Abstract
The principal Afrotropical human malaria vector mosquito, Anopheles gambiae, remains a significant threat to global health. A critical component in the transmission of malaria is the ability of An. gambiae females to detect and respond to human-derived chemical kairomones in their search for blood meal hosts. The basis for host odor responses resides in olfactory receptor neurons (ORNs) that express chemoreceptors encoded by large gene families, including the odorant receptors (ORs) and the variant ionotropic receptors (IRs). While ORs have been the focus of extensive investigation, functional IR complexes and the chemical compounds that activate them have not been identified in An. gambiae. Here we report the transcriptional profiles and functional characterization of three An. gambiae IR (AgIr) complexes that specifically respond to amines or carboxylic acids - two classes of semiochemicals that have been implicated in mediating host-seeking by adult females but are not known to activate An. gambiae ORs (AgOrs). Our results suggest that AgIrs play critical roles in the detection and behavioral responses to important classes of host odors that are underrepresented in the AgOr chemical space.
Similar content being viewed by others
Introduction
At least half the world’s population is at risk for malaria, while an even larger proportion of people are at risk for many other mosquito-borne diseases, including the arboviruses that cause dengue fever, yellow fevers and Zika1. Blood meal host selection by female mosquitoes is one of the most important factors impacting vector-borne disease transmission. Female mosquitoes are behaviorally activated to initiate upwind flights when stimulated by chemical cues, called kairomones, that are emitted by potential host species2. Some members of the An. gambiae s.l. Giles species complex, including An. gambiae s.s. (S form) and An. coluzzii (M form)3 are major sub-Saharan malaria vectors, in part because of their high propensity to acquire blood meals from human hosts2. Although the precise mechanisms underlying mosquito-host interactions have yet to be elucidated, many studies in the past decade have begun to unravel the molecular basis for fly and mosquito olfaction. These processes appear to be at least partly mediated by three families of chemosensory receptors that work together with accessory proteins to provide the mosquito with sensitivity and discrimination across a wide spectrum of odor space4,5. These include a superfamily of odorant receptor encoding genes (Ors), which are the best characterized chemosensory receptors to date6; gustatory receptors (Grs), a subset of which are known to mediate Anopheline responses to carbon dioxide7 and sugars8; and variant ionotropic receptors (Irs), which were first characterized in Drosophila melanogaster and have been shown to respond to amines, carboxylic acids and other environmental cues9,10,11,12,13,14,15. In contrast to the Or and Gr chemoreceptors that show generally high levels of sequence divergence across insect species, the Irs are more highly conserved and respond to amines and potentially other cues in An. gambiae16.
With a single exception, the family of 46 variant ionotropic receptor genes identified in An. gambiae (AgIrs) have not been functionally characterized16. In contrast, much is known about ionotropic receptors in D. melanogaster (DmIrs), where they are expressed in olfactory neurons of sensilla that are morphologically analogous to grooved peg sensilla found in An. gambiae9. Single sensillum recordings from these neurons illustrate a DmIr derived response profile that is distinct from the odor space encoded by DmOrs9,10,11,12,13. As is true for ORs, IRs functionally consist of heteromeric complexes, although unlike ORs, which all solely couple to the highly conserved Orco coreceptor, IR complexes may have one or more co-receptors that function as ligand-gated ion channels9,10,11,12,13,14,15. There is considerable experimental evidence that some DmIrs utilize DmIr8a as a co-receptor while others require both DmIr25a and DmIr76b, but not all three9,10,11,12,13,14,15. In addition to its role as a co-receptor, DmIr25a has recently been shown to function as a thermosensor and play a role control in establishing circadian rhythm in D. melanogaster14. Specifically, DmIr25a and DmIr21a have been shown to mediate exquisite cool temperature avoidance in Drosophila larvae15. Although most of the insect Irs have diverged in terms of gene numbers and coding (i.e. amino acid) sequence within and across species, the candidate co-receptors are highly conserved among insects11,13,16, suggesting that the assembly and function of Irs will be analogous across lineages. We have observed that the AgIrs function independently of AgOrs at the larval stage where AgIr76b, a potential Ir coreceptor, is required for An. gambiae larval responses to butylamine16, but virtually nothing is known about their relationships at the adult stage. This leads us to speculate that butylamine sensitivity in adult An. gambiae also requires the expression of AgIr76b, plus the coreceptor AgIr25a and an as yet identified “tuning” AgIr.
Although several of functional studies have been carried out on the AgOr family of chemoreceptors, very little is known about the roles of AgIrs in odor coding (the relationship between essential features of odor molecules and their transduction into patterns of neural activity) and function. Importantly, butylamine, lactic acid, and other carboxylic acids are activators of olfactory sensory neurons in An. gambiae17,18,19,20,21, are major components of human sweat21,22,23,24, and are known to be powerful attractants to host seeking An. gambiae and other mosquitoes2,25,26,27. Furthermore, functional analyses of AgOr sensitivities revealed a relative lack of responses to these classes of compounds, suggesting that non-AgOr mediated mechanisms likely underlie sensitivity to these important kairomones in An. gambiae4,5.
In the current study we have characterized of a subset of AgIrs that are expressed in the An. coluzzii (hereafter An. gambiae for consistency with gene annotations and the literature) antennae and maxillary palps28. We provide evidence that these AgIrs are expressed in subset(s) of antennal sensilla in An. gambiae that are distinct from AgOr-containing sensilla. In addition, we have characterized AgIr complexes comprised of specific co-receptors in the Xenopus laevis oocyte heterologous system that confer sensitivities to a narrow range of amines and carboxylic acids. Our results confirm that AgIrs function in adult peripheral chemosensory organs where they are likely to form the basis for An. gambiae behavioral responses to these two compound classes, which are critical for mosquito host seeking. Future studies will continue to illuminate the molecular foundations for host odor sensitivity, and specifically the roles that AgIrs play in this process.
Results
Expression of AgIr transcripts in An. gambiae head appendages
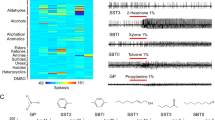
We previously reported the transcript abundances of the AgIr family in An. gambiae adult antennae and maxillary palps based on RNA sequencing28. Here we have adapted that data set to visually display the AgIr transcript profile information for adult female bodies, antennae and maxillary palps (Fig. 1; Supplemental Table S1). The color intensity map in Fig. 1 indicates that the majority of AgIrs are expressed in the adult antennae, with considerably fewer AgIrs expressed in the palps. RPKM values are reported for the co-receptors, AgIr8a, AgIr25a, and AgIr76b as well as for the tuning receptors AgIr41a, AgIr41c, and AgIr75k that have been functionally characterized in this study (Fig. 1; Supplemental Table S1).
Heat map depicting abundances of An. gambiae variant ionotropic receptors (AgIrs) in adult chemosensory tissues (data adapted from28). Scale color intensity indicates Reads Per Kilobase per Million reads (RPKM).
Localization of AgIrs in chemosensory neurons of An. gambiae chemosensory appendages
Based on DmIr studies9,10,11,12,13,14,15 and our own previous observations that AgOrco expression is not observed in ORNs that project into grooved-peg (GP) sensilla29 that are activated by ammonia, a subset of carboxylic acids and other odorants18, we reasoned that signaling in grooved pegs is likely to be transduced by AgIrs. We therefore elected to further examine the relationship between Ir and Orco expression within the antennae of adult female An. gambiae. In order to evaluate the co-expression of AgIr76b and AgOrco in female antenna, double fluorescence in-situ hybridization (FISH) and single FISH, combined with immunohistochemistry (IHC) were conducted. Antennae were probed with pairs of differentially labeled AgIr76b- and AgOrco-specific riboprobes or, alternatively an anti-Orco antibody30. A significant number of AgIr76b and AgOrco labeled cell bodies were identified in the female antenna (Fig. 2). While we noted an apparent increase in the number of labeled neurons along the antenna from proximal to distal flagellomeres, we did not attempt to quantify those differences. This observation may indeed reflect the general apical increase in the number of olfactory sensilla in the female antennae31. Importantly, AgIr76b and AgOrco sense control riboprobes fail to label antennal neurons (Supplemental Figure S1) and double AgIr76b-AgOrco antisense riboprobes clearly label distinct cell bodies, as do the AgIr76b antisense riboprobe and anti-Orco antibody (Fig. 2E–H). Although some small puncta of apparent overlap between AgIr76b and AgOrco expression are present in the Z-stack confocal images (Fig. 2D, yellow arrowheads), we note these signals lie within subtle yet observable cellular outlines. We therefore attribute these signals to be artifacts of image stacking that are due to physical proximity or overlap of distinct cells and are not indicative of AgIr76b and AgOrco co-localization. Taken together we believe these data support the hypothesis that AgIr76b and AgOrco are not co-expressed in antennal neurons in female An. gambiae antennae.
(A–D) Double in situ hybridization using antisense RNA probes showing transcript localizations for the co-receptors AgIr76b (green) and AgOrco (red) are localized within distinct cells within adult female antennae. Yellow arrowheads indicate areas of overlap between green and red signals, due to spatial proximity of distinct cells. (E–H) In situ hybridization using an AgIr76b antisense RNA probe (green) plus immunohistochemical localization of AgOrco using an anti-Orco antibody (red). Scale bars ~50 μm.
While AgIr76b-positive neurons appeared to subtend (Fig. 2A) sensilla that do not express AgOrco, our studies are unable to precisely and unambiguously distinguish their exact sensillar localizations. Nevertheless, while it is possible that AgIr76b may be expressed in ORNs associated with other classes of sensilla, these data are consistent with a model in which AgIr76b is expressed in a distinctset of sensory neurons from the AgOrco-expressing ORNs, which we suggest should be denoted as Ionotropic Receptor Neurons (IRNs).
Functional Characterization of AgIrs in heterologous expression assays
In order to examine AgIr odor coding, we utilized oocytes to heterologously express functional OR and other chemoreceptor complexes. While the majority of AgIr cRNA combinations did not elicit oocyte activation against a panel of over 400 unitary odorants encompassing a broad survey of chemical classes (Supplemental Table S2), two receptors within the AgIR41a clade (Fig. 3) responded to a narrow range of compounds as described below. Coinjections of AgIr41a plus the candidate co-receptors AgIr25a and AgIr76b (hereafter AgIR41a complex) into oocytes conferred responses to amine/imine compounds (Fig. 4A), but not to other chemical classes in our screen. The AgIR41a complex responded to a diverse set of odorants with the nitrogenous heterocyclics 2-methyl-2-thiazoline and pyrrolidine eliciting the greatest magnitude inward currents (Fig. 4A). Control oocytes co-injected with cRNAs encoding only 2 of the 3 AgIrs did not respond to any of the tested compounds (Supplemental Figure S2). These data demonstrate that co-expression of AgIrs in oocytes is sufficient to confer sensitivity to amine/imine compounds and moreover, that AgIr25a and AgIr76b act in this instance as obligate dual co-receptors for AgIr41a responsiveness. Taken together, these data support the hypothesis that AgIrs function in a mechanistically similar manner to DmIrs. Importantly, DmIr41a, the Drosophila homolog of AgIr41a, confers sensitivity to 1,4 diaminobutane (putrescine) in ac2 coeloconic sensilla13. Moreover, DmIr41a and DmIr76b are required for behavioral attraction of fruitflies to putrescine and cadaverine32.
(A) Pairwise amino acid percent identity matrix. (B) Cladograms based on sequence alignments of all AgIRs (circular cladogram) with AgIR41 clade (yellow circle) shown using the N-Methyl-D-Aspartate Receptor 1 (AgNMDAR1) as an outgroup. (C) Alignment of AgIR41a and AgIR41c amino acid sequences using the single letter code. Identical residues are indicated with black boxes over white text. Similar residues are shaded with gray boxes. Numbering indicates aligned positions, including gaps.
(A) cRNAs encoding AgIr41a or AgIr41c (co-injected with AgIr25a and AgIr76b) elicit distinct profiles of chemically-evoked inward currents. Bars indicate mean normalized responses (+/− SEM) for each compound [10−4M] compared with the maximum response to pyrrolidine. Lower case letters indicate significant differences among mean responses (41a complex, P < 0.03 for all comparisons; 41c complex, P < 0.01 for a’-b’ and a’-c’ comparisons, P < 0.07 for b’-c’ comparisons). n = 4 oocytes per trial. (B,C) Concentration-response profiles of AgIR41a (B) or AgIR41c (C) complexes in response to pyrrolidine (closed circles) plotted on log scales. EC50 values (open red circles) are shown. Each data point represents the mean+/− SD (bars). Mean EC50 values are significantly different (t-test; P < 0.001). Representative TEVC traces showing inward current responses to increasing concentrations of pyrrolidine for each AgIR complex (top). n = 4 oocytes each.
Another functional AgIr complex containing AgIr41c, a paralog of AgIr41a (Fig. 3B), plus the AgIr25a/AgIr76b coreceptors conferred sensitivity to a set of amine compounds that were distinct from those inducing responses in the AgIR41a complex (Fig. 4A). In concentration response assays, the AgIR41c complex responded with higher potency to pyrrolidine than the AgIR41a complex (Fig. 4B,C). The calculated EC50 values for pyrrolidine were 110 μM and 3.4 μM for the AgIR41a and AgIR41c complexes, respectively (Fig. 4B,C; Supplemental Table S3). Although AgIRs 41a and 41c are among a handful of paralogous receptors within the AgIR41 clade (Fig. 3B), they are quite divergent at the amino acid level, sharing about 29% identity and 45% similarity overall (Fig. 3C). These results indicate that divergent, yet structurally related AgIrs may be responsible for sensitivities to compounds within specific chemical classes. Moreover, in identifying efficacious ligand for both the AgIR41a and AgIR41c complexes, we have established positive controls for future characterization of mosquito Irs in the TEVC system.
In our screens we also identified a receptor from the AgIR75 clade (Fig. 5) that mediated responses to a second chemical compound class. Oocytes coinjected with cRNAs encoding AgIr75k and AgIr8a (hereafter the AgIR75k complex) responded with high inward current amplitudes to carboxylic acids (Fig. 6A). Oocytes failed to respond to the same compounds when oocytes were co-injected with cRNAs encoding AgIr8a plus other AgIrs (Supplemental Figure S3), including the closely related receptor, AgIr75l (Fig. 5A,C), demonstrating the specificity of the AgIR75k complex response to carboxylic acids. To further examine its functionality, oocytes expressing the AgIR75k complex were tested against a variety of carboxylic acids, among them a series of increasing carbon chain length from hexanoic (6 carbons) to undecanoic (11 carbons) acids. Of these, octanoic and nonanoic acids elicited the greatest inward currents while other structurally dissimilar carboxylic acids failed to elicit responses (Fig. 6A). A concentration response curve was generated for the AgIR75k complex in response to nonanoic acid, producing an EC50 in the low micromolar range (35 μM; Fig. 6B). Our observations with respect to AgIr responses are consistent with the hypothesis that odorant/ligand compound classes can be distinguished by their co-receptor requirements; specifically the coreceptors Ir25a/76b are necessary for amine sensing and the coreceptor Ir8a is necessary for acid sensing. This distinction has been experimentally validated in D. melanogaster10,12,13.
(A) Pairwise amino acid percent identity matrix. (B) Cladograms based on sequence alignments of all AgIRs (circular cladogram) with AgIR75 clade (green) shown using the N-Methyl-D-Aspartate Receptor 1 (AgNMDAR1) as an outgroup. (C) Alignment of AgIR75k and AgIR75l amino acid sequences using the single letter code. Identical residues are indicated with black boxes over white text. Similar residues are shaded with gray boxes. Numbering indicates aligned positions, including gaps.
(A) cRNA encoding AgIr75k (co-injected with AgIr8a) elicits chemically-evoked inward currents in response to short chain carboxylic acids. Bars indicate mean normalized responses (+/− SEM) for each compound [10−4 M] compared with the maximum response to nonanoic acid. Lower case letters indicate significant differences among mean responses (P < 0.01 for all comparisons). n = 13 oocytes per trial. (B) Concentration-response profiles of the AgIR75k complex in response to nonanoic (closed circles) plotted on a log scale. EC50 value (open red circle) is shown. Each data point represents the mean+/− SD (bars). Representative TEVC trace showing inward current responses to increasing concentrations of nonanoic acid for the AgIR75k complex (top). n = 8 oocytes each.
Discussion
We have used RNAseq to identify the widespread expression of AgIrs across adult chemosensory appendages of An. gambiae where we hypothesize they act along with AgOrs and AgGrs to foster the detection and discrimination of semiochemicals. Interestingly, a fine-scale expression analysis employing FISH and IHC on the adult female antennae of the obligate Orco co-receptor and AgIr76b, one of the three potential AgIr co-receptors, does not support IR/OR co-localization but rather suggests cell-autonomous pathways. Indeed, the presence of multiple chemosensory receptor systems acting throughout the mosquito lifecycle, but operating via discrete networks of sensory neurons, is similar to observations made in D. melanogaster and other insects13,33,34. Either finding would be novel compared to Ir expression patterns in D. melanogaster9 and suggest potential functional distinctions between fruit flies and mosquitoes. Conversely, novel AgIr expression could indicate that these genes perform as yet uncharacterized roles as sensors of endogenous chemical cues such as biogenic amines or neuropeptides that are thought to modulate mosquito odor responses35,36,37,38.
We expected that functional AgIr complexes would respond to human kairomones that are known to elicit grooved peg responses, but that have not been correlated with AgOr sensitivity. This would further support the hypothesis that AgIrs underlie functionally distinct chemosensory transduction pathways that are specifically used to locate human hosts for blood meal acquisition by female An. gambiae. By extension, AgIr homologs are likely to underlie similar response pathways in other mosquito species. Such a correlation would be a significant step forward in both enhancing our understanding of the genetic basis of anthropophagy in this organism and would also identify an important new set of molecular targets for the development of novel chemical agents that could interfere with host seeking mosquito behavior, especially to host preference. It is possible that one or more such compounds will become the next generation of personal protective repellents, or surveillance trap-enhancing attractants that can be utilized in integrated vector management programs to help reduce mosquito-borne disease transmission.
In this study, we have defined and functionally characterized the first three AgIr complexes using the oocyte heterologous expression system and two-electrode voltage clamp electrophysiology. The saturated heterocyclic amine pyrrolidine, with a characteristic “spermous” odor39 is a ligand for both the AgIR41a and AgIR41c complexes eliciting robust responses, albeit with differing potencies. This apparent redundancy in sensitivity suggests that either that pyrrolidine may be combinatorially encoded in vivo or that structurally related compound(s) elicit more potent responses in one or both complexes. It is noteworthy that gravid Ae. aegypti females have recently been shown to be attracted to the polyamines putrescine and cadverine in oviposition assays32 supporting a potential role for IRs in mosquito oviposition site selection.
The stimuli that activate the AgIR75k complex are of particular interest as carboxylic acid-containing compounds are a major component of human sweat22,23,24,25 and are likely to be vitally important for female host seeking40. The AgIR75k complex may therefore provide a molecular basis for the host-seeking behavior of this mosquito species. The novel functional data for AgIrs reported and together with a previous study defining a role for AgIr76b in mediating larval responses to butylamine16 further supports the importance this highly conserved gene family in mosquito chemoreception. Interestingly, butylamine did not induce responses in either the AgIR41a or AgIR41c complexes, implying that another AgIr complex is responsible for this sensitivity in larvae.
The ongoing functional characterization of a second major family of chemoreceptors in An. gambiae is an important step forward in our understanding of chemosensation in an insect that continues to have a dramatically negative impact on human health. Of particular significance in this work is the identification of receptor complexes that likely underlie mosquito sensitivities to some of the most important chemical classes that are known to elicit host-seeking behavior. For example, ammonia and lactic acid act synergistically to increase the attractiveness of either odor blends or live hosts to An. gambiae and other vector mosquitoes26,27,41,42,43,44,45,46,47,48,49,50,51,52,53,54. It is noteworthy that DmIrs have been implicated in ammonia and acid sensing in D. melanogaster9,12,13,55. Although we may indeed find that the general principles of Ir function are conserved, the potential differences, subtle or pronounced, would be of interest to many different research communities including evolutionary biology, medical entomology/vector biology, and neurobiology, among others.
Molecular approaches have been successfully used to identify compounds that hyperstimulate ORNs in mosquitoes, thus providing a basis for the development of next generation repellents56,57,58. Indoor residual spraying (IRS) and insecticide treated bednets (ITNs) are the frontline vector control strategies adopted by many of the world’s most malaria endemic countries59,60. The potential negative impacts on human health and residual environmental hazards notwithstanding, insecticides have historically lost their effectiveness as mosquitoes have developed resistance to their toxicities through multiple mechanisms. Currently, pyrethroids are the only class of insecticides in use in many countries, exacerbating the potentially devastating effects of resistance. Thus, the development of alternative vector control methods may become critical components of integrated insect management programs, especially as outbreaks of arboviral diseases such as West Nile, Chikungunya and Zika become increasingly more widespread whilst malaria continues to burden much of the world’s population.
Methods
Statement of Research Ethics
All research was conducted in accordance with the NIH Guidelines for Research Involving Recombinant or Synthetic Nucleic Acid Molecules and the Vanderbilt University Environmental Health and Safety policies. Lab containment practices, equipment and facilities were reviewed and approved by the Vanderbilt Institutional Biosafety Committee (IBC).
Mosquitoes and mosquito rearing
An. coluzzii (SUA 2La/2La) originating from Suakoko, Liberia, previously known as Anopheles gambiae s.s. M form3, was maintained at 27 degrees centigrade, 75% relative humidity under a 12:12 light– dark cycle in the Vanderbilt Insectary as previously described61. As noted in the text, while acknowledging and indeed embracing the recent change in the species nomenclature for An. coluzzii3, we have elected to nevertheless maintain the use of An. gambiae throughout this report in order to maintain consistency with the previously published literature as well as with pre-existing gene annotations.
RNA sequencing and Transcript Abundances
Quantification of AgIr transcripts for adult female antennae and maxillary palps were derived from previously published data28. mRNA isolation and cDNA library preparation were carried out using the Illumina mRNA sequencing kit (Illumina Inc.; San Diego, CA). Libraries were sequenced using an Illumina HiSeq2000. Raw read files were quality checked, uniformly trimmed to account for spurious adapter incorporation (5′end) and for sequencing reaction degeneration (3′-end) and mapped to the An. gambiae genome (http://www.VectorBase.org). Mapping was carried out using seqmap software, configured to allow for a maximum of three mismatches per read. Processed mapping data was then consolidated based upon AGAP number and the results summarized by rseq software. Total weighted reads and AGAP transcript lengths were used to calculate a normalized transcript abundance level in units of Reads Per Kilobase per Million reads mapped (RPKMs)62.
Amino Acid Alignments AgIr Phylogenic Reconstructions
AgIr amino acid sequences were aligned using MUltiple Sequence Comparison by Log-Expectation (MUSCLE63). Phylogenetic trees were constructed using the neighbor-joining method without distance correction64.
Fluorescence in Situ Hybridization with Immunohistochemistry
Full length or partial AgIr clones were generated from cDNA prepared from female An. gambiae appendages via PCR-amplification (Invitrogen, Inc.) and subsequently subcloned TOPO-TA dual promoter vector (Invitrogen, Inc.). Digoxigenin (DIG) or fluorescein (FITC)-labeled fluorescence in situ hybridization (FISH) riboprobes were transcribed from linearized plasmids containing AgIr gene fragments using an RNA labelling mix (Roche Life science, IN, USA) and T7/SP6 RNA polymerase (New England Biolabs, Inc.) under the manufacture’s recommended protocols. Antisense experimental, and sense control, probes were conducted in parallel.
Single riboprobe and antibody studies were performed as described previously65 with the following modifications. Briefly, the antennae were dissected from 4–6 days old female heads and transferred to PBS-T solution immediately (4% paraformaldehyde in 1X RNase-free PBS, 0.1% Triton X-100). Antennae were cryosectioned at 8.0–10.0 μm and collected on Superfrost Plus slides. After fixation with 4% paraformaldehyde in 1X RNase-free PBS for 10 minutes at room temperature, the slides were washed 3 times at room temperature for 2 minutes each in 1X RNase-free PBS, acetylated for 10 minutes (0.1% triethanolamine, 1/400X volume acetic anhydride) at room temperature, and washed in 1X RNase-free PBS at room temperature. Slides were placed in prehybridization solution (50% deionized formamide, 5X SSC, 50 μg/mL heparin sodium salt, 0.1% tween-20) in a 65 °C 5X SSC humidified chamber for 1–2 hours. 40–100 ng antisense or sense riboprobes were diluted into approximately 150 μL of prehybridization solution and heated to 90 °C for 5 minutes, then directly placed on ice. The riboprobes were added to each corresponding slide and a glass coverslip was placed in order to prevent evaporation during hybridization. Slides were placed in a 65 °C 5X SSC humidified chamber for 18–24 hours. Following hybridization, each slide was washed in 5X SSC at 65 °C to facilitate removal of coverslips, followed by a triplicate series of 20-minute high-stringency washes in 0.2X SSC at 65 °C. Slides were washed in TN buffer (0.1% Tris-HCl, 0.15% NaCl) at room temperature for 10 minutes and TNB buffer [0.1% Tris-HCl, 0.15% NaCl, 0.5% Blocking reagent (Perkin Elmer, USA)] for 30–60 min at room temperature.
Following riboprobe hybridization and washing, slides were incubated with 1:500 anti-Digoxigenin-POD (Roche life science, IN, USA) and 1:500 anti-Orco antibody30 diluted in TNB buffer in a humidified chamber overnight at 4 °C and then washed 3 times in TNT buffer (0.1% Tris-HCl, 0.15% NaCl, 0.05% tween-20) for 5 minutes each at room temperature. Subsequently, secondary anti-rabbit cyanine-3 coupled antisera was diluted 1:500 in TNT buffer (0.1% Tris-HCl, 0.15% NaCl, 0.05% Tween-20) and applied for 1 hour at room temperature, then washed 3 times in TNT buffer for 5–10 minutes each with light agitation. Riboprobe signals were developed using tyramide signal amplification (TSA) reagents (PerkinElmer, USA) according to the manufacturer’s instructions for approximately 10 minutes at room temperature in the dark and washed 3 times in TNT buffer for 5–10 minutes each with light agitation. Slides were mounted with Vectashield (Vector Labs, USA) and visualized using either an Olympus BX-60 fluorescence microscope or a Zeiss LSM 510 META Inverted Confocal microscope.
The protocol for double FISH was unchanged with the exception of the inclusion of an additional set of antisense and sense probes to the hybridization mixtures. After riboprobe hybridization and stringency washes as detailed above, slides were incubated with 1:500 anti-Digoxigenin-Peroxidase (POD) (Roche Life science, IN, USA) diluted in TNB buffer in a humidified chamber for 1–3 hours at room temperature and then washed 3 times in TNT buffer with light agitation for 5 minutes each at room temperature. Anti-Digoxigenin-POD signal was developed using Tyramide Signal Amplification (TSA) reagents (Perkin Elmer, USA) according to the manufacturer’s instructions for approximately 10 minutes at room temperature in the dark, then washed 3 times in TNT buffer for 5–10 minutes each with light agitation. The POD enzyme was quenched by placing slides in a 3% H2O2 solution diluted in TNT for 30 minutes at room temperature. The slides were washed 3 times in TNT buffer for 5–10 minutes each with light agitation, placed in TNB for 30 minutes at room temperature and thereafter incubated with anti-fluorescein-POD diluted 1:500 in TNB buffer in a humidified chamber for 1–3 hours at room temperature followed by washing 3 times in TNT buffer with light agitation for 5 minutes each at room temperature. Anti-fluorescein-POD signal was developed using TSA reagents (Perkin Elmer, USA) according to the manufacturer’s instructions for approximately 10 minutes at room temperature in the dark, then washed 3 times in TNT buffer for 5–10 minutes each with light agitation. Slides were mounted with Vectashield and visualized using either an Olympus BX-60 fluorescence microscope or a Zeiss LSM 510 META Inverted Confocal microscope.
cDNA Plasmid Construction and cRNA synthesis
Full-length coding sequences of AgIr cDNAs, based on current gene annotations16, were amplified by RT-PCR from adult An. gambiae antennae. Primers specific for the coding sequences of the AgIrs used were constructed using the predicted sequences as described16 (Supplemental Table S4). The Gateway® cloning system (Invitrogen, Inc.) was used for all AgIr cloning and plasmids were verified by sequencing prior to subcloning into the destination vector, pSP64T, for in vitro transcription and heterologous expression in Xenopus oocytes. In vitro synthesis of 5′ capped complementary RNAs (cRNAs) for AgIr constructs were carried out using the mMESSAGE mMACHINE in vitro expression kits according to the manufacturer’s protocol (Ambion, Inc.).
Xenopus laevis Oocyte Retrieval, Microinjection, and Incubation
All Xenopus laevis protocols were carried out with the approval of the Vanderbilt University Institutional Animal Care and Use Committee. Expression and functional characterization of AgIr complexes were conducted essentially as described for AgOr proteins4. X. laevis ovarian lobes were extracted from fully mature females via laparotomy. An incision less than 1 cm was made ventrally, lateral of the body midline, and along the ovarian wall. Ovarian lobes were extracted and placed in a washing buffer (96 mM NaCl, 2 mM KCl, 5 mM MgCl2, and 5 mM HEPES, pH 7.6) supplemented with 0.1% gentamycin. The ovarian lobes were gently separated via forceps and subsequently treated with 2 mg/mL collagenase S-1 in washing buffer for 1.5 hours at room temperature while on a laboratory shaker. Individual oocytes were incubated in ND96 buffer (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1 mM CaCl2, and 5 mM HEPES, pH 7.5) supplemented with 5% dialyzed horse serum, 50 μg/mL tetracycline, 100 μg/mL streptomycin, and 550 μg/mL sodium pyruvate at 18 °C until microinjection. Each oocyte was injected with 27.6 nL of an AgIr cRNA mixture containing a tuning AgIr and one or more potential co-receptor subunits (i.e. AgIrs 25a + 76b or AgIr8a). After injection, oocytes were incubated separately at 18 °C in supplemented ND96 buffer. Oocytes were allowed to incubate for 3 to 5 days until assayed by two-electrode voltage clamp electrophysiology.
Two-Electrode Voltage Clamp (TEVC) Electrophysiology
Whole-cell currents were recorded from injected oocytes using the OC-725C feedback amplifier (Warner Instruments). This amplifier was connected to a computer interface through the Axon Digidata 1440 A Digitizer; data acquisition and analysis were carried out using pCLAMP 10 software (Axon Instruments). The current-injecting and voltage-sensing electrodes were made of a Ag/AgCl wire housed in a pulled borosilicate glass capillary containing 3 M KCl. Each oocyte was clamped at a holding potential of −80 mV while being perfused with buffer.
Chemical Stimuli
Approximately 400 unitary compounds of the highest commercial grade were tested in this study (Table S1). One molar (1 M) stocks of each compounds were prepared in DMSO and subsequently diluted in ND96. For initial compound screens, mixtures of 10–12 compounds of the same functional class were prepared at 1 × 10−4M each and pipetted in the TEVC perfusion system. Compounds from mixtures that elicited oocyte responses were tested individually to identify ligand-receptor pairings. A minimum threshold of 50 nA was used as a cutoff for positive responses to screened compounds, roughly equal to the noise level in the system. For the most efficacious compounds, concentration response profiles were generated using multiple oocytes.
Statistical Treatment of Data
Descriptive statistics, including arithmetic means, standard deviations and standard errors of the mean were calculated using Microsoft Excel. The R statistical package (www.r-project.org) was used for subsequent analyses, except where otherwise indicated. Oocyte inward currents were normalized by dividing individual responses by the maximum response per trial, followed by calculating the arithmetic mean of normalized responses for each compound. Assumptions of normality and equal variance were confirmed with the Shapiro-Wilk and F-tests, respectively, and arithmetic means for each treatment in the oocyte TEVC functional assays were analyzed for statistical significance by single-factor ANOVA; significant results were followed up with Tukey-Kramer post-tests to distinguish among group means. Concentration response curves and EC50 values were calculated using Prism 4 (GraphPad Software, Inc.). The Student’s t-test was applied to mean EC50 values for AgIr41a and AgIr41c pyrrolidine responses to evaluate statistical significance.
Additional Information
How to cite this article: Pitts, R. J. et al. Variant Ionotropic Receptors in the Malaria Vector Mosquito Anopheles gambiae Tuned to Amines and Carboxylic Acids. Sci. Rep. 7, 40297; doi: 10.1038/srep40297 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Tropical Infectious Diseases. 1 & 2 (Elsevier Publishing, 2006).
Takken, W. & Verhulst, N. O. Host Preferences of Blood-Feeding Mosquitoes. Annu Rev Entomol 58, 433–53 (2011).
Coetzee, M., Hunt, R. H. & Wilkerson, R. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619, 246–274 (2013).
Wang, G., Carey, A. F., Carlson, J. R. & Zwiebel, L. J. Molecular basis of odor coding in the malaria vector mosquito Anopheles gambiae . Proceedings of the National Academy of Sciences 107, 4418–4423 (2010).
Carey, A. F., Wang, G., Su, C.-Y., Zwiebel, L. J. & Carlson, J. R. Odorant reception in the malaria mosquito Anopheles gambiae . Nature 464, 66–71 (2010).
Suh, E., Bohbot, J. D. & Zwiebel, L. J. Peripheral olfactory signaling in insects. Current Opinion in Insect Science 6, 86–92 (2014).
Lu, T. et al. Odor Coding in the Maxillary Palp of the Malaria Vector Mosquito Anopheles gambiae . Curr Biol 17, 1533–1544 (2007).
Freeman, E. G., Wisotsky, Z. & Dahanukar, A. Detection of sweet tastants by a conserved group of insect gustatory receptors. Proceedings of the National Academy of Sciences 111, 1598–1603 (2014).
Benton, R., Vannice, K., Gomezdiaz, C. & Vosshall, L. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 136, 149–162 (2009).
Abuin, L. et al. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69, 44–60 (2011).
Croset, V., Rytz, R., Cummins, S., Budd, A. & Brawand, D. Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. PLoS Genet 6, e1001064 (2010).
Ai, M. et al. Acid sensing by the Drosophila olfactory system. Nature 468, 691–695 (2010).
Silbering, A. F. et al. Complementary Function and Integrated Wiring of the Evolutionarily Distinct Drosophila Olfactory Subsystems. Journal of Neuroscience 31, 13357–13375 (2011).
Chen, C. et al. Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520 (2015).
Ni, L. et al. The Ionotropic Receptors IR21a and IR25a mediate cool sensing in Drosophila. eLife 5, e13254 (2016).
Liu, C. et al. Distinct Olfactory Signaling Mechanisms in the Malaria Vector Mosquito Anopheles gambiae . PLoS Biol 8 (2010).
Qiu, Y. T., van Loon, J. J. A., Takken, W., Meijerink, J. & Smid, H. M. Olfactory Coding in Antennal Neurons of the Malaria Mosquito, Anopheles gambiae . Chem Senses 31, 845–863 (2006).
Meijerink, J. & van Loon, J. Sensitivities of antennal olfactory neurons of the malaria mosquito, Anopheles gambiae, to carboxylic acids. J Insect Physiol 45, 365–373 (1999).
Meijerink, J., Braks, M. A. & Van Loon, J. J. Olfactory receptors on the antennae of the malaria mosquito Anopheles gambiae are sensitive to ammonia and other sweat-borne components. J Insect Physiol 47, 455–464 (2001).
Meijerink, J. et al. Identification of Olfactory Stimulants for Anopheles gambiae from Human Sweat Samples. J Chem Ecol 26, 1367–82 (2000).
Kwon H. W., Lu T., Rützler M. & Zwiebel L. J. Olfactory responses in a gustatory organ of the malaria vector mosquito Anopheles gambiae . Proc Natl Acad Sci USA 103, 13526–31 (2006).
Cork, A. & Park, K. C. Identification of electrophysiologically-active compounds for the malaria mosquito, Anopheles gambiae, in human sweat extracts. Med Vet Entomol 10, 269–276 (1996).
Bernier, U. R., Booth, M. M. & Yost, R. A. Analysis of Human Skin Emanations by Gas Chromatography/Mass Spectrometry. 1. Thermal Desorption of Attractants for the Yellow Fever Mosquito (Aedesaegypti) from Handled Glass Beads. Anal. Chem. 71, 1–7 (1999).
Bernier, U. R., Kline, D. L., Barnard, D. R., Schreck, C. E. & Yost, R. A. Analysis of Human Skin Emanations by Gas Chromatography/Mass Spectrometry. 2. Identification of Volatile Compounds That Are Candidate Attractants for the Yellow Fever Mosquito (Aedesaegypti). Anal. Chem. 72, 747–756 (2000).
Bernier, U. R. et al. Synergistic attraction of Aedes aegypti (L.) to binary blends of L-lactic acid and acetone, dichloromethane, or dimethyl disulfide. J Med Entomol 40, 653–656 (2003).
Smallegange, R. C., Qiu, Y. T., van Loon, J. J. A. & Takken, W. Synergism between ammonia, lactic acid and carboxylic acids as kairomones in the host-seeking behaviour of the malaria mosquito Anopheles gambiae sensu stricto (Diptera: Culicidae). Chem Senses 30, 145–152 (2005).
Smallegange, R. C., Verhulst, N. O. & Takken, W. Sweaty skin: an invitation to bite? Trends Parasitol 27, 143–8 (2011).
Pitts, R. J., Rinker, D. C., Jones, P. L., Rokas, A. & Zwiebel, L. J. Transcriptome profiling of chemosensory appendages in the malaria vector Anopheles gambiae reveals tissue- and sex-specific signatures of odor coding. BMC Genomics 12, 271 (2011).
Pitts, R. J., Fox, A. N. & Zwiebel, L. J. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae . Proc Natl Acad Sci USA 101, 5058–5063 (2004).
Benton, R., Sachse, S., Michnick, S. W. & Vosshall, L. B. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo . PLoS Biol 4, e20 (2006).
Pitts, R. J. & Zwiebel, L. J. Antennal sensilla of two female anopheline sibling species with differing host ranges. Malar J 5, 26 (2006).
Hussain A. et al. Ionotropic Chemosensory Receptors Mediate the Taste and Smell of Polyamines. PLoS Biol 14, e1002454 (2016).
Silbering, A. F. & Benton, R. Ionotropic and metabotropic mechanisms in chemoreception:‘chance or design’? EMBO Rep 11, 173–79 (2010).
Missbach, C. et al. Evolution of insect olfactory receptors. eLife 3, e02115–e02115 (2014).
Siju, K., Hansson, B. & Ignell, R. Immunocytochemical localization of serotonin in the central and peripheral chemosensory system of mosquitoes. Arthropod Structure and Development 37, 248–259 (2008).
Siju, K. P., Bräcker, L. B. & Grunwald Kadow, I. C. Neural mechanisms of context-dependent processing of CO 2avoidance behavior in fruit flies. Fly (Austin) 8, 68–74 (2014).
Siju, K. P. Neuromodulation in the chemosensory system of mosquitoes-neuroanatomy and physiology. Doctoral Thesis: Swedish University of Agricultural Sciences (2009).
Siju, K. P. et al. Neuropeptides in the antennal lobe of the yellow fever mosquito, Aedes aegypti. J Comp Neurol 522, 592–608 (2013).
Amoore, J. E., Forrester, L. J. & Buttery, R. G. Specific anosmia to 1-pyrroline: The spermous primary odor. J Chem Ecol 1, 299–310 (1975).
Smallegange, R. C., Qiu, Y. T., Bukovinszkiné-Kiss, G., Loon, J. J. A. & Takken, W. The Effect of Aliphatic Carboxylic Acids on Olfaction-Based Host-Seeking of the Malaria Mosquito Anopheles gambiae sensu stricto. J Chem Ecol 35, 933–943 (2009).
Jawara, M. et al. Optimizing odor-baited trap methods for collecting mosquitoes during the malaria season in The Gambia. PLoS ONE 4, e8167 (2009).
Jawara, M. et al. Field Testing of Different Chemical Combinations as Odour Baits for Trapping Wild Mosquitoes in The Gambia. PLoS ONE 6, e19676 (2011).
Qiu, Y. T., Smallegange, R. C., van Loon, J. J. A. & Takken, W. Behavioural responses of Anopheles gambiae sensu stricto to components of human breath, sweat and urine depend on mixture composition and concentration. Med Vet Entomol 25, 247–55 (2011).
Allan, S. A., Bernier, U. R. & Kline, D. L. Laboratory evaluation of lactic acid on attraction of Culex spp. (Diptera: Culicidae). J Vector Ecol 35, 318–324 (2010).
Smallegange, R. C., Knols, B. G. J. & Takken, W. Effectiveness of Synthetic Versus Natural Human Volatiles as Attractants for Anopheles gambiae (Diptera: Culicidae) Sensu Stricto. J Med Entomol 47, 338–344 (2010).
Okumu, F. O. et al. Development and field evaluation of a synthetic mosquito lure that is more attractive than humans. PLoS ONE 5, e8951 (2010).
Bernier, U. R., Kline, D. L., Allan, S. A. & Barnard, D. R. Laboratory comparison of Aedes aegypti attraction to human odors and to synthetic human odor compounds and blends. J Am Mosquito Contr 23, 288–293 (2007).
Qiu, Y. T. et al. Attractiveness of MM-X Traps Baited with Human or Synthetic Odor to Mosquitoes (Diptera: Culicidae) in The Gambia. J Med Entomol 44, 970–983 (2013).
Williams, C. R. et al. Laboratory and field assessment of some kairomone blends for host-seeking Aedes aegypti. J Am Mosquito Contr 22, 641–647 (2006).
Dekker, T., Steib, B., Carde, R. & Geier, M. L-lactic acid: a human-signifying host cue for the anthropophilic mosquito Anopheles gambiae . Med Vet Entomol 16, 91–98 (2002).
Murphy, M., Dunton, R., Perich, M. & Rowley, W. Attraction of Anopheles (Diptera: culicidae) to volatile chemicals in Western Kenya. J Med Entomol 38, 242–244 (2001).
Steib, B. M., Geier, M. & Boeckh, J. The effect of lactic acid on odour-related host preference of yellow fever mosquitoes. Chem Senses 26, 523–528 (2001).
Geier, M., Bosch, O. J. & Boeckh, J. Ammonia as an attractive component of host odour for the yellow fever mosquito, Aedes aegypti . Chem Senses (1999).
Kline, D. L., Takken, W., Wood, J. R. & Carlson, D. A. Field studies on the potential of butanone, carbon dioxide, honey extract, 1-octen-3-ol, L-lactic acid and phenols as attractants for mosquitoes. Med Vet Entomol 4, 383–391 (1990).
Silbering, A. F. et al. Ir40a neurons are not DEET detectors. Nature 534, E5–E7 (2015).
Jones, P. L., Pask, G. M., Rinker, D. C. & Zwiebel, L. J. Functional agonism of insect odorant receptor ion channels. Proceedings of the National Academy of Sciences 108, 8821–8825 (2011).
Turner, S. L. et al. Ultra-prolonged activation of CO2-sensing neurons disorients mosquitoes. Nature 474, 87–91 (2011).
Pask, G. M., Romaine, I. M. & Zwiebel, L. J. The Molecular Receptive Range of a Lactone Receptor in Anopheles gambiae . Chem Senses 38, 19–25 (2012).
The malERA Consultative Group on Vector Control. A Research Agenda for Malaria Eradication: Vector Control. PLoS Med 8, e1000401 (2011).
World Health Organization. 1–242 (2014).
Suh, E., Choe, D. H., Saveer, A. M. & Zwiebel, L. J. Suboptimal Larval Habitats Modulate Oviposition of the Malaria Vector Mosquito Anopheles coluzzii . PLoS One 11, e0149800 (2016).
Mortazavi, A., Williams, B. A., Mccue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Meth 5, 621–628 (2008).
Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nuc Acids Res 32, 1792–7 (2004).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4, 406 (1987).
Pinaud, R., Mello, C. V., Velho, T. A., Wynne, R. D. & Tremere, L. A. Detection of two mRNA species at single-cell resolution by double-fluorescence in situ hybridization. Nature Protoc 3, 1370–1379 (2008).
Acknowledgements
We thank Zhen Li for mosquito rearing, oocyte preparation and injections. Access to a Leica CM1900 Cryostat was generously provided by the laboratory of Dr. James Patton (Vanderbilt University). We acknowledge The Vanderbilt Cell Imaging Shared Resource for training and use of the LSM 510 confocal microscope and the Vanderbilt Mass Spectrometry Research Center for compound analysis. This work was conducted with the support of Vanderbilt University and a grant from the National Institutes of Health (NIAID, AI056402) to LJZ.
Author information
Authors and Affiliations
Contributions
Conceived the experiments: R.J.P., S.L.D. and L.J.Z. Conducted the experiments: R.J.P., S.L.D. and Z.Z. Analyzed the results: R.J.P., S.L.D., Z.Z. and L.J.Z. Performed statistical analyses: R.J.P. Wrote the first draft of the manuscript and figures: R.J.P. Revised and edited the manuscript: R.J.P., S.L.D., Z.Z. and L.J.Z. Approved the final manuscript: R.J.P., S.L.D., Z.Z. and L.J.Z.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Pitts, R., Derryberry, S., Zhang, Z. et al. Variant Ionotropic Receptors in the Malaria Vector Mosquito Anopheles gambiae Tuned to Amines and Carboxylic Acids. Sci Rep 7, 40297 (2017). https://doi.org/10.1038/srep40297
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40297
This article is cited by
-
Olfactory sensitivity differentiates morphologically distinct worker castes in Camponotus floridanus
BMC Biology (2023)
-
Ionotropic receptors in the turnip moth Agrotis segetum respond to repellent medium-chain fatty acids
BMC Biology (2022)
-
The antennal transcriptome of Triatoma infestans reveals substantial expression changes triggered by a blood meal
BMC Genomics (2022)
-
Mosquito host-seeking diel rhythm and chemosensory gene expression is affected by age and Plasmodium stages
Scientific Reports (2022)
-
Differentiation of action mechanisms between natural and synthetic repellents through neuronal electroantennogram and proteomic in Aedes aegypti (Diptera: Culicidae)
Scientific Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.