were developed after a minimum follow-up time of 10 years with HCV infection.
Statistical analysis
Chi-square test, Chi-square test for trend (linear by linear association), Student’s t-test, and multivariate logistic regression analysis were used to investigate the relationship among Mx1 and OAS polymorphisms and severe liver disease. We included the SNP with the Enter algorithm (Forced Entry) and the covariables with the Stepwise algorithm (at each step, factors are considered for removal or entry: a p-value for entry and exit of 0.15 and 0.20, respectively). Thus, each logistic regression test was only adjusted by the most significant co-variables associated with each one of the outcome variables, avoiding the over-fitting of the regression. The covariables used were gender, age, alcohol intake at biopsy study, BMI, HOMA, nadir CD4+ T-cells, AIDS, undetectable HIV-RNA (<50 copies/ml), CD4+ T-cells, time on cART, type of cART, HCV-RNA ≥500,000 IU/ml, time of HCV infection, and HCV genotype. Besides, we also used data of the PNPLA3 rs738409 polymorphism and the IL28B rs12980275 polymorphism, which have already been published by us and showed significant association with liver fibrosis in HIV/HCV coinfected patients36,37. The genetic association study was carried out according to the genetic model that best fit our data (additive, recessive, dominant, codominant, and overdominant). These analyses were tested according to the goodness of fit evaluated by Akaike information criterion (AIC) value and Bayesian information criterion (BIC). These analyses were performed by using the IBM SPSS Statistics for Windows, Version 21.0 (IBM Corp, Chicago, Armonk, NY, USA).
Hardy-Weinberg equilibrium (HWE) for all SNPs was assessed by a Chi-square test, considering equilibrium when p > 0.05. In addition, pair-wise linkage disequilibrium (LD) analysis was computed to detect the inter-marker relationship using the standardized D′ and r2 values by Haploview 4.2 software. Haplotype-based association testing was performed using Plink software (http://pngu.mgh.harvard.edu/~purcell/plink/) comparing each haplotype with the rest of haplotypes (there was no reference category). All p-values were two-tailed and statistical significance was defined as p < 0.05.
The in silico analysis for possible functional implication of each polymorphism was evaluated by using two web-tools: a) VarioWatch (http://genepipe.ncgm.sinica.edu.tw/variowatch/); b) rSNABase (http://rsnp.psych.ac.cn/).
Additional Information
How to cite this article: García-Álvarez, M. et al. Mx1, OAS1 and OAS2 polymorphisms are associated with the severity of liver disease in HIV/HCV-coinfected patients: A cross-sectional study. Sci. Rep. 7, 41516; doi: 10.1038/srep41516 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Heim, M. H. & Thimme, R. Innate and adaptive immune responses in HCV infections. Journal of hepatology 61, S14–S25, doi: 10.1016/j.jhep.2014.06.035 (2014).
Mengshol, J. A., Golden-Mason, L. & Rosen, H. R. Mechanisms of Disease: HCV-induced liver injury. Nature clinical practice. Gastroenterology & hepatology 4, 622–634, doi: 10.1038/ncpgasthep0961 (2007).
Lo Re, V. 3rd et al. Hepatic decompensation in antiretroviral-treated patients co-infected with HIV and hepatitis C virus compared with hepatitis C virus-monoinfected patients: a cohort study. Annals of internal medicine 160, 369–379, doi: 10.7326/m13-1829 (2014).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annual review of immunology 32, 513–545, doi: 10.1146/annurev-immunol-032713-120231 (2014).
Barkhash, A. V. et al. Variability in the 2′–5′-Oligoadenylate Synthetase Gene Cluster Is Associated with Human Predisposition to Tick-Borne Encephalitis Virus-Induced Disease. Journal of Infectious Diseases 202, 1813–1818, doi: 10.1086/657418 (2010).
Lim, J. K. et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS pathogens 5, e1000321, doi: 10.1371/journal.ppat.1000321 (2009).
Thamizhmani, R. & Vijayachari, P. Association of dengue virus infection susceptibility with polymorphisms of 2′−5′-oligoadenylate synthetase genes: a case-control study. The Brazilian journal of infectious diseases: an official publication of the Brazilian Society of Infectious Diseases 18, 548–550, doi: 10.1016/j.bjid.2014.03.004 (2014).
Hamano, E. et al. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochemical and biophysical research communications 329, 1234–1239, doi: 10.1016/j.bbrc.2005.02.101 (2005).
Chen, L. B. et al. Relationship between SNP rs10774671 on OAS-1 gene and spontaneous HBeAg seroconversion in chronic HBV infection. Zhonghua shi yan he lin chuang bing du xue za zhi = Zhonghua shiyan he linchuang bingduxue zazhi = Chinese journal of experimental and clinical virology 23, 35–37 (2009).
Bader El Din, N. G. et al. Association of Myxovirus Resistance Gene Promoter Polymorphism with Response to Combined Interferon Treatment and Progression of Liver Disease in Chronic HCV Egyptian Patients. Journal of interferon & cytokine research: the official journal of the International Society for Interferon and Cytokine Research, doi: 10.1089/jir.2014.0137 (2015).
Knapp, S. et al. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun 4, 411–419, doi: 10.1038/sj.gene.6363984 (2003).
Hijikata, M., Ohta, Y. & Mishiro, S. Identification of a Single Nucleotide Polymorphism in the MxA Gene Promoter (G/T at nt –88) Correlated with the Response of Hepatitis C Patients to Interferon. Intervirology 43, 124–127 (2000).
Suzuki, F. et al. Single nucleotide polymorphism of the MxA gene promoter influences the response to interferon monotherapy in patients with hepatitis C viral infection. Journal of viral hepatitis 11, 271–276, doi: 10.1111/j.1365-2893.2004.00509.x (2004).
Haller, O., Staeheli, P., Schwemmle, M. & Kochs, G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends in microbiology 23, 154–163, doi: 10.1016/j.tim.2014.12.003 (2015).
Fernandez, M. et al. In vivo and in vitro induction of MxA protein in peripheral blood mononuclear cells from patients chronically infected with hepatitis C virus. The Journal of infectious diseases 180, 262–267, doi: 10.1086/314859 (1999).
Sadler, A. J. & Williams, B. R. G. Interferon-inducible antiviral effectors. Nat Rev Immunol 8, 559–568 (2008).
Zhao, Y., Kang, H., Ji, Y. & Chen, X. Evaluate the relationship between polymorphisms of OAS1 gene and susceptibility to chronic hepatitis C with high resolution melting analysis. Clinical and experimental medicine 13, 171–176, doi: 10.1007/s10238-012-0193-6 (2013).
Li, C.-Z. et al. Polymorphism of OAS-1 determines liver fibrosis progression in hepatitis C by reduced ability to inhibit viral replication. Liver International 29, 1413–1421, doi: 10.1111/j.1478-3231.2009.02061.x (2009).
El Awady, M. K. et al. Single nucleotide polymorphism at exon 7 splice acceptor site of OAS1 gene determines response of hepatitis C virus patients to interferon therapy. Journal of gastroenterology and hepatology 26, 843–850, doi: 10.1111/j.1440-1746.2010.06605.x (2011).
Barkhash, A. V., Kochneva, G. V., Chub, E. V., Mikhailova, S. V. & Romaschenko, A. G. Association between polymorphisms in OAS2 and CD209 genes and predisposition to chronic hepatitis C in Russian population. Microbes and infection/Institut Pasteur 16, 445–449, doi: 10.1016/j.micinf.2014.02.004 (2014).
Rueger, S. et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut, doi: 10.1136/gutjnl-2014-306997 (2014).
Patin, E. et al. Genome-wide association study identifies variants associated with progression of liver fibrosis from HCV infection. Gastroenterology 143, 1244-1252 e1241–1212, doi: 10.1053/j.gastro.2012.07.097 (2012).
Park, J. H. et al. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc. Natl. Acad. Sci. USA 108, 18026–18031, doi: 10.1073/pnas.1114759108 (2011).
Perneger, T. V. What’s wrong with Bonferroni adjustments. BMJ 316, 1236–1238 (1998).
Sterne, J. A. & Davey Smith, G. Sifting the evidence-what’s wrong with significance tests? BMJ 322, 226–231 (2001).
Panel de expertos de Gesida Plan Nacional sobre el Sida y Asociación Española para el Estudio del Hígado. [Recommendations of Gesida/PNS/AEEH for the management and treatment of the adult patient co-infected with HIV and hepatitis A, B and C virus]. Enfermedades infecciosas y microbiologia clinica 28, 31 e31–31, doi: 10.1016/j.eimc.2009.12.001 (2010).
Ghany, M. G., Strader, D. B., Thomas, D. L. & Seeff, L. B. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49, 1335–1374, doi: 10.1002/hep.22759 (2009).
Thorpe, L. E. et al. Risk of hepatitis C virus infection among young adult injection drug users who share injection equipment. American journal of epidemiology 155, 645–653 (2002).
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Ascione, A. et al. Peginterferon alfa-2a plus ribavirin is more effective than peginterferon alfa-2b plus ribavirin for treating chronic hepatitis C virus infection. Gastroenterology 138, 116–122, doi: 10.1053/j.gastro.2009.10.005 (2010).
Jimenez-Sousa, M. A. et al. IL28RA polymorphism is associated with early hepatitis C virus (HCV) treatment failure in human immunodeficiency virus-/HCV-coinfected patients. J. Viral Hepat. 20, 358–366, doi: 10.1111/jvh.12041 (2013).
Resino, S. et al. An artificial neural network improves the non-invasive diagnosis of significant fibrosis in HIV/HCV coinfected patients. The Journal of infection 62, 77–86, doi: 10.1016/j.jinf.2010.11.003 (2011).
Bedossa, P. & Poynard, T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology (Baltimore, Md.) 24, 289–293, doi: 10.1002/hep.510240201 (1996).
Hong, E. P. & Park, J. W. Sample size and statistical power calculation in genetic association studies. Genomics & informatics 10, 117–122, doi: 10.5808/GI.2012.10.2.117 (2012).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74, doi: 10.1038/nature15393 (2015).
Jimenez-Sousa, M. A. et al. Impact of patatin-like phospholipase domain-containing 3 gene polymorphism (rs738409) on severity of liver disease in HIV/hepatitis C virus-coinfected patients. Aids 30, 465–470, doi: 10.1097/QAD.0000000000000908 (2016).
Guzman-Fulgencio, M. et al. IL28B polymorphisms are associated with severity of liver disease in human immunodeficiency virus (HIV) patients coinfected with hepatitis C virus. The Journal of infection 66, 170–178, doi: 10.1016/j.jinf.2012.10.025 (2013).
Acknowledgements
This work has been supported by grants given by Fondo de Investigación de Sanidad en España (FIS) [Spanish Health Founds for Research] [grant numbers PI14/01094, PI14CIII/00011] and Red Española de Investigación en SIDA (RIS) [AIDS Research Network] [grant numbers RD16CIII/0002/0002RD16 and RD16/0025/0017]. This work has been (partially) funded by the RD12/0017 project as part of the Plan Nacional R + D + I and cofinanced by ISCIII- Subdirección General de Evaluación y el Fondo Europeo de Desarrollo Regional (FEDER). J.B. is an investigator from the Programa de Intensificación de la Actividad Investigadora en el Sistema Nacional de Salud (I3SNS). M.G.A., D.P.T. and M.A.J.S. are supported by “Instituto de Salud Carlos III” [grant numbers CD12/00442, CM12/00043, CD13/00013, respectively]. The authors thank the Spanish National Genotyping Center (CeGen) for providing SNP genotyping services (http://www.cegen.org).
Author information
Authors and Affiliations
Contributions
M.G.A. and S.R. performed all statistical analysis, interpretation of the data and wrote the manuscript. J.B. and S.R. participated in the study concept and design. J.B., T.A.E., C.D., and F.T., participated in patient selection, collection of samples and acquisition of data. D.P.T., M.A.J.S., and S.V.M. participated in sample preparation, DNA isolation and genotyping pre-procedure, and contributed with critical revision of the manuscript. S.R. supervised the study. All authors revised the manuscript from a draft by S.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
García-Álvarez, M., Berenguer, J., Jiménez-Sousa, M. et al. Mx1, OAS1 and OAS2 polymorphisms are associated with the severity of liver disease in HIV/HCV-coinfected patients: A cross-sectional study. Sci Rep 7, 41516 (2017). https://doi.org/10.1038/srep41516
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep41516
This article is cited by
-
Differential transcriptome response following infection of porcine ileal enteroids with species A and C rotaviruses
Virology Journal (2023)
-
Molecular Mechanisms of ZIKV-Induced Teratogenesis: A Systematic Review of Studies in Animal Models
Molecular Neurobiology (2023)
-
A comprehensive genome-wide profiling comparison between HBV and HCV infected hepatocellular carcinoma
BMC Medical Genomics (2019)
-
Consequence of HIV and HCV co-infection on host immune response, persistence and current treatment options
VirusDisease (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.