Abstract
Despite the unique nature of endemic species, their origin and population history remain poorly studied. We investigated the population history of 28 coral reef fish species, close related, from the Gambier and Marquesas Islands, from five families, with range size varying from widespread to small-range endemic. We analyzed both mitochondrial and nuclear sequence data using neutrality test and Bayesian analysis (EBSP and ABC). We found evidence for demographic expansions for most species (24 of 28), irrespective of range size, reproduction strategy or archipelago. The timing of the expansions varied greatly among species, from 8,000 to 2,000,000 years ago. The typical hypothesis for reef fish that links population expansions to the Last Glacial Maximum fit for 14 of the 24 demographic expansions. We propose two evolutionary processes that could lead to expansions older than the LGM: (a) we are retrieving the signature of an old colonization process for widespread, large-range endemic and paleoendemic species or (b) speciation; the expansion reflects the birth of the species for neoendemic species. We show for the first time that the demographic histories of endemic and widespread reef fish are not distinctly different and suggest that a number of processes drive endemism.
Similar content being viewed by others
Introduction
The existence of endemic coral reef fish species is a challenge for evolutionary biologists to explain given tropical waters are widely connected. The highest levels of endemism on coral reefs are observed near islands at the peripheries of the Indo-Malay-Philippines Archipelago (IMPA), global hotspot of reef fish species diversity1,2,3,4,5. As examples from the Pacific Ocean, the percentage of reef fish that are endemic in the Hawaiian archipelago is 25%6, 22% in Easter Island7, and 14% in the Marquesas Islands8. Even if actual patterns of distribution of coral reef fish species are now well depicted, the evolution and processes underlying the establishment and maintenance of endemic species remains unclear. Despite their unique nature and their potentially higher risk of extinction, the origin and population history of endemic species is poorly studied.
The Pleistocene era (c. 1.8–0.01 Ma), which affected the distribution and demographic history of both terrestrial and costal marine species9, was characterized by glacial cycles and sea level fluctuations up to 150 m below present sea level10,11. Large parts of continental shelves were exposed during low sea level, altering shallow water habitat and likely reducing coral reef area12,13,14. These dramatic changes to the environment influence the demographic history of populations, leaving a footprint in the pattern of genetic diversity15 that will vary depending on the range extent of the species (i.e., how much of the global species was affected by the event). Population bottlenecks and expansions have often been retrieved in marine populations and shown to coincide with the last major sea level changes impacting population dynamics9,16,17,18.
Species biology can affect how reef fish species respond in term of population size variation to major sea level changes such as through varying the larval phase or habitat19,20,21,22. However, very few studies have explored how species with different range size respond to major climatic events23,24. Endemic species have by definition a limited distribution25 so are widely expected to be highly vulnerable to environmental changes that are locally disrupting26,27. This is particularly the case for coral reef fishes forming metapopulations where connectivity occurs only during the larval stage, i.e. colonization of new suitable habitats and migrant exchanges among already established populations. Populations of widespread species even at the edges of their geographic range can still be considered as part of a large metapopulation. The exchange of migrants through larval connectivity, even if very infrequent, can enable recovery from major environmental changes28. In contrast, endemic species with a limited geographical distribution cannot rely on outcrossing with source populations, being either a newly established species (neoendemism) or the remnant of an ancestral widespread species (paleoendemism). In summary, two main differences characterize endemic vs widespread species: the size of the habitat and the degree of connectivity. We therefore expect to see major differences in the demographic histories of endemic and widespread species.
We investigate the genetic diversity and demographic history of 28 reef fish species from the Gambier and Marquesas archipelagos, representing five major reef fish families and selected to include both endemic and widespread species of the same genus. In particular, we examine whether range size is a determinant of the demographic history we retrieve; i.e., do demographic histories vary with range size? Examining multiple pairs of close related species allows us to infer if co-distributed species shared the same demographic history, and test the potential influence of their range distribution. We used both mitochondrial and nuclear DNA sequences to examine demographic history. The use of several independent markers provides a replicate of the coalescent process and improves demographic estimates by reducing the coalescent variance29,30,31. Historically, mitochondrial markers have been widely used in population genetics because of the availability of universal primers (at least for groups of related species) and because of the lack of intralocus recombination, which may bias demographic inferences. However, nuclear markers can now be more easily typed and can also be used to examine demographic history. Using both marker types can enhance our ability to depict complex evolutionary history of species32. This study represents the first large-scale comparison of the demographic history of endemic versus widespread marine species. We resolve the demographic histories of the subject species and discuss the potential underlying evolutionary processes that led to present day patterns.
Results
Genetic diversity
The 28 species revealed a wide range of genetic variability (Table 1). Of the four factors used as predictors of haplotype and nucleotide diversity for the 4 genes, the only significant predictors were archipelago for nucleotide diversity of the GnRH intron (W = 82, p-value < 0.05; higher values for Gambier Islands) and families for the S7 intron (H4 = 10.2945, p-value < 0.05; with higher values for Serranidae and lower for Chaetodontidae). Haplotype and nucleotide diversity were also not significantly different when analyzed by congener pairs made up of an endemic and widespread species (COI: h: V = 21, p = 0.32; π: V = 14, p = 0.34; cytb: h: V = 29, p = 0.50; π: V = 11, p = 1; GnRH: h: V = 34, p = 0.56; π: V = 21, p = 0.73; S7: h: V = 21, p = 0.91; π: V = 19, p = 0.72). Overall haplotype diversity was higher for cytochrome b (0.754 ± 0.197) and S7 intron (0.773 ± 0.231) than for COI (0.598 ± 0.227) and GnRH intron (0.432 ± 0.255). Nucleotide diversity was higher for S7 (0.008 ± 0.007) than for cytochrome b (0.003 ± 0.003), COI (0.002 ± 0.002) or GnRH intron (0.002 ± 0.002).
Past population size changes
Overall, we retrieved population expansion for almost all species, regardless of their range size or locality. Indeed, we found population expansions for 24 species and a constant effective size for 4 species (Tables 2 and 3).
Neutrality tests and Extended Bayesian Skyline Plot (EBSP) analysis allowed us to retrieve population expansion for 24 species. For 19 species, we found a population expansion with significant neutrality test FS for at least one gene for both type of marker, EBSP rejecting a constant size model in favor of one demographic change and a clear graphical expansion (Table 2, Figs 1 and S1). For 5 species, we retrieved a population expansion only from the mitochondrial markers while nuclear markers indicated a signal of constant population. Approximate Bayesian Computation (ABC) analyses were concordant with negative growth rates retrieved, indicating population expansions (Table 2). The species for which expansions were found (19 of 28 with two types of marker, 24 of 28 with one type of marker) are from both archipelagos and include both endemic (5 large-range, 8 small -range) and widespread species (8 species), 4 of the 5 families (except Chaetodontidae), and all types of reproduction (Table 2).
We find a constant effective population size only for 4 species (Tables 2 and 3). For these, neutrality tests and EBSP analysis indicated a constant population size for either both type of markers (Plectroglyphidodon lacrymatus) or most of the analyses (Chaetodon declivis, C. citrinellus and Plectroglyphidodon leucozonus) (Tables 2 and 3). All species are from Marquesas; three are widespread species and one is a large-range endemic species. They belong to two different families (Chaetododontidae and Pomacentridae) that differ in their reproduction strategies (pelagic eggs released vs. benthic spawners species).
Timing of the demographic events
We found overall mostly signal of population expansion (24/28 species), with expansion times that varied widely among species, ranging from 8,000 YBP to 2,000,000 YBP, no matter the method used or the genes screened.
For mitochondrial markers, EBSP analysis shows that expansion time varied from 15,000 YBP to 440,000 YBP (Table 3, Fig. 1 and Figs S1 and S2). The timing of expansions retrieved does not differ significantly among species. None of the tested factors were significant: family (H3 = 2.8578, p = 0.41), archipelago (W = 55.5, p = 0.62), range-size classification (W = 57.5; p = 0.57 and H2 = 0.5568, p = 0.76), and reproductive strategy (H2 = 2.6737, p = 0.26) (Fig. 2 and Fig. S3). For nuclear markers, EBSP analysis shows that expansion times varied more widely among species than was found for mitochondrial markers, ranging from 8,000 YBP to 2,000,000 YBP (Table 3, Figs S1 and S2). As it was the case with the mitochondrial markers, the timing of expansions retrieved for nuclear markers does not differ significantly among species. Again, none of the tested factors were significant: family (H3 = 5.5257, p = 0.14), archipelago (W = 38, p = 0.43), range-size classification (W = 30, p = 1; and H2 = 0.0473, p = 0.98), and reproductive strategy (H2 = 4.4632, p = 0. 11) (Fig. 2 and Fig. S3).
We compared the population expansion timing retrieved with the EBSP (TEBSP) for the two types of marker. We found a concordant time (within 25,000 years) of expansion for some species (e.g. Epinephelus irroratus, Ostorhinchus relativus, or Stegastes aureus). For others, TEBSP varied greatly between marker types within the same species (e.g. Acanthurus nigricans, Pristiapogon kallopterus or Chromis fatuhivae) (Table 3, Fig. 3 and Fig. S2). Considering the 24 species displaying population expansion, we found TEBSP that varied by a factor of 26 for the mitochondrial marker and 250 for the nuclear marker. When narrowed to family, TEBSP varied for Pomacentridae by a factor from 1.7 (Acanthuridae) to 26.7 (Pomacentridae) for the mitochondrial markers and from 2.27 (Apogonidae) to 250 (Pomacentridae) for the nuclear markers. Finally, comparisons between closest relatives (i.e. same genus) revealed both very similar TEBSP and very divergent ones, with a variation by a factor of 1.04 (Pseudogramma) to 13.33 (Chromis) for mitochondrial markers and 1.6 (Epinephelus) to 12.14 (Acanthurus) for nuclear markers.
The shape of the dots denotes the method (ABC vs EBSP) applied on each type of marker (mt: mitochondrial; nuc: nuclear). Colors denote potential demographic scenarios associated to the expansions retrieved: Last Glacial Maximum (LGM), colonization of the archipelagos by the species through connectivity (Colonization) or birth of species (Birth of the sp).
Expansion times and credible intervals provided by the ABC analyses (TABC + CI) relaxed the differences observed for each species considering solely the EBSP expansion time. We retrieved distribution of expansion time (i.e. TEBSP with EBSP and TABC + CI with the ABC) concordant in general but not overlapping for 7 species, mostly due to values retrieved with the EBSP for nuclear marker.
Discussion
Population expansions dominate the demographic history of endemic and widespread Pacific reef fish. The timing of the population expansions we retrieved varies greatly, between 15,000–440,000 YBP for mitochondrial markers and between 8,000–2,000,000 YBP for nuclear markers (Table 3 and Fig. 3). There are no consistent patterns among the predictor variables with respect to when the expansions likely occurred. Population expansion timing even varied both among and within the endemic and widespread species, which was unexpected. Such expansions have usually been associated with Pleistocene interglacial periods, when sea level variations likely profoundly affected habitat distribution10,33. Indeed, correlations between population expansion and such Pleistocene sea-level changes have been reported for many reef fish families in the literature: Acanthuridae19,34,35, Chaetodontidae and Pomacentridae19, Gobiidae36, Holocentridae37, Lutjanidae38, or Scaridae39,40, and also for other marine organisms such as gastropods41,42. Nine glacial cycles have been recorded in the last 800,000 years, all of which resulted in major sea level variations43,44 that could have produced departures from genetic equilibrium conditions. However, if Pleistocene climate changes drive species demography, we should observe both expansions and population contractions following climatic oscillation. Strikingly, we found two clear patterns only in our survey; (i) population expansion; and (ii) a constant effective size, with the former largely dominating the demographic history of endemic and widespread Pacific reef fish (Table 2). Neither neutrality tests nor EBSP provided any evidences of population contraction for any markers in any species. The statistical power to detect a bottleneck is dependent on the number of markers used45. Unfortunately, reconstructing complex demography can be a very difficult task even when analyzing whole genome data (see for example Boitard et al.46). Using relatively few markers like in this study, it is possible that no coalescence survived the bottleneck preceding the last expansion, erasing all traces of more ancient expansions47.
We expected then to recover only the most recent expansion, occurring after the Last Glacial Maximum (LGM), i.e., between 26,500 and 19,000–20,000 years ago48. Therefore, we considered that a species expanded because of the climatic change after the LGM if the estimated expansion time was <26,500 years B.P. When looking at TEBSP we found only 5 species for which at least one of the two marker types (i.e., mitochondrial or nuclear) was strictly consistent with an expansion after the LGM (Abudefduf conformis, Chromis flavapicis, Epinephelus irroratus, Pristiapogon kallopterus, and Stegastes emeryi; Table 3 and Fig. 3). TEBSP is a point estimates inferred visually and a posteriori after the likelihood based analysis performed with BEAST. We therefore performed an ABC estimate of the expansion time to compute its 95% credible interval. We found the distribution of TABC consistent with the LGM for 9 more species with at least one of the two marker types (Abudefduf sordidus, Chromis agilis, Chrysiptera galba, C. glauca, Dascyllus strasburgi, Epinephelus fasciatus, Ostorhinchus relativus, Stegastes aureus, S. fasciolatus; Table 3 and Fig. 3). These 14 species (5 detected with TEBSP and 9 with TABC) probably experienced variations of their population size during the Pleistocene interglacial periods and started expanding soon after their habitats were restored.
However, we retrieved expansion times for both endemic and widespread species older than the LGM for almost half of the species. These results are totally in agreement with previous phylogeographic studies based on mitochondrial markers. Population expansions have been found for reef fish in Marquesas Islands of 15,400–23,900 YBP for Chaetodon ornatissimus49; 180,000–370,000 YBP for Lutjanus kasmira and 120,000–240,000 YBP for L. fulvus38. We propose two other scenarios as potential explanations of the large range we find in the timing of population expansions and the lack of differences between endemic and widespread species.
-
a
Signals of population expansion observed before the LGM correspond to the colonization of the archipelagos by the species through connectivity. This scenario seems to be more dominant as most expansions were older than the LGM (14/24 expansions retrieved considering expansion and mixed categories (Table 3)). This may apply to widespread species like Chromis agilis (mtDNA: 140,000 YBP, nucDNA: 1,400,000 YBP), or Ostorhinchus apogonoides (mtDNA: 440,000 YBP) and large-range endemic species like C. bami (mtDNA: 200,000 YBP, nucDNA: 350,000 YBP). For these species, migrants coming from other demes of the metapopulation may have recolonized new empty habitat. This scenario may also apply to the small-range endemic species Chromis fatuhivae (mtDNA: 400,000 YBP, nucDNA: 2,000,000 YBP), a species presenting an important genetic divergence (11% with cytochrome b, roughly 11Mya with a 1% divergence rate) with its only close relative known to date, C. bami50. Small-range endemic species can be paleoendemic, i.e. these are the remnants of species that once had a much larger range size.
-
b
The expansions correspond to the actual success of local speciation. This scenario is only likely for relatively young endemic species (i.e. neoendemics). Several endemic species from these regions have demonstrated a young evolutionary history and show incomplete lineage sorting when distance trees are computed with their closest sister species (e.g. Canthigaster criobe51, Kuhlia petiti52, Mulloidichthys mimicus53, or Acanthurus reversus54). Divergence time constitutes the upper bound of the speciation process and the initiation of the differentiation of species. This scenario may apply to the Marquesan endemic species Acanthurus reversus (mtDNA: 175,000 YBP, nucDNA: 70,000 YBP), Plectroglyphydodon sagmarius (mtDNA: 220,000 YBP, nucDNA: 650,000 YBP), and Chromis abrupta (mtDNA: 110,000 YBP, nucDNA: 1,600,000 YBP). All of them are the sibling species of widespread species that they replace in Marquesas (respectively A. olivaceus55, P. imparipennis and C. margaritifer50) and present a young evolutionary history. A. reversus shows incomplete lineage sorting with A. olivaceus54 and so does C. abrupta and C. margaritifer (many C. abupta COI haplotypes matches (100% means identity) C. margaritifer COI haplotypes (GenBank numbers: FJ583159; FJ583158; and FJ583162)) while P. sagmarius present little genetic divergence with its sibling species, less than 2% (P. imparipennis COI sequence number JQ350225 (GenBank)), equivalent to a divergence date of less than 2 Mya. The expansion retrieved for these species would then correspond to their birth as endemic species.
All our results are based on the hypothesis that we correctly specified the mutation rate for all our species and all the genes here considered. Rates may vary greatly from one species to another and the estimates obtained rely heavily on the statistical methods used to calibrate the clock56,57. Nonetheless, molecular clock rates for COI and cytochrome b for reef fish are well known and we used the 1–2% divergence rate generally used as a consensus in the literature23,49,56,58,59. Concerning nuclear markers, we used a divergence rate for S7 calibrated on reef fishes60 and polar fishes20. They constitute the only calibration so far obtained for this intron while no calibration has been proposed yet for the GnRH intron. However, evolution rates for nuclear DNA are usually slower than those for mitochondrial DNA57,61 and such rate seems to be in accordance with rates of mitochondrial markers we used, though those rates can vary greatly in vertebrates56,62,63. We acknowledge that variability in the molecular rate of evolution among our species may exist and inflate the differences we have observed in the estimated expansion times. Nevertheless, if we consider TEBSP, we note that it varies between the youngest and the oldest estimate by a factor of 26 for the mitochondrial markers and 250 for the nuclear markers. Such wide distribution was confirmed when looking at closely related species: expansion times varied for example for Pomacentridae by a factor of 19 for the mitochondrial markers and 200 for the nuclear markers and for Chromis genus by a factor of 13,3 for the mitochondrial markers and 5,7 for the nuclear markers. Similar values are found when considering the mode of TABC. In summary, the width of the distribution of those estimated expansion time (both TEBSP and TABC) for both mitochondrial and nuclear markers is so large that it cannot be explained by the uncertainty of the molecular rate or the variance associated with the estimation process. In summary, we cannot reconcile all the dates to a single environmental change. It is much more likely that each species (or at least several groups of species) was affected differently by successive specific events.
The use of two types of molecular markers allowed us to reconstruct the demographic history of reef fishes from two remote archipelagos hosting high level of endemism. We find that the demographic history of most Pacific reef fish species, despite their variety of range size and life traits, is dominated by population expansions. Consistently with the absence of difference in the demographic signature, we find that the genetic diversity of endemic and widespread species is similar. This matches the conclusions of our previous study, which was only based on one mitochondrial marker (cytochrome b)64, but see also Eble, et al.23 and Hobbs, et al.65. We strengthen this result here by adding another mitochondrial locus and two unlinked nuclear loci. Genetic diversity and signature of effective population size change are therefore not correlated with the range-size distribution of reef fish species. More loci will be needed to refine the demographic trajectories of each species, both to describe events that could have been missed due to a lack of statistical power and to improve the parameter estimation process. Overall, this study highlights that demographic histories of endemic species do not differ from widespread species; both have complex and similar histories.
Methods
Specimen collection
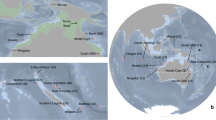
The Gambier archipelago (centered on 23°S, 134°W) is southeast of the Tuamotu Archipelago; it includes 11 high islands spreading only over 40 km and enclosed by a wide barrier reef. The Marquesas archipelago spread over 500 km between 8°–11°S and 141°–138°W and includes 12 high volcanic islands surrounded by fringing reefs66; these are the northeastern-most islands of French Polynesia. We sampled a total of 1,244 reef fishes using polespears or anesthetic while SCUBA diving and exploring all islands of the Gambier Islands and of the Marquesas Islands respectively in 2010 and 2011. This dataset is composed of 28 species (~45 individuals per species), with 20 and 8 co-distributed species respectively in Marquesas and in Gambier Islands (Fig. 4a), and includes a wide range of life history traits (Table 1). Species range distribution (Fig. 4b) varied from widespread (range size > 12 000 km, 13 species), to large-range endemic (1000–8000 km, 6 species) and small-range endemic (less than 500 km, 9 species, see justification in Delrieu-Trottin et al.64). Five different families are represented: Acanthuridae, Apogonidae, Chaetodontidae, Pomacentridae and Serranidae. Several reproductive strategies are represented meaning larval dispersion potential varies greatly (Table 1). Difference of sampling between the archipelagos relates to the uniqueness of Marquesas Islands. Third highest region of endemism for coral reef fishes in the Indo-Pacific8, they host many more cases of endemism.
(a) Species sampled in Marquesas (20) and Gambier Islands (8) (~45 individuals per species) with their range size. (b) The maximum extent of each species is reported with the location itself (e.g. Marquesas) or the westward and eastward know location to date (e.g. Cocos Keeling–Galapagos). The map was produce using R82, R package ‘maps’90 and modified using Adobe Illustrator CS5 v 15.0.2. http://www.adobe.com/products/illustrator.html.
Ethics Statement
The study protocol was approved by the National Center for Scientific Research and is in accordance to the laws of the French Republic and of the collectivity of French Polynesia.
Laboratory procedures and genetic analyses
We extracted whole genomic DNA from fin tissues preserved in 96% EtOH at ambient temperature using QIAxtractor (QIAGEN, Crawley) according to manufacturer’s protocols. Fragments of the cytochrome b for 1,237 individuals (Cyt b, 739–999 bp) and cytochrome C oxydase subunit 1 for 1,005 individuals (COI, 571–688 bp), both parts of mitochondrial genome, were amplified using PCR. These were sequenced using the universal primers GLUDGL-CB3H of Palumbi et al.67 and different combinations of primers from Ward et al.68. In addition, part of the third intron in the Gonadotropin-Releasing Hormone gene for 912 individuals (GnRH, 267–417 bp) and part of the first intron of the S7 ribosomal protein gene for 973 individuals (S7, 485–783 bp) were amplified. For these, we used primers GnRH3.3F-GnRH3.3R69 and primers S7RPEX1F-S7RPEX2R70. Fragments were amplified using PCR protocols and sequencing as described by Williams et al.51. Differences in the number of samples are due to our inability to amplify cytochrome b for 3 species (Epinephelus irroratus, Epinephelus fasciatus and Ostorhinchus relativus), GnRH intron for 7 species (Dascyllus strasburgi, Chromis flavapicis, Chromis abrupta, Apogon lativittatus, Ostorhinchus apogonoides, Ostorhinchus relativus and Pristiapogon kallopterus) and S7 intron for 3 species (Apogon lativittatus, Epinephelus fasciatus and Ostorhinchus apogonoides). Overall, we worked with a dataset comprised of at least one mitochondrial marker and one nuclear marker for 26 species. Four markers were available for 19 species, 3 markers for 5 species and 2 markers for 4 species. Only mitochondrial data were available for 2 species (Table 1).
Sequences were edited using GENEIOUS PRO v.6.1.7 (Biomatters) and aligned with Clustal W71 as implemented in GENEIOUS. Alignments were unambiguous with no indels or frameshift mutations. For the nuclear markers, allelic state from sequences with multiple heterozygous single nucleotide polymorphisms (SNPs) was estimated using PHASE v.2.1.1 as implemented in DnaSP72,73,74,75. We tested for recombination using SBP and GARD methods implemented online on the Datamonkey webserver76,77. The GARD and SBP tests failed to find recombination for any of the nuclear markers of either species.
Haplotype diversity (h) and nucleotide diversity (π) were calculated using the software package DnaSP v.5.175. To detect departures from a neutral Wright–Fisher model, we used Fu’s FS neutrality test78. Assuming selective neutrality, significant negative values of FS indicate population growth while significant positive values are a signature of either genetic subdivision or population contraction. The test was implemented in DnaSP v.5.177 and its significance was determined from 1000 coalescent simulations.
Changes in effective population size (Ne) through time were estimated using the Extended Bayesian Skyline Plot (EBSP) implemented in BEAST v.1.779,80. EBSP is a non-parametric model that does not specify any prior hypothesis on the tempo and mode of changes of the Ne. EBSP allows the analysis of multiple loci, reducing the stochastic variance of the coalescent process and improving the reliability of demographic inferences47. Mitochondrial and nuclear markers have different mechanisms of evolution and inheritance, which are also affected by reproductive strategies and sex-biased processes. The species investigated use a range of reproductive strategies (e.g. sequential hermaphrodites vs gonochoristic species, couple vs harem) so we analyze mtDNA and nuclear markers separately in the EBSP analyses.
The two nuclear loci are unlinked so have different genealogies, substitution models and clock rates but the same underlying demography. As the mitochondrial genes are in linkage by structure, we used different substitution models and clock rates but the same genealogy. Sequence divergence estimates for the two mitochondrial markers (COI and Cyt b) in reef fish range from 1% to 2% per million years56,58,59,81. We set a strict clock with a uniform prior distribution for the clock rate with an upper and lower mutation rate ranging from 0.5 × 10−8 to 1 × 10−8 per site per year. We retrieved various sequence divergence estimates for S7 intron from 0.28% to 1.7%20,56,60 while no divergence rate was available in the literature for the third intron in the Gonadotropin-Releasing Hormone (GnRH). For these two introns, we set a strict clock with a uniform prior distribution for the clock rate with an upper and lower mutation rate ranging from 0.14 × 10−8 to 0.85 × 10−8 per site per year. To take into account possible site-specific variation in the mutation rate we used the HKY + G model of mutation for all genes. We ran 10 million MCMC iterations with a thinning interval of 1,000. We checked convergence by visually inspecting the trace and computing the effective sample size (which was always higher than 100) for each parameter in two independent runs using the program TRACER v.1.5 (http://tree.bio.ed.ac.uk/software/tracer/). EBSP were plotted using R v.3.2.382 (R Development Core Team 2015) setting the burn-in to 10%.
Generation time is unknown for most species selected for this study and it potentially varies greatly among them as we selected a large range of families. To remove that uncertainty, we therefore set the mutation rate in units per site per year to obtain time points expressed in calendar years; independent of the generation time80. That method allows comparing potential expansion times between close related species but also different families. The effective size being scaled by the generation time (unknown for most species), only the timing of expansions between species will be discussed in the present work.
Finally, we compute the posterior distribution of the number of demographic changes occurring along the gene genealogies. This method formally tests how many times Ne changed through the history of the gene genealogy, allowing to reject a constant size population model. We rejected a constant population size model if the lower bound of the 95% high posterior density (HPD) of this distribution was higher than zero. Expansion times were identified from the skyline output as the point when the Ne started to increase.
The data did not meet assumptions of normality so we computed the non-parametric tests Wilcoxon-Mann-Whitney and Kruskal-Wallis to examine differences in genetic diversity (haplotype and nucleotide) and demography (TMRCA, time of expansion) among four factors: range size (endemic vs. widespread and all three range types), archipelagos, families, and reproductive strategies. We also computed Wilcoxon signed-rank test to compare the genetic diversity indices for endemic species that had a widespread congener (i.e. same genus) caught in the same archipelago. All statistical analyses were performed on R, using the vegan package83, and ggplot2 for graphics84.
To compute the full posterior distribution of the expansion time (T) for each species we developed an Approximate Bayesian Computation (ABC) approach85. ABC summarizes the probability of the data through a vector of observed summary statistics and it is therefore very flexible as it can investigate any demographic model provided it can be simulated86. EBSP showed that all species were either expanding (following approximately an exponential growth) or constant (see results). EBSP uses a likelihood function to compute the probability of the observed data and it is therefore to be preferred over methods which approximate the likelihood. However, the EBSP does not compute explicitly the Texp, which is inferred visually a posteriori and has thus no credible interval associated. We therefore implemented the following three parameters model with ABC: an ancestral constant population characterized by an effective population size (Nanc) starts to increase (or decrease) exponentially at Texp to reach at time 0 the modern effective population size (Nmod). We assigned the following uniform priors to the three parameters: Nanc and Nmod: {1,000 ÷ 10,000,000}; Texp: {100 ÷ 2,000,000}. Mutation rate was set in years as in the EBSP; we used a value of 10−8 per site per year for the mtDNA genes and 8 * 10−9 for the nuclear genes. We performed 500,000 coalescent simulations with parameter values extracted from prior distributions using fastsimcoal2 v.2.5.187. We computed as summary statistics the number of haplotype, the number of segregating sites, the mean pairwise difference and Fu’s Fs using alrsumstat88. Parameters were estimated from the 5,000 simulations closest to the observed dataset using a local linear regression according to Beaumont et al.85 as implemented under the R environment in the library abc89. Analyses were performed separately for the whole mtDNA (one genealogy) and the two nuclear genes (two independent genealogies with the same underlying demography) as in the EBSP.
Additional Information
Accession codes: Sequences have been deposited in GenBank; GenBank accession number KM455125–KM455538 for cytochrome b (unique sequences); KY207629–KY207951 for COI (unique sequences); KY207952–KY208907 for GnRH intron and KY208908–KY209890 for S7 intron.
How to cite this article: Delrieu-Trottin, E. et al. Population expansions dominate demographic histories of endemic and widespread Pacific reef fishes. Sci. Rep. 7, 40519; doi: 10.1038/srep40519 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Bellwood, D. & Wainwright, P. The history and biogeography of fishes on coral reefs In Coral Reef Fishes: Dynamics and Diversity in a Complex Ecosystem (ed. Sale, P. F. ) 5–32 (Academic Press, San Diego, 2002).
Jones, G., Munday, P. & Julian Caley, M. (2002) Rarity in coral reef fish communities. In: Coral Reef Fishes: Dynamics and Diversity in a Complex Ecosystem (ed. Sale, P. F. ) 81–101 (Academic Press, San Diego, 2002).
Connolly, S., Bellwood, D. & Hughes, T. Indo-Pacific biodiversity of coral reefs: deviations from a mid-domain model. Ecology 84, 2178–2190 (2003).
Allen, G. Conservation hotspots of biodiversity and endemism for Indo-Pacific coral reef fishes. Aquat. Conserv. 556, 541–556 (2008).
Briggs, J. C. Diversity, endemism and evolution in the coral triangle. J. Biogeogr. 36, 2008–2010 (2009).
Randall, J. E. Reef and Shore Fishes of the South Pacific (University of Hawai’i Press, Honolulu, Hawai’i 2005).
Randall, J. E. & Cea, A. Shore fishes of Easter Island (University of Hawai’i Press, Honolulu, Hawai’i 2011).
Delrieu-Trottin, E. et al. Shore fishes of the Marquesas Islands, an updated checklist with new records and new percentage of endemic species. Checklist 11, 1758 (2015).
Hewitt, G. M. Ice ages: their impact on species distributions and evolution InEvolution on Planet Earth (eds. Rothschild, L. J. & Lister, A. M. ) 339–361 (Academic Press 2003).
Lambeck, K. & Chappell, J. Sea level change through the last glacial cycle. Science 292, 679–86 (2001).
Siddall, M., Rohling, E. & Almogi-Labin, A. Sea-level fluctuations during the last glacial cycle. Nature 423, 853–858 (2003).
Shulman, M. J. & Bermingham, E. Early life histories, ocean currents, and the population genetics of Caribbean reef fishes. Evolution 49, 897–910 (1995).
Nelson, J., Hoddell, R., Chou, L., Chan, W. & Phang, P. Phylogeographic structure of false clownfish, Amphiprion ocellaris, explained by sea level changes on the Sunda shelf. Mar. Biol. 37, 727–736 (2000).
Hickerson, M. & Ross, J. Post-glacial population history and genetic structure of the northern clingfish (Gobbiesox maeandricus), revealed from mtDNA analysis. Mar. Biol. 138, 407–419 (2001).
Hewitt, G. M. The genetic legacy of the Quaternary ice ages. Nature 405, 907–913 (2000).
Bard, E. et al. Deglacial sea-level record from Tahiti and the timing of global meltwater discharge. Nature 382, 241–244 (1996).
Hewitt, G. M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc B. 359, 183–195 (2004).
Woodroffe, C. D. et al. Response of coral reefs to climate change: Expansion and demise of the southernmost Pacific coral reef. Geophys. Res. Lett. 37, L15602 (2010).
Fauvelot, C., Bernardi, G. & Planes S. Reductions in the mitochondrial DNA diversity of coral reef fish provide evidence of population bottlenecks resulting from Holocene sea-level change. Evolution 57, 1571–1583 (2003).
Janko, K. et al. Did glacial advances during the Pleistocene influence differently the demographic histories of benthic and pelagic Antarctic shelf fishes? Inferences from intraspecific mitochondrial and nuclear DNA sequence diversity. BMC Evol. Biol. 7, 220 (2007).
Ramon, M. L., Nelson, P. A., De Martini, E., Walsh, W. J. & Bernardi, G. Phylogeography, historical demography, and the role of post-settlement ecology in two Hawaiian damselfish species. Mar. Biol. 153, 1207–1217 (2008).
Ludt, W. B., Bernal, M. A., Bowen, B. W. & Rocha, L. A. Living in the past: phylogeography and population histories of Indo-Pacific wrasses (genus Halichoeres) in shallow lagoons versus outer reef slopes. Plos One 7, e38042 (2012).
Eble, J. A., Toonen, R. J. & Bowen, B. W. Endemism and dispersal: comparative phylogeography of three surgeonfishes across the Hawaiian Archipelago. Mar. Biol. 156, 689–698 (2009).
Drew, J. A. & Barber, P. H. Comparative phylogeography in fijian coral reef fishes: a multi-taxa approach towards marine reserve design. Plos One 7, e47710 (2012).
Anderson, S. Area and endemism. Q. Rev. Biol. 69, 451–471 (1994).
Templeton, A. R., Shaw, K., Routman, R. & Davis, S. K. The genetic consequences of habitat fragmentation. Ann. Mo. Bot. Gard. 77, 13–27 (1990).
Ellstrand, N. C. & Elam, D. R. Population genetic consequences of small population size: Implications for plant conservation. Annu. Rev. Ecol. Syst. 24, 217–242 (1993).
Hughes, T. P., Bellwood, D. R., Folke, C., Steneck, R. S. & Wilson, J. New paradigms for supporting the resilience of marine ecosystems. Trends Ecol. Evol. 20, 380–386 (2005).
Felsenstein, J. Accuracy of coalescent likelihood estimates: do we need more sites, more sequences, or more loci? Mol. Biol. Evol. 23, 691–700 (2006)
Brito, P. H. & Edwards, S. V. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica 135, 439–55 (2009).
Wakeley, J. Coalescent Theory: An Introduction. (ed. Greenwood Village, Colorado: Roberts & Company Publishers 2009).
Fagundes, N. J. et al. Statistical evaluation of alternative models of human evolution. Proc. Natl Acad. Sci. USA 104, 17614–17619 (2007).
Lambeck, K., Yokoyama, Y. & Purcell, T. Into and out of the Last Glacial Maximum: sea-level change during Oxygen Isotope Stages 3 and 2. Quat. Sci. Rev. 21, 343–360 (2002).
Klanten, O. S., Choat, J. H. & Herwerden, L. Extreme genetic diversity and temporal rather than spatial partitioning in a widely distributed coral reef fish. Mar. Biol. 150, 659–670 (2006).
Horne, J. B., van Herwerden, L., Choat, J. H. & Robertson, D. R. High population connectivity across the Indo-Pacific: Congruent lack of phylogeographic structure in three reef fish congeners. Mol. Phylogenet. Evol. 49, 629–638 (2008).
Thacker, C. Population structure in two species of the reef goby Gnatholepis (Teleostei: Perciformes) among four South Pacific island groups. Coral Reefs, 23, 357–366 (2004).
Craig, M. T., Eble, J. A., Bowen, B. W. & Robertson, D. R. High genetic connectivity across the Indian and Pacific Oceans in the reef fish Myripristis berndti (Holocentridae). Mar. Ecol. Prog. Ser. 334, 245–254 (2007).
Gaither, M. R., Toonen, R. J., Robertson, D. R., Planes, S. & Bowen, B. W. Genetic evaluation of marine biogeographical barriers: perspectives from two widespread Indo-Pacific snappers (Lutjanus kasmira and Lutjanus fulvus). J. Biogeogr. 37, 133–147 (2010).
Bay, L., Choat, J., Herwerden, L. & Robertson, D. High genetic diversities and complex genetic structure in an Indo-Pacific tropical reef fish (Chlorurus sordidus): evidence of an unstable evolutionary past? Mar. Biol. 144, 757–767 (2004).
Winters, K. L., Herwerden, L., Choat, J. H. & Robertson, D. R. Phylogeography of the Indo-Pacific parrotfish Scarus psittacus: isolation generates distinctive peripheral populations in two oceans. Mar. Biol. 157, 1679–1691 (2010).
Hellberg, M. E., Balch, D. P. & Roy, K. Climate-driven range expansion and morphological evolution in a marine gastropod. Science 292, 1707–1710 (2001).
Crandall, E. D., Frey, M., Grosberg, R. K. & Barber, P. H. Contrasting demographic history and phylogeographical patterns in two Indo-Pacific gastropods. Mol. Ecol. 17, 611–626 (2008).
EPICA. Eight glacial cycles from antarctic ice core. Nature 429, 623–628 (2004).
McManus, J. Palaeoclimate: A great grand-daddy of ice cores. Nature 429, 611–612 (2004).
Hoban, S. M., Gaggiotti, O. E. & Bertorelle, G. The number of markers and samples needed for detecting bottlenecks under realistic scenarios, with and without recovery: a simulation‐based study. Mol. Ecol. 22, 3444–3450 (2013).
Boitard, S., Rodríguez, W., Jay, F., Mona, S. & Austerlitz, F. Inferring population size history from large samples of genome-wide molecular data-an approximate Bayesian computation approach. PLoS Genet 12, e1005877 (2016).
Heled, J. & Drummond, A. J. Bayesian inference of population size history from multiple loci. BMC Evol. Biol. 8, 289 (2008).
Clark, P. U. et al. The last glacial maximum. Science 325, 710–714 (2009).
DiBattista, J. D., Rocha, L. A., Craig, M. T., Feldheim, K. A. & Bowen, B. W. Phylogeography of two closely related Indo-Pacific butterflyfishes reveals divergent evolutionary histories and discordant results from mtDNA and microsatellites. J. Hered. 103, 617–629 (2012).
Randall, J. E. Four New Damselfishes (Perciformes: Pomacentridae) from the Marquesas Islands. Copeia 2001, 92–107 (2001).
Williams, J. T., Delrieu-Trottin, E. & Planes, S. A new species of Indo-Pacific fish, Canthigaster criobe, with comments on other Canthigaster (Tetraodontiformes: Tetraodontidae) at the Gambier Archipelago. Zootaxa 3523, 80–88 (2012).
Feutry, P. et al. Evolution of diadromy in fish: insights from a tropical genus (Kuhlia species). Am Nat. 181, 52–63 (2013).
Lessios, H. & Robertson, D. Speciation on a round planet: phylogeography of the goatfish genus Mulloidichthys. J. Biogeogr. 40, 2373–2384 (2013).
Gaither, M. R., Bernal, M. A. & Coleman, R. R. Genomic signatures of geographic isolation and natural selection in coral reef fishes. Mol. Ecol. 24, 1543–1557 (2015).
Randall, J. E. & Earle, J. L. Acanthurus reversus, a new species of surgeonfish (Perciformes: Acanthuridae) from the Marquesas Islands. Proceedings of the California Academy of Sciences 51, 473–481 (1999).
Lessios, H. A. The Great American Schism: divergence of marine organisms after the rise of the Central American Isthmus. Annu Rev Ecol Syst 39, 63–91 (2008).
Eytan, R. I. & Hellberg, M. E. Nuclear and mitochondrial sequence data reveal and conceal different demographic histories and population genetic processes in Caribbean reef fishes. Evolution 64, 3380–97 (2010).
Bowen, B. W., Bass, A. L., Rocha, L. A., Grant, W. S. & Robertson, D. R. Phylogeography of the trumpetfishes (Aulostomus): ring species complex on a global scale. Evolution 55, 1029–1039 (2001).
Reece, J. S., Bowen, B. W., Joshi, K., Goz, V. & Larson, A. Phylogeography of two moray eels indicates high dispersal throughout the Indo-Pacific. J. Hered. 101, 391–402 (2010).
Bernardi, G. & Lape, J. Tempo and mode of speciation in the Baja California disjunct fish species Anisotremus davidsonii . Mol. Ecol. 14, 4085–4096 (2005).
Lin, C. P. & Danforth, B. N. How do insect nuclear and mitochondrial gene substitution patterns differ? Insights from Bayesian analyses of combined datasets. Mol. Phylogenet. Evol. 30, 686–702 (2004).
Nabholz, B., Glemin, S. & Galtier, N. Strong variations of mitochondrial mutation rate across mammals–the longevity hypothesis. Mol. Biol. Evol. 25, 120–130 (2008).
Lanfear, R., Welch, J. J. & Bromham, L. Watching the clock: studying variation in rates of molecular evolution between species. Trends Ecol. Evol 25, 495–503 (2010).
Delrieu-Trottin, E., Maynard, J. & Planes, S. Endemic and widespread coral reef fishes have similar mitochondrial genetic diversity. Proc. R. Soc. B 281, 20141068 (2014).
Hobbs, J.-P. A., van Herwerden, L., Jerry, D. R., Jones, G. P. & Munday, P. L. High genetic diversity in geographically remote populations of endemic and widespread coral reef angelfishes (genus: Centropyge). Diversity 5, 39–50 (2013).
Clouard, V. & Bonneville, A. Ages of seamounts, islands, and plateaus on the pacific plate. Geol. Soc. Spec. Pap. 388, 71–90 (2005).
Palumbi, S. R. et al. The simple fool’s guide to PCR (Honolulu: Department of Zoology, University of Hawaii 1991).
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R. & Hebert, P. D. N. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc B. 360, 1847–1857 (2005).
Hassan, M., Lemaire, C., Fauvelot, C. & Bonhomme, F. Seventeen new exon primed intron crossing polymerase chain reaction amplifiable introns in fish. Mol. Ecol. Resour. 2, 334–340 (2002).
Chow, S. & Hazama, K. Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol. Ecol. 7, 1255–6 (1998).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Stephens, M., Smith, N. J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001).
Stephens, M. & Donnelly, P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 73, 1162–1169 (2003).
Stephens, M. & Scheet, P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 76, 449–462 (2005).
Librado, P. & Rozas, J. Dnasp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Pond, S. & Frost, S. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 (2005).
Pond, S., Posada, D., Granevor, M., Woelk, C. & Frost, S. GARD: a genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098 (2006).
Fu, Y. X. New statistical tests of neutrality for DNA samples from a population. Genetics 143, 557–70 (1996).
Drummond, A. J., Rambaut, A., Shapiro, B. & Pybus, O. G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 22, 1185–92 (2005).
Drummond, A. J. & Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (2007).
Bermingham, E., McCafferty, S. & Martin, A. P. Fish biogeography and molecular clocks: perspectives from the Panamanian isthmus In Molecular systematics of fishes (eds. Kocher, T. D. & Stepien, C. A. ) 113–128 (Academic Press, New York, 1997).
R. Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See http://www.R-project.org/(2015).
Oksanen, J., Kindt, R., Legendre, P. & O’Hara, R. B. Vegan: community ecology package. R package 2.0–2. See http://cran.rproject.org/web/packages/vegan/index.html (2011).
Wickham, H. ggplot2: elegant graphics for data analysis. (ed. Springer, NY 2009).
Beaumont, M. A., Zhang, W. & Balding, D. J. Approximate Bayesian Computation in population genetics. Genetics 162, 2025–2035 (2002).
Bertorelle, G., Benazzo, A. & Mona, S. ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol. Ecol. 19, 2609–2625 (2010).
Excoffier, L., Dupanloup, I., Huerta-Sanchez, E., Sousa, V. C. & Foll, M. Robust demographic inference from genomic and SNP data. Plos Genet. 9, e1003905 (2013).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567 (2010).
Csillery, K., Francois, O. & Blum, M. G. B. abc: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 3, 475–479 (2012).
Becker, R. A. & Wilks, A. R. Original S Code et al. maps: Draw Geographical Maps. R package version version 3.1.1 (2008).
Acknowledgements
This work was supported by grants from the French National Agency for Marine Protected Area, which funded the sampling expedition ‘Pakaihi I Te Moana’, and the ANR IMODEL and Contrat de Projet Etat-Territoire in French Polynesia, funders of the ‘CORALSPOT’ expedition. Additional funding was provided by the IFRECOR in French Polynesia and the TOTAL Foundation. We are grateful to T. Frogier, P. Mery, P. Sasal, T. Cribb, R. Galzin, M. Kulbicki, J. Mourier and X. (Pipapo) Curvat for their field assistance in the Gambier and the Marquesas Islands, along with the crew of the Claymore II and Braveheart. We are grateful to the Genotoul bioinformatics platform Toulouse Midi-Pyrenees (Bioinfo Genotoul) for providing computing resources. Finally, we thank the two anonymous reviewers for providing constructive reviews of earlier versions of the manuscript.
Author information
Authors and Affiliations
Contributions
E.D.-T., S.P. and M.V. conceived the study. E.D.-T., S.P. and M.V. conducted field work. E.D.-T. and V.N. generated the sequences. E.D.-T. cleaned the sequences. E.D.-T. and S.M. ran and analyzed the population genetic analyses. All authors contributed to writing and revising the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Delrieu-Trottin, E., Mona, S., Maynard, J. et al. Population expansions dominate demographic histories of endemic and widespread Pacific reef fishes. Sci Rep 7, 40519 (2017). https://doi.org/10.1038/srep40519
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40519
This article is cited by
-
Comparative phylogeography and demographic histories of five widely distributed tropical eastern Pacific fishes
Marine Biology (2023)
-
Clarifying the taxonomy of some cryptic blennies (Blenniidae) in their native and introduced range
Scientific Reports (2022)
-
Genetic connectivity in Sparisoma aurofrenatum (redband parrotfish): an unexpected journey
Hydrobiologia (2022)
-
Phylogeography, population connectivity and demographic history of the Stoplight parrotfish, Sparisoma viride (Teleostei: Labridae), in the Greater Caribbean
Coral Reefs (2022)
-
Strong genetic isolation of the black-lipped pearl oyster (Pinctada margaritifera) in the Marquesas archipelago (French Polynesia)
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.