Abstract
Approaches enabling efficient phosphorus utilization in crops are of great importance. In cereal crop like wheat, utilization of inorganic phosphate (Pi) is high and mature grains are the major sink for Pi utilization and storage. Research that addresses the importance of the Pi homeostasis in developing grains is limited. In an attempt to understand the Pi homeostasis in developing wheat grains, we identified twelve new phosphate transporters (PHT), these are phyologentically well distributed along with the members reported from Arabidopsis and rice. Enhanced expression of PHT1-subfamily genes was observed in roots subjected to the Pi starvation suggesting their active role in Pi homeostasis. Differential expression patterns of all the PHT genes during grain filling stages suggested their importance in the filial tissues. Additionally, high accumulation of Pi and total P in aleurone correlates well with the expression of TaPHTs and other phosphate starvation related genes. Tissue specific transcript accumulation of TaPHT1.1, TaPHT1.2, TaPHT1.4 in aleurone; TaPHT3.1 in embryo and TaPHT4.2 in the endosperm was observed. Furthermore, their transcript abundance was affected in low phytate wheat grains. Altogether, this study helps in expanding the knowledge and prioritize the candidate wheat Pi-transporters to modulate the Pi homeostasis in cereal grains.
Similar content being viewed by others
Introduction
Phosphorus (P) is an essential mineral nutrient for plant growth and development1. Seeds of cereal and legume crops are major “sink” for P over-accumulation which consequently results in over-consumption of total P fertilizer used worldwide2. Due to low phytoavailability of inorganic or free phosphate (Pi), in a sustainable agricultural production, it is important to consider the contribution offered by the stored seed P. In general Pi acquisition occurs through the roots of the plants and subsequently transported to the stem, leaves and other tissue. Remobilization of Pi takes place in the foliar parts of the plants and subsequently majority of it gets stored into the developing grains as phytic acid (PA)3. Therefore, any variation in Pi acquisition, distribution and redistribution during plant development will significantly alter the seed total P during cereal grain development4. Due to the absence of symplasmic linkage between maternal and filial generations, once remobilized Pi reaches seed apoplasm, subsequent Pi transport totally depends on filial tissue functions which may impact net seed total P5,6.
Pi transport from the rhizosphere to the different plant tissues, primarily involves multiple phosphate transporters (PHTs) belonging to either PHT1, PHT2, PHT3 or PHT4 sub-families7,8,9,10,11. Most of the studies till date addressed the efficient utilization of soil P and its uptake by the roots via these transporters12,13,14. The regulation of PHT1 candidates involves the Pi starvation response regulators (PHR1) through their binding to cis-element P1BS (PHR1 binding sequence), SPX-proteins and PHO2 regulation15,16,17. Studies in rice (Oryzae sativa) and Arabidopsis (Arabidopsis thaliana) have suggested the role of PHT1 family genes in remobilization of P from senescing tissues to the actively developing tissues and their role in embryo development18,19. In addition to that, Pi transport within the plant tissues is also contributed by PHT3 and PHT4 family members by regulating ‘intracellular Pi starvation’ signalling20,21. Earlier, it has been proposed that the P translocation and subsequent loading in grains is a demand-driven process22. However, researchers have to yet explore the mechanism involved for sensing the Pi loading in seed tissue during grain-filling.
In cereals during the early stages of maturation, Pi is transported in the seeds and rapidly converted to the bound form, commonly referred as PA2,23. The acidic nature of PA enables chelation of important micronutrients in form of reservoir generally referred as phytin bodies24,25. In grains, Pi is transported through pericarp, containing vascular compartment embedded in ground tissues. Subsequently, it is delivered to the tissues surrounding the developing seed such as, nucellar projection cells in wheat (Triticum aestivum) and barley (Hordeum vulgare)4. Once Pi gets deposited into the endosperm cavity (especially in case of wheat), the uptake process at the maternal/filial interface is adapted by specialized filial cells, such as “endosperm/aleurone transfer cells” or the cells of the embryo-surrounding region5.
Wheat is an agronomical important crop and a major source of nutrition in the developing countries, but it utilizes large amount of exogenous P during cultivation. P in addition to its role in the agronomic quality of grain also promotes uniform heading and faster maturity26. The acquired soil P, is vital for plant development that supports the germination of wheat seedling and vegetative growth till maturity. The high accumulation of PA in grains also suggests the presence of a controlled regulatory mechanism for Pi-PA homeostasis that could be active during seed development stages27,28. In order to develop Pi-efficient wheat, it is therefore important to understand the process that involves the Pi homeostasis and allocation in the developing grains of wheat. Despite the presence of clues related to the role of seed Pi, limited efforts were undertaken to address the same22,29. Therefore, it is important to understand the step-wise regulatory mechanism for the P accumulation in the wheat seed tissues. In this study, we identified 23 wheat PHT including 12 new members that span all the four sub-families of transporters and were further characterized for their expression in filial tissues during the grain filling stages. Expression of wheat PHTs was also studied in low phytate wheat, that suggested Pi-PA homeostasis in developing grains.
Results
Pi concentration in developing wheat seeds and tissues
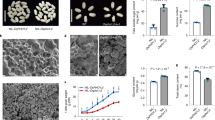
To compare the Pi accumulation in the developing grains, Pi and total P estimation was performed in the wheat grains. The concentration of total P throughout seed development remained stable with the exception for 7 DAA where enhanced accumulation was observed (Fig. 1A). Our analysis showed a slight increase in total Pi accumulation from 7 to 28 DAA of grain development (Fig. 1B). During this duration of grain filling, the concentration of Pi ranges from ~ 0.9–1.3 μmol/mg fresh weight. After 28 DAA, Pi level was slightly decreased at the maturation stage. This constant accumulation of Pi during the grain maturation reinforce that the developing grains are the sink tissue for phosphate storage. Next, Pi was analyzed in the filial tissues of 14 DAA wheat grains. Pi accumulation in aleurone tissue was 2-fold higher when compared to endosperm and embryo (Fig. 1C). The total P concentration was also 3-fold higher in aleurone in comparison with endosperm (Supplementary Fig. 1). This analysis suggests an existence of a Pi gradient in the filial tissues of developing wheat grains with over-accumulation of Pi in aleurone as compared to other tissue. These results also support our speculation for the possible role of PHTs those could be involved in the phosphate related homeostasis during grain development.
Estimation of inorganic phosphate (Pi) and total P in wheat seeds.
Measurement of Pi and total phosphorous in wheat seeds. (A) Total P content in developing seeds. (B) Pi content in developing wheat seeds mainly, 7, 14, 21, 28, 35 DAA and mature seed (MS) (C) Pi content in 14 DAA aleurone (Al), endopserm (En) and embryo (Em). Each bar represents the mean ± SE of at least three biological replicates.
In-silico identification and structural analysis of wheat PHTs
Previously, only eight wheat PHTs were reported30. To identify additional wheat PHTs, Arabidopsis (PHT1–19) and rice (PHT1-26) sequences were used as query to perform tBLASTN analysis. Subsequently, sequence alignment and unigene BLAST analysis was used to assemble 492 wheat ESTs into 23 different wheat genes (Supplementary Table 1). Their predicted intron-exon structure are represented in Supplementary Fig. 2. The nomenclature of identified 23 wheat PHTs was based on the sequence similarity to closest rice homologs. The analysis resulted in the identification of 12 additional wheat PHTs those cover all the subgroups of transporters.
Phylogenetic tree developed from the protein sequence alignments of PHT family members distributed the wheat PHTs into four families (PHT1–PHT4) like in Arabidopsis8 and Rice10. Cluster I contains 13 wheat genes (TaPHT1.1-1.13) from PHT1 family; cluster II contains previously reported TaPHT2.1 from PHT2 family; cluster III have 3 wheat genes (TaPHT3.1-3.3) of PHT3 family; and lastly cluster IV consisted of 6 wheat genes (TaPHT4.1-4.6) belonging to the PHT4 family (Fig. 2). Transport classification database (www.tcdb.org/analyze.php) revealed the presence of 4-12 transmembrane domains (TMs) on the predicted 23 TaPHT proteins (Supplementary Table 1 and Fig. 3). Similar to rice and Arabidopsis, almost all wheat PHT1 transporters having 12 TM domains clustered together (cluster I) except for TaPHT1.10 (9 TMs) and TaPHT1.12 (10 TMs; Supplementary Fig. 3). Distribution of TaPHT2.1 in cluster II containing 13 TM domains was in agreement with the previous reports9 (Supplementary Fig. 4). Whereas, the members of TaPHT3 transporters (cluster III) were found to have only 5 TM regions (Supplementary Fig. 5). Lastly, in cluster IV, TaPHT4 family members contains 12 TM domains, whereas, members from rice and Arabidopsis have 11–12 and 10–12 TM domains respectively (Supplementary Fig. 6). Our analysis suggests that the protein architecture of TaPHT sub-family members was conserved among the sub-family (Fig. 3).
Phylogenetic relationship between the identified wheat PHT proteins along with PHT proteins from Arabidopsis thaliana and Oryzae sativa.
Analysis was performed by Neighbour-joining tree analyses conducted using Mega558. Bootstrap values were calculated as a percentage of 1000 trial. The analysis involved 68 protein sequences. The GeneBank accession numbers of the protein are listed in Supplementary Table 3.
Genomic location and expression of wheat PHTs during Pi starvation
Chromosomal locations for all the TaPHT members were identified using IWGSC database (Supplementary Table 1). The wheat PHTs were distributed mainly on chromosome 4 (8 PHT genes), chromosome 5 (6 PHT genes), chromosomes 3 and 6 (3 PHT genes on each) and chromosomes 1 and 2 (2 PHT genes each) (Supplementary Table 1). In an attempt to get insight for the regulation of these wheat PHTs, the 1-kb upstream regions of all the wheat PHT genes were analyzed for the presence of cis elements. As expected, the searches revealed presence of multiple phosphate, hormone, sugar and stress responses related cis elements (Supplementary Table 2). Among these 82 putative cis-elements, 14 cis-elements grouped into Pi-related responsiveness, hormone-response, sugar-response and stress-response. Interestingly, 11 of 23 wheat PHT genes were found to contain the P1BS DNA motif (GNATATNC) on their promoter region (Supplementary Table 2). Among wheat PHT1 sub-family genes only six members have the phosphate responsive domain. The promoter of only three members of TaPHT3 sub-family, TaPHT2.1 and TaPHT4.2 contains a P1BS binding motif.
The expression pattern of wheat PHT1 subfamily was examined in roots at two different time points (15 and 20 days) of Pi starvation (Fig. 4A). qRT-PCR analysis were carried out using gene specific conserved primers targeting the conserved sequence of transcripts arising from multiple genomes (A,B or D). During our experiment no significant changes in the expression of wheat PHTs were observed in the control plants at 15 and 20 days; therefore, the fold change in expression of wheat PHTs was calculated w.r.t. 15 days control samples. Firstly, expression response of phosphate starvation response genes (PSR) was tested for the Pi starvation in roots (Fig. 4B). Results suggested high expression of PSR related genes in roots suggesting a starvation response from the plants. Next, the expression of all the PHT1 sub-family genes was studied (Fig. 4C). In roots, wheat PHT1 subfamily of genes showed a distinct expression pattern that could be divided into three types of responses. Most of the genes followed, type-I response that was characterized by an incessant increase in the gene transcript at both 15 and 20 days of starvation. This expression pattern was observed for eight wheat PHTs including TaPHT1.1, TaPHT1.2, TaPHT1.3, TaPHT1.5, TaPHT1.8, TaPHT1.11, TaPHT1.12 and TaPHT1.13. Second,type-II expression was characterized by induction at specific time point. For example, TaPHT1.6 and TaPHT1.7 were induced at 15 days, whereas, TaPHT1.9 and TaPHT1.10 were induced at 20 days after starvation. Lastly, in the type-III response, induction level of was similar at both the time points as in case of TaPHT1.4. The wheat Pi-transporters were also differentially expressed in shoots (data not shown). Summarizing the expression analysis, it was observed that members of wheat PHT1 subfamily showed transcriptional changes under Pi starvation condition.
Quantitative real-time PCR expression analysis of putative wheat phosphate transporter genes and PSR marker genes under phosphate starvation.
Expression was performed for PHT1 family (TaPHT1.1-1.13) and PSR marker genes (TaIPS1, TaPHR1, TaSPX1 and TaPHO2) under 15 days and 20 days of Pi starvation. (A) Growth of wheat plant and concentration of Pi under Pi-starvation. (B) Transcript abundance of PSR marker genes. (C) Relative expression of TaPHT1 family members under Pi-limiting condition. Each bar indicates the mean of four to five replicates with the indicated standard deviation of the mean.
Expression of wheat PHTs in developing grains and tissue
Expression of few Pi-homeostasis related genes, including PHTs was earlier reported in wheat seedlings subjected to limited Pi conditions31. Therefore, one could anticipate the presence of Pi and PA in developing grains that might influence the expression patterns of these genes. In the current study, we analyzed the expression at different stages of seed development (7, 14, 21 and 28 DAA). Our analysis suggested differential expression of these genes during grain maturation (Fig. 5). For example, at the early stage of development, i.e. 7 DAA, only TaPHT3.1 and TaPH4.2 showed significantly higher expression compared to other transporters. Similarly, at 14 DAA transcript abundance of TaPHT3.1, TaPH4.2 and TaPHT1.1 was higher compared to other transporters. At 21 DAA, multiple wheat PHTs have a high expression, including TaPHT1.4, TaPHT3.1, TaPH3.2, TaPHT4.2 and TaPHT4.4. At the 28 DAA, TaPHT1.1, TaPHT1.2, TaPHT1.4, TaPHT3.1, TaPHT3.2, TaPHT4.2 and TaPHT4.4 were highly expressed. These data suggested active expression of wheat PHTs during grain development. In addition to that, at 28 DAA, maximum number of wheat PHTs were up-regulated. The differential expression patterns of wheat PHTs and PA pathway genes32, suggested interlink between Pi transport and PA biosynthesis.
Relative transcript levels of 23 TaPHTs during seed development.
The cDNA templates were prepared from 2 μg of DNA free RNA isolated from different time point of seed maturation of 7, 14, 21 and 28 DAA. qRT-PCR expression was performed for TaPHT1.1-1.13, TaPHT2.1, TaPHT3.1-3.3 and TaPHT4.1-4.6 during grain filling stages. Each bar indicates the mean of four to five replicates with the indicated standard deviation of the mean.
To gather the evidence for the role of PHTs in the Pi-allocation within the seed compartments, highly expressed wheat PHTs (TaPHT1.1, TaPHT1.4, TaPHT1.2, TaPHT3.1, TaPHT3.2, TaPHT4.2 and TaPHT4.4) were further selected. Tissue specific expression analysis of these genes was performed in glume, rachis, aleurone, endosperm and embryo of 14 DAA seed. Result suggested specific expression patterns of wheat PHTs (Fig. 6). For instance, expression of TaPHT1.2 and TaPHT1.3 was high in aleurone tissue, whereas, TaPHT3.1 mRNA was significantly expressed in embryo and rachis. Unlike others, transcript abundance of TaPHT4.2 and TaPHT4.4 was highest in endosperm. The above expression analysis suggested the possibility of divergent function of PHTs in a tissue-specific manner.
Transcript accumulation of candidate TaPHTs in 14 DAA spike and seed tissues.
Relative expression of TaPHT1.1, TaPHT1.2, TaPHT1.4, TaPHT3.1, TaPHT3.2, TaPHT4.2 and TaPHT4.4 was analyzed in Glume (Glu), Rachis (Ra).Aleurone (Al), endosperm (En) and embryo (Em) of the 14 DAA old tissues. Each bar indicates the mean of four to five replicates with the indicated standard deviation of the mean.
Expression response of PSR related genes in wheat filial tissue
The tissue specific expression patterns and functionality of the wheat PHTs, suggested the coordinated response of these transporters. Our analysis also suggested high accumulation of Pi in aleurone tissue as compared to embryo and endosperm. Therefore, we speculate that the PSR related genes might also participate in controlling the accumulation of Pi in wheat tissue. Expression of the PSR related genes was tested in different tissues of wheat grains (Fig. 7). Expression characterization was performed for known PSR related genes, i.e., TaPHR1, TaSPX1, TaPHO2, TaPHO85 and TaIPS1. qRT-PCR revealed that the relative transcript levels of TaSPX1, TaPHO2, TaPHO85 and TaIPS1 were significantly higher in aleurone in comparison to endosperm and embryo. Surprisingly, transcript level of TaPHR1 was suppressed in the aleurone and embryo, but was significantly higher in endosperm of 14 DAA grains. Such contrasting, expression of PSR related genes in the wheat filial tissue suggested the importance of Pi-starvation signaling events and interplay of wheat PHTs in seed tissue.
Influence of seed development on transcript levels of phosphate starvation response (PSR) related genes.
The cDNA templates were prepared from 2 μg of DNA free RNA isolated from different tissues of 14 DAA wheat grain. Expression was studied for representative PSR related genes i.e. TaPHR1, TaSPX1, TaIPS1 andTaPHO2 at grain filling stage. Each bar indicates the mean of four to five replicates with the indicated standard deviation of the mean. **Indicates significant differences at p < 0.05 w.r.t. other tissue (Annova, Origin 6.1).
Lowering in phytic acid affects the expression of wheat PA biosynthesis genes and PHTs
In an attempt to gain insight for the Pi-PA homeostasis in wheat grains, we utilized the previously developed low phytate transgenic wheat33. The expression of the selected PA biosynthesis genes and Pi-transporters were performed in the 14 DAA seeds of two different low phytic acid lines (K4G3-5-1 and K1B4-2-5). Perturbation in the expression patterns of the selected genes was observed as compared to the non-transgenic seeds of C306. The transcripts of TaPHT1.1, TaPHT1.4 and TaPHT4.2 were largely affected (Fig. 8). Higher expression of these genes was observed in transgenic seeds. For the genes involved in PA biosynthesis, TaIPK1 and TaITPK2 did not show any significant difference in the expression level, whereas, TaITPK1 showed high expression in both the transgenic plants. These experiments suggest that lowering of PA selectively affects the transcript level of genes involve in Pi transport and PA biosynthesis.
Relative expression levels of selected wheat Pi-transporters (TaPHT1.1, TaPHT1.4, TaPHT3.1 and TaPHT4.2) and PA biosynthesis genes.
The cDNA templates were prepared from 2 μg of DNA free RNA isolated from different tissues of 14 DAA wheat grain of C306 (non-transgenic) and two lines of transgenic wheat with low PA (K4G3-1-5-1 and K1B4-2-5). Each bar indicates the mean of three replicates with the indicated standard deviation of the mean. **Indicates significant differences at p < 0.05 w.r.t. to C306 (Annova, Origin 6.1).
Discussion
Wheat utilizes a substantially large amount of Pi, which is stored in seeds as PA. Cereal grains are an important reservoir of P that could be exploited for plant development. Not many reports related to the tissue specific compartmentalization of Pi in wheat seed tissues have been addressed. In order to study this, we report here first comprehensive analysis of wheat PHT transporter gene family with emphasis upon the tissue specificity and drawing clues regarding their possible function. Given the close proximity with the Arabidopsis and rice homologs, we expect that the wheat PHTs should also perform similar functions. Herein, we identified seed-specific Pi transporters, those could be the potential target to modulate the influx in developing cereal grains.
For wheat, the major source of total grain P is directly influenced by Pi transport from vegetative organs34. The translocation of Pi into the developing grains fulfills the requirement for cellular function and further storing as a reservoir in the form of phytate to be utilized for nutritional requirement during germination35. Previous studies in developing rice seed suggested that total P is concomitantly accumulated during 6 to 14 DAA and remained stable till maturation, whereas concentrations of Pi remains same throughout seed development with a slight decrease towards the maturation36. In our study, total P concentration was highest at 7 DAA and after a slight decrease it remained stable till maturation. This suggests a maximum utilization of incoming Pi during the early developmental stage. In general, the concentration of Pi during wheat seed development remained invariably constant. The Pi accumulation pattern during rice grain development was also shown to be constantly low36. Therefore, one can speculate that during wheat seed development, constant lower levels of Pi till 28 DAA is maintained by continuous translocation of Pi from vegetative tissues into developing seeds. However, the source tissue for providing available Pi to rice and wheat grains during development might differ due to difference in senescence rate of vegetative tissues34. Pi is a potent inhibitor of ADP-Gluc pyrophosphorylase, a key regulatory enzyme in starch biosynthesis37,38. Thus, during seed development high starch levels in wheat endosperm must be correlated with low concentrations of Pi.
For cereal grains, limited reports are available that describe the co-relation between Pi concentrations and its subsequent movement into the filial tissues19,22. In wheat, two principal filial tissues, aleurone and endosperm are involved in a biochemical and transcriptional reprogramming during grain filling to facilitate nutrient transport39,40.The total P concentration was found to be very high in aleurone as compared to endosperm of the 14 DAA seed, the stage when aleurone is discrete (Fig. 1C and Supplementary Fig. 1). Our analysis suggested that the aleurone accumulate more Pi than endosperm. The reason of differential Pi gradient in these tissues is still unknown, however, two inferences can be more acceptable on the basis of previous studies. Firstly, at 14 DAA, favorable translocation of more Pi in aleurone is required for actively accumulating phytic acid; secondly the unfavorable translocation of Pi in the endosperm is to promote starch biosynthesis to keep ADP-Gluc pyrophosphorylase active. Therefore, this comprehensive expression analysis of TaPHTs and Pi-homeostasis related genes in these tissues may provide better insight about Pi-transport and regulation mechanism during grain filling. During our study, we observed that aleurone accumulated more Pi with high expression of TaPHT genes (especially PHT1 members) and PSR genes. Thus, basis of TaPHTs expression in aleurone is uncertain, as it accumulates more phosphorous than other tissue. Does Pi starvation responses govern the Pi transport in seed tissues similar to root and shoot? This remains an open question. High phytic acid content and expression of wheat PA pathway genes in aleurone32 are major factors for its physiological difference from roots and shoots w.r.t. Pi homeostasis. Overexpression of PA biosynthesis genes in rice resulted in an enhanced influx of P from vegetative organs into seeds41. These observations support the hypothesis for the continuous uptake of Pi that is translocated to aleurone layer, where it is rapidly utilized for the biosynthesis of PA.
Previous studies, have reported the identification of only eleven wheat PHTs (mainly, TaPT1-8; TaPHT2.1) and their expression profiles under Pi-starvation9,30,42. In this study, additional members of PHTs were included and their expression patterns were studied during seed development (mainly 7, 14, 21 and 28 DAA). To our knowledge, no homologs of PHT3 and PHT4 genes were previously reported from wheat. Recent reports provided the functional clues for multiple rice PHTs, suggesting that certain Pi-transporter (OsPT1, OsPT8, OsPT14 and OsPT18) modulate the Pi transport within the seed tissues22. Based on these evidences and our results, the high expression response of TaPHT1.1, TaPHT1.2, TaPHT1.4, TaPHT3.1, TaPHT3.2, TaPHT4.2 and TaPHT4.4 suggests the importance of wheat transporters in the specific seed tissue (Fig. 6). Knockdown of OsPT8, a rice phosphate transporter; restricted the Pi allocation into embryo and affected the total P concentrations43. These observations also reinforce the possibility to target specific PHTs for gene silencing to modulate Pi balance in wheat. Analysis of aleurone and endosperm transcriptome data from previous reports39 showed several fold induction of PHT genes belonging primarily to PHT1 and PHT3 family. This supports our observations about the high abundance of Pi transporters in aleurone. Membrane-related transport activity has been found to be important in the wheat seed development specially near crease region and nucellar projections after phloem unloading44,45. In-situ PCR for TaPHT1.1 transcript near chalazal region show importance of this tissue interface in transport of Pi within filial tissues (Supplementary Fig. 7). In our study, high transcript abundance of TaPHT1.1, TaPHT1.2, TaPHT1.4 and TaPHT3.2 was observed in aleurone (including crease region). Thus, it seems that the increased expression of multiple PHTs, might be as a consequence of enhanced accumulation of Pi in aleurone tissue. During grain development, accumulated Pi gets utilized for the synthesisof PA through a series of enzymes like inositol phosphate kinases in a lipid dependent or independent manner3,32. Therefore, maintaining the flux of Pi for phosphorylation series of myo-inositol moieties may be one of the probable roles of PHT members in the aleurone.
Endosperm being a storage tissue involved actively in starch synthesis46. Pi, PPi, ATP and ADP tightly regulate both starch synthesis and degradation in wheat endosperm cell47,48. In this study, we found the expression of PHT4.2 and PHT3.1 in wheat endosperm and embryo, respectively. Thus, we could hypothesize that the low Pi concentration regulated by PHTs plays important role in regulating starch biosynthesis in the endosperm. Although not conclusive, further experiments are required to confirm this hypothesis. Similarly, wheat PHT3 could have an involvement in the stress-related or other metabolic related functions in the embryo49.
lpa mutants have provided few clues for the Pi accumulation in grains. However, the link between inositol signalling pathway and Pi transport in seed tissues is still unknown4. Our results suggest that lowering of PA in wheat grains, affects the expression of PA biosynthesis and Pi-transporter related genes in wheat (Fig. 8). Previous studies on wheat has shown that lowering of PA eventually decreases the total P in the aleurone with increased total P in the inner endosperm50. In our case, the changes in the expression of wheat PHTs in lpa wheat grains could be as a result of varying P fractions forms in the grain tissues. These observations were supported from the recent studies that showed the involvement of genes related to Pi signalling; sulfur metabolism and sulphate transport; and in PA metabolism51. Therefore, the co-relation between Pi-PA homeostasis is of high importance in developing cereal grains.
The present study provides the comprehensive genomic analysis and spatio-temporal expression characterization of wheat PHTs and further explored their possible role in the filial tissue. The identified Pi-transporters could therefore be a potential candidate for gene targeting to modulate the gradient of Pi and biosynthesis of PA in seeds. Our data also contribute in laying the foundation towards understanding the molecular basis of Pi homeostasis in wheat developing grains. Overall, this study substantiates the role of Pi-transporters in uptake and regulating the optimal balance of Pi and thereby participates in controlling the Pi-PA homeostasis in wheat seed compartments.
Materials and Methods
Identification of PHT genes in wheat genome and sequence analysis
The in-silico identification of PHT genes from wheat was adapted from the approach reported previously52. The protein sequences of PHT family genes (PHT1, 2, 3 and 4) from Arabidospsis and rice (Supplementary Table 3) were queried against wheat expressed sequence tags (EST) database in tBLASTn program at NCBI (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi). All the different EST accessions were queried in Unigene (http://www.ncbi.nlm.nih.gov/unigene) for assigning unique Unigene ID to particular set of ESTs. These ESTs were mapped individually on cereal data base (DB) (http://www.cerealsdb.uk.net/cerealgenomics/CerealsDB/search_reads.php), ensemble plants (http://plants.ensembl.org/common/Tools/Blast?db=core) and the International Wheat Genome Sequencing Consortium (IWGSC) sequence databases (http://wheat-urgi.versailles.inra.fr/Seq-Repository/BLAST) to obtain genome contigs for each EST. The IWGSC genome contig sequences were used to obtain CDS/transcript sequence using FGENESH annotation program. The transcript sequences were submitted to Pfam database for respective conserved domain corresponding to PHT1 (Pi:H+ symporter), PHT2 (Pi:H+ symporter), PHT3 (Pi:H+ symporter or Pi/OH- antiporter) and PHT4(Na+-dependent Pi cotransporter) family genes. For wheat PHTs identified with incomplete coding sequence, complete CDS and genomic structure were derived by genome assemblies using cereal DB, ensemble plants and IWGSC survey sequence databases and subsequent BLASTX analysis at NCBI. Information on chromosomal localization was obtained for all genes from IWGSC database. The naming of the wheat PHT genes were based on sequence similarity with rice orthologues.
The protein sequence alignment and construction of neighbour-joining phylogenetic tree was performed using MEGA 553. Bootstrap values were calculated as a percentage of 1000 trials. The prediction of potential motifs in the wheat PHT family gene sequences was done using Multiple Em (Expectation maximization) for Motif Elicitation (MEME) program version 3.5.454. Promoter sequences (1 kb upstream of start codon) for all TaPHTs were retrieved from IWGSC genomic contig sequences and screened for cis-elements using plant cis-acting regulatory DNA elements (PLACE) algorithm (http://www.dna.affrc.go.jp/PLACE/signalscan.html).
Plant material
A bread wheat (Triticum aestivum) variety, C306 was grown in three replicates with adequate amount of nutrients at the research farm of National Agri-Food Biotechnology Institute (NABI). The main individual spikes of the biological replicates were tagged at the first days after anthesis (DAA). The tagged spikes were harvested at four main developmental stages i.e. 7, 14, 21, and 28 DAA and frozen in liquid nitrogen for RNA extraction. Seeds from transgenic wheat with lower phytic acid were used from the previous study33. To compare the expression of genes in aleurone (includes crease area), endosperm and embryo, these tissues were manually separated from 14 DAA seeds on dry ice and were frozen for further processing. For studying gene expression in different parts of wheat, tissues were collected from seeds, roots, shoots, leaves and flag leaf of wheat plants at the stage of 14 DAA.
For Pi starvation experiment, wheat plants with three biological replicates (each having 10–12 seedlings of 7 days old) for each mentioned conditions were grown using hydroponic culture carried out in growth chamber set at 20 ± 1 °C, 50–70% relative humidity and photon rate of 300 μmol quanta m−2 s−1 with 16 h day/8 h night cycle. Hydroponic system with Hoagland medium solution was used for phosphate starvation experiments. For high P and low P conditions, 1 mM KH2PO4and 10 μM KH2PO4 (additional 190 μM KCl) was used respectively. In each PhytaboxTM containing the growing seedlings nutrient medium was refreshed every day. For evaluating P uptake and gene expression, plants were grown in high and low P medium for 15 and 20 days, and subsequently roots and shoots were harvested for further analysis16.
RNA isolation
Total RNA was isolated from 50–100 mg tissue from various tissue samples. RNA was isolated from the multiple stages of wheat seed development (7, 14, 21 and 28 DAA) and 14 DAA plant tissues (root, leaf, stem, flag leaf, rachis, glume, aleurone, endosperm and embryo) was done using the guanidine thiocyanate extraction method with TrizolTM reagent (Invitrogen). For the seed tissue-specific study, 14 DAA seeds were subjected to manual dissection in the presence of dry ice with careful removal of embryo with a little cut on dorsal side of seed, followed by peeling off the aleurone and allowing embryo-less starchy endosperm to separate out. The collected aleurone tissue also contained crease region. As nucellar epidermis and integuments were too small, no clear observation was made concerning their fate. For whole-seed tissue collections, a minimum of fifteen seeds from three different plants were pooled together. The tissues were subsequently snap-frozen in liquid nitrogen. The trace amount of genomic DNA was eliminated by DNase I treatment using an RNase free kit (Ambion, USA).
Quantitative real time PCR (qRT-PCR)
Two micrograms of DNA-free RNA was used for the first strand cDNA synthesis using the Transcriptor First Strand cDNA Synthesis Kit RT-PCR (Roche, USA) with random hexamer primers following the manufacturer’s guidelines. The qRT-PCR reactions were performed using gene-specific primers (Supplementary Table 4) by using QuantiTect SYBR Green RT-PCR Master mix (Qiagen) for 45 cycles on ABI 7700 Sequence Detector (Applied Biosystems, Foster City, CA, USA). The Ct values obtained were normalized against ELF-α, ARF and 18 S rRNAbecause their expression was shown to be consistent. At least two or three separate RNA preparations from the biological samples were used for the transcript expression analysis with four to five technical replicates for each cDNA sample. Target-specific product amplification was verified by melting curve analysis after every run. The relative transcript levels were determind by 2−∆∆CT method55 for every cDNA samples.
Assay of free phosphate and total phosphate in wheat seed tissues
Pi concentration in wheat seeds (7, 14, 21 and 28 DAA), 14 DAA seed tissues (aleurone, endosperm and embryo) and starvation samples (roots and shoots) was measured by molybdate-blue colorimetric method56. The 0.5 g of tissues in three biological replications was extracted in 0.5 ml of extraction buffer (10 mM Tris, 1 mM EDTA, 100 mM NaCl, 1 mM β-mercaptoethanol, and 1 mM phenylmethylsulfonyl fluoride, pH 8.0) as described by earlier57. Further, 0.7 ml of assay solution (Ascorbic acid, 10% and 0.42% ammonium molybdate in 1 N H2SO4) was added to 0.3 ml of sample solution (or to 0.3 ml of water for the blank) and incubated at 42 °C for 40 minutes. Pi concentrations were detected at the absorbance of 820 nm. For the standard preparations, 1 mM of KH2PO4 solution was used at different volumes (0, 5, 15, 20, 25, 30, 35, 40, 45 μL).
For total P analysis, approximately 100 mg powder was digested with nitric acid in the Microwave Reaction System (Mars 6, CEM Corporation, USA). P concentration was estimated in the digested samples using inductively coupled plasma mass spectrometry (ICP-MS; 77006AgilentTechnologies, Santa Clara, CA), following standard protocol.
Additional Information
How to cite this article: Shukla, V. et al. Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci. Rep. 6, 39293; doi: 10.1038/srep39293 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Bucher, M. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytologist. 173, 11–26 (2007).
Lott, J. N. A., Ockenden, I., Raboy, V. & Batten, G. D. Phytic acid and phosphorus in crop seeds and fruits: A global estimate. Seed Science Research. 10, 11–33 (2000).
Raboy, V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Science. 177, 281–296 (2009).
Raboy, V. et al. Barley (Hordeum vulgare L.) Low Phytic Acid 1-1: An Endosperm-Specific, Filial Determinant of Seed Total Phosphorus. Journal of Heredity. 105, 656–665 (2014).
Patrick, J. W. & Offler, C. E. Compartmentation of transport and transfer events in developing seeds. Journal of Experimental Botany. 52, 551–564 (2001).
Olsen, O. A. Nuclear Endosperm Development in Cereals and Arabidopsis thaliana. Plant Cell. 16, S214–S227 (2004).
Raghothama, K. G. Phosphate Acquisition. Annual Review of Plant Physiology and Plant Molecular Biology. 50, 665–693 (1999).
Rausch, C. & Bucher, M. Molecular mechanisms of phosphate transport in plants. Planta. 216, 23–37 (2002).
Guo, C. et al. Function of wheat phosphate transporter gene TaPHT2;1 in Pi translocation and plant growth regulation under replete and limited Pi supply conditions. Planta. 237, 1163–1178 (2013).
Liu, F., Chang, X. J., Ye, Y., Xie, W. B., Wu, P. & Lian, X. M. Comprehensive Sequence and Whole-Life-Cycle Expression Profile Analysis of the Phosphate Transporter Gene Family in Rice. Molecular Plant. 4, 1105–1122 (2011).
Miyaji, T. et al. AtPHT4; 4 is a chloroplast-localized ascorbate transporter in Arabidopsis. Nature Communication. 6, 5928 (2015).
Shin, H., Shin, H. S., Dewbre, G. R. & Harrison, M. J. Phosphate transportin Arabidopsis: Pht1;1and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant Journal. 39, 629–642 (2004).
Nagy, R. et al. Differential regulation of five Pht1 phosphate transporters from maize (Zea mays L.). Plant biology (Stuttgart, Germany). 8, 186–197 (2006).
Paszkowski, U., Kroken, S., Roux, C. & Briggs, S. P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences USA 99, 13324–13329(2002).
Zhou, J. et al. Osphr2 is Involved in Phosphate-Starvation Signaling and Excessive Phosphate Accumulation in Shoots of Plants. Plant Physiology. 146, 1673–1686 (2008).
Wang, J. et al. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Annals of Botany. 6, 1139–1153 (2013).
Wang, Z. Y. et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proceedings of the National Academy of Sciences USA 41, 14953–14958 (2014d).
Nagarajan, V. K., Jain, A., Poling, M. D., Lewis, A. J., Raghothama, K. G. & Smith, A. P. Arabidopsis Pht1; 5 Mobilizes Phosphate Between Source and Sink Organs and Influences the Interaction Between Phosphate Homeostasis and Ethylene Signaling. Plant Physiology. 156, 1149–1163 (2011).
Zhang, F. et al. Involvement of OsPht1; 4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant Journal. 82, 556–569 (2015).
Hassler, S. et al. Lack of the Golgi phosphate transporter PHT4; 6 causes strong developmental defects, constitutively activated disease resistance mechanisms and altered intracellular phosphate compartmentation In Arabidopsis. The Plant Journal 72, 732–744 (2012).
Arpat, A. B., Magliano, P., Wege, S., Rouached, H., Stefanovic, A. & Poirier, Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant Journal. 71, 479–491(2012).
Wang, F., Rose, T., Jeong, K., Kretzschmar, T. & Wissuwa, M. The knowns and unknowns of phosphorus loading into grains, and implications for phosphorus efficiency in cropping systems. Journal of Experiment Botany 67, 1221–1229 (2016).
Raboy, V. myo-Inositol-1, 2, 3, 4, 5, 6-hexakisphosphate. Phytochemistry 64, 1033–43 (2003).
Bohn, L., Meyer, A. S. & Rasmussen, S. K. Phytate: impact on environment and human nutrition. A challenge for molecular breeding. Journal of Zheijang University Science. B9, 165–191 (2008).
Loewus, F. A. & Murthy, P. P. N.myo-Inositol metabolism in plants. Plant Science. 150, 1–19 (2000).
Peng, Z. & Li, C. Transport and partitioning of phosphorus in wheat as affected by P withdrawal during flag-leaf expansion. Plant Soil. 268, 1 (2005).
Rae, A., Cybinski, D. H., Jarmey, J. M. & Smith, F. W. Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Molecular Biology. 53, 27–36 (2003).
Belgaroui, N. et al. Over-expression of the bacterial phytase US417 in Arabidopsis reduces the concentration of phytic acid and reveals its involvement in the regulation of sulfate and phosphate homeostasis and signaling. Plant Cell Physiology. 55, 1912–1924 (2014).
Redekar, N. R., Biyashev, R. M., Jensen, R. V., Helm, R. F., Grabau, E. A. & Maroof, M. A. S. Genome-wide transcriptome analyses of developing seeds from low and normal phytic acid soybean lines. BMC Genomics. 16, 1074 (2015).
Davies, T. G. E., Ying, J., Xu, Q., Li, Z. S., Li, J. & Gordon-Weeks, R. Expression analysis of putative high-affinity phosphate transporters in Chinese winter wheats. Plant, Cell & Environment. 25, 1325–1339 (2002).
Oono, Y. et al. Characterisation of the wheat (triticum aestivum L.) transcriptome by de novo assembly for the discovery of phosphate starvation-responsive genes: gene expression in Pi-stressed wheat. BMC Genomics. 14, 77 (2013).
Bhati, K. K. et al. Differential expression of structural genes for the late phase of phytic acid biosynthesis in developing seeds of wheat (Triticum aestivum L.). Plant Science. 224, 74–85 (2014).
Bhati, K. K., Alok, A., Kumar, A., Kaur, J., Tiwari, S. & Pandey, A. K. Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 67(14), 4379–4389 (2016).
Rose, T. J., Rengel, Z., Ma, Q. & Bowden, J. W. Differential accumulation patterns of phosphorus and potassium by canola cultivars compared to wheat. Journal of Plant Nutrition and Soil Science 170, 404–411 (2007).
White, P. J. & Veneklaas, E. J. Nature and nurture: the importance of seed phosphorus. Plant and Soil. 357, 1–8 (2012).
Iwai, T., Takahashi, M., Oda, K., Terada, Y. & Yoshida, K. T. Dynamic Changes in the Distribution of Minerals in Relation to Phytic Acid Accumulation during Rice Seed Development. Plant Physiology. 160, 2007–2014 (2012).
Preiss, J. Regulation of the biosynthesis and degradation of starch. Annu Rev Plant Physiol 33, 431–454 (1982).
Plaxton, W. C. & Preiss, J. Purification and Properties of Nonproteolytic Degraded ADP glucose Pyrophosphorylase from Maize Endosperm. Plant Physiology 83(1), 105–112 (1987).
Gillies, S. A., Futardo, A. & Henry, R. J. Gene expression in the developing aleurone and starchy endosperm of wheat. Plant Biotechnology Journal. 10, 668–679 (2012).
Zhang, W.-H., Zhou, Y., Dibley, K. E., Tyerman, S. D., Furbank, R. T. & Patrick, J. W. Review: Nutrient loading of developing seeds. Functional Plant Biology. 34, 314–331 (2007).
Tagashira, Y., Shimizu, T., Miyamoto, M., Nishida, S. & Yoshida, K. T. Overexpression of a Gene Involved in Phytic Acid Biosynthesis Substantially Increases Phytic Acid and Total Phosphorus in Rice Seeds. Plants. 4(2), 196–208 (2015).
Aziz, T., Finnegan, P. M., Lambers, H. & Jost, R. Organ-specific phosphorus-allocation patterns and transcript profiles linked to phosphorus efficiency in two contrasting wheat genotypes. Plant Cell & Environment. 37, 943–960 (2014).
Li, Y. et al. Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Science. 230, 23–32 (2015).
Wang, H. L., Offler, C. E. & Patrick, J. W. Nucellar projection transfer cells in the developing wheat grain. Protoplasma. 182, 39–52 (1994).
Weschke, W. et al. Sucrose transport into barley seeds: molecular characterization of two transporters and implications for seed development and starch accumulation. Plant J. 21, 455–467 (2000).
Munck, L. Improvement of nutritional value in cereals. Heredita. 72, 1–128 (1972).
Emes, M. J., Bowsher, C. G., Hedley, C., Burell, M. M., Scrase-Field, E. S. F. & Tetlow, I. J. Starch synthesis and carbon partitioning in developing endosperm. Journal of Experimental Botany. 54, 569–575 (2003).
Jenner, C. F., Ugalde, T. & Aspinall, D. The Physiology of Starch and Protein Deposition in the Endosperm of Wheat. Australian Journal of Plant Physiology. 18, 211–226 (1991).
Cao, H. et al. Distinct metabolic changes between wheat embryo and endosperm during grain development revealed by 2D-DIGE-based integrative proteome analysis. Proteomics. 16, 1515–1536 (2016).
Guttieri, M., Bowen, D., Dorsch, J. A., Raboy, V. & Souza, E. Identification and characterization of a low phytic acid wheat. Crop Sci. 44, 418–424 (2004).
Zhao, H. et al. Disruption of OsSULTR3; 3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytologist. 211, 926–939 (2016).
Bhati, K. K. et al. Genome-wide identification and expression characterization of ABCC-MRP transporters in hexaploid wheat. Frontiers in Plant Science. 6, 488 (2015).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. Mega4: Molecular Evolutionary Genetics Analysis (Mega) software version 4.0. Molecular Biology Evolution. 24, 1596–1599 (2007).
Bailey, T. L., Williams, N., Misleh, C. & Li, W. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Research. 34, 369–373 (2006).
Schmitteng, L. Analysis of Relative Gene Expression Data Using Real Time Quantitative PCR and the 2-(Delta Delta C (T)) Method. Methods. 25, 402–408 (2001).
Chen, P. S., Toribara, T. Y. & Warner, H. Microdetermination of phosphorus. Analytical Chemistry. 28, 1756–1758 (1956).
Chiou, T., Aung, K., Lin, S., Wu, C., Chiang, S. & Su, C. Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell. 18, 412–421 (2006).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology Evolution. 28, 2731–2739 (2011).
Acknowledgements
The authors would like to thank Executive Director, NABI for facilities and support. This research was funded by the NABI core grant to A.K.P. Fellowship for VS was supported by CSIR-SRF grant. S.A. and M.K. thank ICMR and UGC-CSIR for scholarships respectively.K.K.B. acknowledges DBT for Senior Research Fellowships.
Author information
Authors and Affiliations
Contributions
V.S. and A.K.P. designed the study. V.S., M.K., S.A. performed the experiments. S.M. and K.K.B. helped in the Bioinformatics work. V.S., K.K.B., J.K., A.K.P. analyzed the results and wrote the manuscript. A.K.P. conceived, supervised and provided financial support for this study. All the authors read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Shukla, V., Kaur, M., Aggarwal, S. et al. Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci Rep 6, 39293 (2016). https://doi.org/10.1038/srep39293
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39293
This article is cited by
-
Unraveling Metabolic Profile of Wheat Plants Subjected to Different Phosphate Regimes
Journal of Soil Science and Plant Nutrition (2023)
-
A tale of two players: the role of phosphate in iron and zinc homeostatic interactions
Planta (2022)
-
Phosphorus homeostasis: acquisition, sensing, and long-distance signaling in plants
Molecular Biology Reports (2022)
-
Wheat inositol pyrophosphate kinase TaVIH2-3B modulates cell-wall composition and drought tolerance in Arabidopsis
BMC Biology (2021)
-
Genome-wide molecular characterization of Phosphate Transporter 1 and Phosphate Starvation Response gene families in Elaeis guineensis Jacq. and their transcriptional response under different levels of phosphate starvation
Acta Physiologiae Plantarum (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.