Abstract
Phosphoenolpyruvate carboxylase (PEPC) plays an important role in assimilating atmospheric CO2 during C4 and crassulacean acid metabolism photosynthesis, and also participates in various non-photosynthetic processes, including fruit ripening, stomatal opening, supporting carbon–nitrogen interactions, seed formation and germination, and regulation of plant tolerance to stresses. However, a comprehensive analysis of PEPC family in Glycine max has not been reported. Here, a total of ten PEPC genes were identified in soybean and denominated as GmPEPC1-GmPEPC10. Based on the phylogenetic analysis of the PEPC proteins from 13 higher plant species including soybean, PEPC family could be classified into two subfamilies, which was further supported by analyses of their conserved motifs and gene structures. Nineteen cis-regulatory elements related to phytohormones, abiotic and biotic stresses were identified in the promoter regions of GmPEPC genes, indicating their roles in soybean development and stress responses. GmPEPC genes were expressed in various soybean tissues and most of them responded to the exogenously applied phytohormones. GmPEPC6, GmPEPC8 and GmPEPC9 were significantly induced by aluminum toxicity, cold, osmotic and salt stresses. In addition, the enzyme activities of soybean PEPCs were also up-regulated by these treatments, suggesting their potential roles in soybean response to abiotic stresses.
Similar content being viewed by others
Introduction
The enzyme phosphoenolpyruvate (PEP) carboxylase (PEPC; EC 4.1.1.31) catalyzes the irreversible β-carboxylation of PEP in the presence of bicarbonate (HCO3−) and Mg2+ or Mn2+ to yield oxaloacetate (OAA) and inorganic phosphate (Pi)1. It is widely distributed in archaea, bacteria, cyanobacteria, green algae, protozoa, and vascular plants, but is absent from animals and fungi1,2. In plants, the reaction catalyzed by PEPC is the primary fixation step of photosynthetic CO2 assimilation in C4 photosynthesis and crassulacean acid metabolism (CAM). In most nonphotosynthetic tissues and in the leaves of C3 plants, PEPC plays the anaplerotic role of replenishing tricarboxylic acid (TCA) cycle with intermediates that are consumed for a variety of biosynthetic pathways and nitrogen assimilation1,2. Therefore, besides its fundamental role in the initial fixation of atmospheric CO2 during C4 and CAM photosynthesis, PEPC has a wide range of non-photosynthetic roles including supporting carbon–nitrogen interactions2, seed formation and germination2,3, fruit ripening2, guard cell metabolism during stomatal opening4, and provision of malate as a respiratory substrate for symbiotic N2-fixing bacteroids in legume root nodules5. Recently, accumulating evidence has confirmed that a large number of PEPC genes are induced by abiotic and biotic stresses and play important roles in regulation of plant tolerance to stress2,6,7. For example, an up-regulated PEPC expression in response to salinity or drought stress has been well documented in C3, C4 and CAM plants6,7,8,9,10. Meanwhile, overexpression of PEPC gene in transgenic plants enhanced their tolerance to drought or salt stress11, whereas knockdown of PEPC leading to increased sensitivity to osmotic stress12.
Plants have two types of PEPCs that belong to different evolutionary lineages, namely, plant-type PEPC (PTPC) isozyme and bacterial-type PEPC (BTPC) isozyme types1,3,13. Previous studies showed PTPC genes encode highly closely related 100–110 kDa polypeptides that contain a conserved N-terminal seryl-phosphorylation site and typically exist as homotetrameric Class-1 PEPCs1,2. It is suggested that all PTPCs evolved from a common ancestral gene and display a high degree of conservation at the genetic level14. BTPC genes encode larger 116–118 kDa polypeptides that exhibit low (40%) sequence identity with PTPCs, and contain a prokaryotic-like (R/K) NTG C-terminal tetrapeptide3. Although key residues and domains involved in the catalytic reaction and binding of substrates and inhibitors are conserved in both PTPCs and BTPCs, BTPCs lack the distinctive N-terminal serine phosphorylation motif of PTPCs, and appear to only exist as catalytic and regulatory subunits of extraordinary heteromeric complexes (Class-2 PEPCs)3.
Since the release of whole-genome sequences of many plant species, numerous PEPC proteins have been successfully identified and investigated in plants including Arabidopsis thaliana13, Hordeum vulgare15, Lotus japonicus5, Solanum lycopersicum16, Solanum tuberosum17, and Triticum aestivum18. For instance, four PEPC genes (PEPC1- PEPC4) were characterized in A. thaliana13. Three dicot C4 PEPC (ppc-A, ppc-B, ppc-C) genes were analyzed in Flaveria (Asteraceae), with the ppc-A gene being identified as the gene recruited for use in the C4 photosyntehic pathway19. In grasses, there are four PEPC genes that have been predominantly characterized, and ppc-C4 and ppc-B2 are often recruited for use in C4 photosynthesis20. In sedges, there are five PEPC genes that have been predominantly characterized, with the ppc-1 gene being recruited for use in C4 photosynthesis21.
Soybean (Glycine max) is one of the most important crops for vegetable protein and oil worldwide, and its capacity to fix atmospheric nitrogen through symbioses with soil-borne microorganisms would help sustainable agriculture development. Previously, five PEPC genes including GmPEPC122 (also termed GmPEPC1523), GmPEPC423, GmPEPC723, GmPEPC1624, and GmPEPC1725 have been identified in soybean. The release of the complete soybean genome sequences makes it possible to identify genome-wide PEPC genes in soybean26. To get a more comprehensive understanding of the PEPC gene family in soybean, a genome-wide identification and characterization of soybean PEPCs was performed. Their phylogenetic relationships and protein motifs were analyzed together with the PEPCs from 12 other plant species. In addition, their expression patterns in deferent organs and in response to various abiotic stresses and hormones were also investigated, including aluminum (Al), cold, salt, and osmotic stresses as well as exogenously applied abscisic acid (ABA), aminocyclopropane carboxylic acid (ACC), gibberellic acid (GA), and jasmonic acid (JA). To our knowledge, this is the first study on genome-wide analysis of the soybean PEPC gene family, which would provide valuable information for further functional investigations of soybean PEPC.
Results
Identification and phylogenetic analysis of soybean PEPC genes
Blast searches of the soybean genome (G. max var. Williams 82) were performed by using all of the four Arabidopsis PEPC sequences as baits, and a total of 10 putative PEPC genes were identified (Table 1). Their deduced proteins were subjected to domain analysis using Pfam and SMART databases. These 10 non-redundant genes were confirmed as PEPC and denominated as GmPEPC1-GmPEPC10, according to the soybean nomenclature based on physical locations. The detailed information of GmPEPC gene family members, including gene name, gene model name, protein ID, chromosomal position, length of open reading frame (ORF), as well as the size (number of amino acids), molecular weight (MW), isoelectric point (pI), and proposed subcellular localization of the deduced protein, is shown in Table 1. These 10 GmPEPC genes locate on six chromosomes in soybean, with three GmPEPC genes on chromosome 12, two GmPEPC genes on chromosome 6 and chromosomes 13, respectively, and one GmPEPC on Chromosome 2 and 12 each (Table 1).
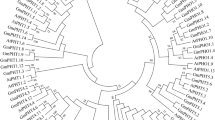
To investigate the evolutionary relationship between soybean PEPC proteins and known PEPCs from other species, a neighbor-joining (NJ) phylogenetic tree was constructed using 75 PEPC proteins from 13 different plant species (Fig. 1). The dendrogram showed that these PEPCs could be classified into two subfamilies (PTPC and BTPC), based on their sequence similarity. PTPC subfamily contains 61 members and is further separated into five groups (PTPC I to PTPC V) (Fig. 1). BTPC subfamily is further separated into two groups (BTPC I and BTPC II) (Fig. 1). The 10 GmPEPCs were assigned to three groups with high bootstrap values: GmPEPC4, GmPEPC7, and GmPEPC10 in PTPC IV group; GmPEPC3, GmPEPC6, GmPEPC8, and GmPEPC9 in PTPC V group; and GmPEPC1, GmPEPC2, and GmPEPC5 in BTPC II group (Figs 1 and 2). Similar group distribution patterns of PEPCs were also found in Gossypium raimondii, Medicago truncatula and Phaseolus vulgaris (Table 2). The total number of PEPCs in soybean is the largest compared with the other 12 plant species (Table 2). Phylogenetic analysis also identified some closely related orthologous PEPCs between G. max and P. vulgaris (Fig. 1; Supplementary Figure S1): GmPEPC7, GmPEPC10 and PvPEPC3, GmPEPC4 and PvPEPC5, in PTPC IV group; GmPEPC3, GmPEPC6 and PvPEPC6, GmPEPC8, GmPEPC9 and PvPEPC2, in PTPC V group; GmPEPC1, GmPEPC2 and PvPEPC1, GmPEPC5 and PvPEPC4, in BTPC II group, suggesting that an ancestral set of PEPCs existed prior to the divergence of G. max and P. vulgaris.
Phylogenetic analysis of PEPC proteins in soybean and other plant species.
The full-length amino acid sequences of 75 PEPC proteins from 13 different plant species were used to construct the neighbor-joining tree using ClustalX 2.0 and MEGA 6.0 with 1000 bootstrap replicates. Branches with less than 50% bootstrap support were collapsed. The PEPCs were classified into two subfamilies (PTPC and BTPC). Subfamily PTPC is further separated into five clades from PTPC I to PTPC V, whereas subfamily BTPC is separated into two clades including BTPC I and BTPC II. Different plant species were distinguished by different colors.
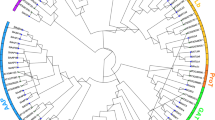
The phylogenetic relationship and exon-intron structures of PEPC family in soybean.
Exon-intron structure was analyzed by online tool Gene Structure Display Server (GSDS). Lengths of exons and introns of GmPEPC genes were exhibited proportionally as indicated by the scale on the bottom. The classification of soybean PEPCs was indicated by the phylogenetic relationship on the left.
Gene structure and conserved motif of PEPC family
To obtain further insights into the possible structural evolution of PEPC genes, their exon-intron organizations were compared. As shown in Fig. 2, GmPEPC genes of different subfamilies exhibit different exon-intron structures. GmPEPC genes in PTPC V and PTPC IV contain 10 exons and 9 introns, while GmPEPC genes in BTPC II contain 20 exons and 19 introns. During the evolution of multigene families, diversification of gene structure is a common phenomenon, which may facilitate evolutionary cooption of genes for new functions to adapt to environmental changes. Different exon-intron structures between subfamilies were also observed for PEPC genes in both C3 (such as G. max and Oryza sativa) and C4 (such as Zea mays and Sorghum bicolor) plant species (Supplementary Figure S1), indicating conserved features of the gene structure of PEPC family. Meanwhile, the numbers of exons and introns are also conserved within each subfamily (Supplementary Figure S1). For example, most genes from PTPC subfamily usually contain 10 exons and 9 introns; most genes from BTPC subfamily contain 20 exons and 19 introns (Supplementary Figure S1). However, the sizes of plant PEPC genes vary dramatically, while the lengths of exons are similar within each subfamily, which suggests that the sizes of the genes depend largely on the sizes of the introns.
The GmPEPC protein sequences show highly conserved features of plant PEPC proteins (Fig. 3). The multiple alignments of the 10 soybean PEPC amino acid sequences showed 70.23% identical positions. The average identity scores of soybean PTPC proteins and soybean BTPC proteins were 92.30% and 92.44%, respectively. We predicted 10 most conserved motifs of plant PEPCs from all tested plant species using MEME software (Table 3; Fig. 4; Supplementary Figure S2). Results showed that PTPC proteins contain 10 motifs, and BTPC proteins contain 9 motifs (lacking motif 8, Supplementary Figure S2), which supported the phylogenetic relationship and classification of plant PEPCs. Notably, seven (1–5, 8 and 10) out of 10 motifs were annotated as key domains involved in the catalytic reaction and in the binding of substrates and inhibitors, which are conserved in both PEPC types (PTPC and BTPC). Additionally, we also analyzed PEPC motifs taking locations in the simulated structure into consideration. As shown in Fig. 4, five motifs contain PEP binding sites, including motif 1 to motif 5. These five PEPC motifs in all analyzed plant species showed the following patterns: motif 1 (SWMGGDRDGNP[RN]VT) has a highly conserved residue PEP binding site W; motif 2 (GKQEVM[IV]GYSDSGKDAGR[LF][ST]AAW) contains a PEP binding site M, also with a D774 that constitutes the binding site for Mg2+ and with a K777 that constitutes the binding site for HCO3−; Motif 3 (GI[EG][ST]LRAIPW[IY]F[AS]WTQ) has a PEP binding site R, also with a S/A that constitutes the binding site for S/A 755 site; Motif 4 (G[TS]YGRGGGP[TC][HY]L) and Motif 5 ([RI][ML]NIGSRP[SA]) only has a PEP binding site R. Motif 8 (EMVFAKG[DN]PG) and Motif 10 ([LI][REQ][LSIN]R[DLEN][SAP][YF]) only has an aspartate binding site K or R, respectively. Whereas another three unknown functional motifs of 6, 7 and 9 were found in all 75 plant PEPCs (Table 3; Fig. 4; Supplementary Figure S2).
cis-elements in the promoter regions of GmPEPC genes
It has been reported that a large number of PEPC genes are induced by abiotic and biotic stresses and play important roles in the regulation of plant tolerance to stresses2,6,7. To investigate the possible roles of PEPCs identified in the soybean genome, the 1500 bp sequences upstream of the start codon of GmPEPC genes were used to analyze cis-elements in their promoter regions by PlantCARE (Fig. 5; Supplementary Table S4). Nineteen putative cis-elements responsive to biotic stresses [including EIRE (elicitor-responsive element27), S-box (pathogen-related cis-element28), W-box29, TC-rich repeats (defense and stress-responsive element30), and WUN-motif (wound-responsive element31)], abiotic stresses [ARE (anaerobic-responsive element), HSE (heat stress-responsive element32), LTR (low-temperature responsive element33), MBS (MYB binding site34)], and phytohormones such as ABA [abscisic acid, ABRE (ABA responsive element35)], auxin (TGA-element36; AuxRR-core37), ET [ethylene, ERE (ethylene-responsive element38 and GCC-box39)], GA (gibberellin, GARE40), JA (jasmonic acid, CGTCA-motif41 and TGACG-motif42), SA (salicylic acid, TCA-element43) and endosperm development (P-box44) were present in the promoters of GmPEPC genes (Fig. 5, Supplementary Table S4). There are other cis-elements (Supplementary Table S4) such as light responsive elements (Box I45, GATA-motif46 and GT1-motif47) were also present in the promoter regions of GmPEPC genes. These results provide further support for the diverse roles of GmPEPC genes in soybean developmental processes, as well as response to biotic and abiotic stresses.
Predicted cis-elements in the promoter regions of GmPEPC genes.
The 1500 bp promoter regions of 10 GmPEPC genes were analyzed to predict the cis-elements, which were presented as colored ellipses: ABA responsive element (ABRE), anaerobic responsive element (ARE), auxin responsive element (TGA-element, AuxRR-core), light-responsive element (G-box), gibberellin responsive element (GARE-motif), ethylene responsive element (ERE, GCC-box), heat stress responsive element (HSE), low-temperature-responsive element (LTR), MYB binding site (MBS), pathogen-related cis-element (S-box), defense and stress-responsive element (TC-rich repeats), salicylic acid responsive element (TCA-element), jasmonic acid responsive element (TGACG-motif, CGTCA-motif), wound-responsive element (WUN-motif), endosperm development (P-box), and WRKY binding site (W-box). The numbers on the top indicate the relative positions to the start codon.
Expression patterns of GmPEPC genes in different soybean tissues
The expression patterns of GmPEPC genes in soybean different tissues were analyzed by qRT-PCR. The results showed that these 10 GmPEPC genes were expressed with different tissue-specific patterns (Fig. 6). For example, GmPEPC4 was expressed higher in stem, leaf, flower, and younger seed but lower in the seed of 15 days after flowering (DAF) and 50 DAF. Both GmPEPC3 and GmPEPC8 were expressed at lower levels in soybean leaf, but GmPEPC3 showed relatively higher expression levels during seed development, while GmPEPC8 expressed higher in root. Both GmPEPC1 and GmPEPC5 showed higher expression in soybean leaf but low in seed at 35 DAF stage. GmPEPC6, GmPEPC7 and GmPEPC9 showed high expression levels in all tested soybean tissues except seed of 35 DAF. The expression levels of GmPEPC2 and GmPEPC10 were low in all tested tissues (Fig. 6).
Expression patterns of GmPEPC genes in different soybean tissues.
The expression patterns of 10 GmPEPC genes in 14 soybean tissues were investigated by qRT-PCR, using soybean GmRP15 gene as the internal control. Root, stem, leaf, flower, and seeds from different development stages were subjected to analysis. DAF: day after flowering. The experiments were repeated three times, log 2 based value was used to create the heat map with clustering of genes. The expression levels are shown from the lowest (blue) to highest (yellow) in heat colors as indicated by the scale on the top.
Expression profiles of GmPEPC genes in response to abiotic stresses and phytohormones
The presences of stress-responsive cis-elements in the promoter regions of the GmPEPC genes suggest their involvement in soybean response to different stresses. To further investigate the possible functions of soybean PEPC genes, the transcriptional expression of 10 GmPEPC genes in soybean plants were analyzed by qRT-PCR analysis after exogenous application of ABA, ACC, GA, and JA as well as under Al toxicity, cold stress, osmotic stress, and salt stress (Fig. 7; Supplementary Figures S3 and S4). Almost all GmPEPC genes responded to the exogenously applied hormones and stress treatments, with the majority being upregulated most of the time, except GmPEPC2, GmPEPC4 and GmPEPC10, which were non-responsive or downregulated at most of the tested time points under these treatments (Fig. 7; Supplementary Figures S3 and S4). In soybean leaves (Fig. 7A), under ABA treatment, the relative expression levels of GmPEPC1, GmPEPC3, GmPEPC6, and GmPEPC8 were induced at least two time points, while GmPEPC4, GmPEPC5 and GmPEPC10 showed decreased transcripts at least two time points. After ACC treatment, GmPEPC1, GmPEPC5, GmPEPC6, GmPEPC8, and GmPEPC9 were substantially upregulated. The increase in their levels started as early as 3 h after ACC stress and continued till 24 h of stress. On the other hand, several GmPEPC genes (GmPEPC2, GmPEPC4, and GmPEPC7) were significantly downregulated during ACC stress. With GA treatment, except GmPEPC2 and GmPEPC10 were significantly downregulated at 6 h, all of the rest GmPEPC genes were induced at least two time points. The expression profile of soybean PEPC genes under JA treatment is similiar with GA stress, except GmPEPC2 and GmPEPC10, which were down-regulated, and all of the rest genes were induced at least two time points. Under Al stress, the expression of GmPEPC1, GmPEPC3, GmPEPC5, GmPEPC6 and GmPEPC8 was induced at most of the time points, while the expression of GmPEPC9 was only induced at 6 h. The relative expression of GmPEPC4 and GmPEPC7 were inhibited under Al stress. Most GmPEPC genes, except for GmPEPC2, GmPEPC3 and GmPEPC10, were upregulated during cold stress treatment. Under salt stress, the transcript abundance of GmPEPC3, GmPEPC6, GmPEPC8, and GmPEPC9 increased in at least three time points, while GmPEPC10 decreased across all time points (Fig. 7A; Supplementary Figure S3). Water-deficit conditions were imposed by transferring the soybean seedlings to 20% PEG solution and the expression levels of GmPEPC genes were examined during stress. GmPEPC3, GmPEPC5, GmPEPC6, GmPEPC8, and GmPEPC9 showed induced expression in at least two time points whereas GmPEPC2 and GmPEPC10 showed downregulation across all time-points. The cluster analysis showed that GmPEPC6, GmPEPC8, and GmPEPC9 were grouped together and showed more responsive to abiotic stresses.
Expression profiles of PEPC genes in leaves (A) and roots (B) of soybean in response to ABA, ACC, GA, JA, Al, cold, salt (NaCl) and osmotic (PEG) treatments. The relative expression levels of the 10 PEPC genes were quantified by qRT-PCR, using soybean GmRP15 gene as the internal control. Leaves and roots of 14-d soybean seedlings are used to investigate the changes in PEPC expression under different treatments, including 100 μM ABA (Abscisic acid), 100 μM ACC (aminocyclopropane carboxylatesythase), 100 μM GA (Gibberellin), 100 μM JA (Jasmonic acid), 25 μM AlCl3 (pH 4.3), 4 °C cold, 200 mM NaCl, and 20% PEG6000. The experiments were repeated three times, log2 based value (fold change) was used to create the heat map with clustering of genes. All data were normalized to the expression level of control (0 h). The scale represents the relative expression levels from low (blue) to high (red).
We also analyzed the expression patterns of GmPEPC genes in soybean roots under phytohormone and abiotic treatments (Fig. 7B; Supplementary Figure S4). The transcript levels of four GmPEPC genes (GmPEPC1, GmPEPC3, GmPEPC5, and GmPEPC6) can be significantly induced in response to exogenous application of ABA, while all other genes especially GmPEPC10 are downregulated by ABA. GmPEPC1, GmPEPC6 and GmPEPC9 were highly upregulated by ACC. However, the expression levels of GmPEPC2 and GmPEPC3 declined in at least three time points. With GA treatment, GmPEPC5, GmPEPC8, and GmPEPC9 were induced while GmPEPC7 was downregulated during the whole treatment. Under JA treatment, the transcript levels of four GmPEPC genes (GmPEPC3, GmPEPC5, GmPEPC8, and GmPEPC9) can be significant induced, while GmPEPC2 showed significant downregulation across all time points. Under Al stress, except for GmPEPC4 and GmPEPC7, the expression levels of all other eight GmPEPC genes were increased. Under cold stress, the mRNA levels of most GmPEPC genes (except GmPEPC10) exhibited significantly upregulated in most time points. Under salt stress, the transcript levels of eight GmPEPC genes were upregulated except GmPEPC1 and GmPEPC10. When treated with 20% PEG, the expression of GmPEPC6, GmPEPC8, and GmPEPC9 was induced significantly, while GmPEPC2 and GmPEPC10 showed down-regulation (Fig. 7B; Supplementary Figure S4). Three genes, GmPEPC6, GmPEPC8, and GmPEPC9 showed significant induced expression in soybean roots under abiotic stresses and were cluster together, which is consistent with their expression patterns in soybean leaves (Fig. 7).
PEPC activity in response to abiotic stresses and phytohormones
To further investigate the potential functions of GmPEPC genes in soybean response to abiotic stresses, the enzyme activities of GmPEPCs in soybean leaves and roots under different phytohormones and abiotic treatments were assayed. As shown in Fig. 8, ABA and ACC treatments significantly increased the PEPC activity compared with control in soybean leaves. JA treatment resulted in a significant increased PEPC activity in both leaves and roots. Under abiotic stresses, the PEPC activity in leaves was significantly increased by cold, PEG, and salt stresses, especially PEG treatment. In soybean roots, the PEPC activity was significantly upregulated by cold and Al stresses.
PEPC activities in soybean leaves and roots subjected to different abiotic and phytohormone treatments.
Leaves and roots of 14-d soybean seedlings are used to investigate the changes in PEPC activities under different treatments, including 100 μM ABA (Abscisic acid), 100 μM ACC (aminocyclopropane carboxylatesythase), 100 μM GA (Gibberellin), 100 μM JA (Jasmonic acid), 25 μM AlCl3 (pH 4.3), 4 °C cold, 200 mM NaCl, and 20% PEG6000. Data shown are means ± SD of three independent experiments. Statistical significance of differences between control and treated groups was analyzed using Student’s t-test (*indicates P < 0.05).
Discussion
PEPC plays an important role in assimilating atmospheric CO2 during C4 and CAM photosynthesis, and also participates in various non-photosynthetic processes. However, a comprehensive analysis of GmPEPC genes in soybean has not been reported. In the present study, a total of ten PEPCs were identified in the soybean genome from Phytozome database48. The numbers of PEPC genes in soybean were slightly more than that in other plant species, such as A.thaliana (4 PEPC genes), Brachypodium distachyon (6 PEPC genes), G. raimondii (6 PEPC genes), M. truncatula (5 PEPC genes), O. sativa (6 PEPC genes), Panicum virgatum (8 PEPC genes), P. vulgaris (6 PEPC genes), Ricinus communis (2 PEPC genes), S. tuberosum (5 PEPC genes), S. bicolor (6 PEPC genes), T. aestivum (5 PEPC genes), and Z. mays (6 PEPC genes) (Table 2). As the soybean genome is larger than the genomes of rice and Arabidopsis, it is possible that the soybean genome contains more PEPC genes than them. Interestingly, the genome size of soybean is smaller than maize and sorghum, but it also contains more PEPC genes. This might be due to the two ancient genome duplication events happened in soybean around 59 and 13 million years ago, which resulted in multiple copies of around 75% of its genes26.
Phylogenetic analyses of higher plant PEPC genes has been previously carried out in families of Cyperaceae, Poaceae, and Molluginaceae to determine which PEPC gene is recruited for use in C4 biochemistry20,49,50. Plants have two types of PEPCs including plant-type PEPC (PTPC) and bacterial-type PEPC (BTPC)1,3,13. In the core eudicots, there are two primary PEPC gene lineages that have been studied to date: ppc-1(PTPC) and ppc-2 (BTPC)50. For example, in Arabidopsis, there are four PEPC genes, including three PTPC and one BTPC13. In rice, five rice PEPC genes (Osppc1, 2a, 2b, 3, and 4) encode the PTPCs and the other (Osppc-b) encodes a BTPC. In addition, all plant genomes sequenced to date, including that of ancestral green algae, contain at least one BTPC gene2. In this study, soybean PEPC genes were also distributed into two subfamilies (PTPC and BTPC), with seven genes (GmPEPC3, GmPEPC4, GmPEPC6, GmPEPC7, GmPEPC8, GmPEPC9, and GmPEPC10) in PTPC subfamily, and three genes (GmPEPC1, GmPEPC2, and GmPEPC5) in BTPC group. Moreover, the gene structures of PTPC and BTPC genes were different: PTPC genes have a highly conserved genomic structure composed of approximately ten exons, whereas BTPC genes have a very different and more complex structure with approximately twenty exons1. Similar results were also observed in this study, where GmPEPCs in PTPC subfamily contain 10 exons, and GmPEPCs in BTPC subfamily contain 20 exons (Fig. 2). We predicted 10 most conserved motifs in 75 PEPCs from 13 different species (Fig. 4; Supplementary Figure S3), and the ten PEPCs in soybean also possessed 10 conserved motifs (Table 3; Fig. 4). These data, together with previous studies, suggest similar origins and evolution patterns of the PEPC genes in different species.
PEPC genes have been reported in a wide range of tissues, including cell cultures and seedlings of A. thaliana51, seeds of H. vulgare and R. communis15, root nodules of L. japonicus and G. max5,52, cell cultures of S. lycopersicum16, seedlings of S. tuberosum17 and leaves of T. aestivum18. Besides, PEPC has a wide range of non-photosynthetic roles including seed formation and germination, fruit ripening, guard cell metabolism during stomatal opening2. Former study showed that the soybean bacterial-type gene GmPEPC17 (termed GmPEPC5 in this study) was expressed mainly in aboveground organs25. We also noticed that a higher expression level of GmPEPC5 in stem and leaf, but not in the flower and root. Besides, the other two GmBTPC genes (GmPEPC1 and GmPEPC2) have no or low expression in flowers (Fig. 6), which is consistent with the previous report that Arabidopsis BTPC transcripts were detected at low levels in siliques and flowers13. It is noteworthy that almost no detectable expression level of GmPEPC2 was found in the examined tissues (Fig. 6). On the other side, three GmPTPC genes (GmPEPC6, GmPEPC7 and GmPEPC9) expresses at relatively higher levels in most tissues and showed a similar expression pattern under normal condition (Fig. 6), whereas these genes expressed differently as reported before25. In addition, GmPEPC8 is expressed at low levels in the tissues tested, except a relative higher level in root (Fig. 6), whereas the broad but low-level expression pattern of this gene has been reported25.
Extensive studies suggested PEPC genes play important roles in plant response to abiotic stresses. For example, studies have shown that PEPC overexpressing transgenic rice has a relatively higher photosynthetic rate under high light and temperature conditions53. In addition, abiotic stresses, such as salt and chilling injury, can induce PEPC gene expression in wheat, Arabidopsis and sorghum8,10,54. In this study, we showed that the promoter regions of most GmPEPC genes contained 19 stress-responsive cis-regulated elements such as ABA, cold, drought, heat, salt, and wound-related elements, indicating their potential roles in soybean response to phytohormone and stresses (Fig. 5; Supplementary Table S4). Previous reports also demonstrated the crucial roles of PEPC genes in various stress responses and in hormone signaling transduction2,7,8,9,10, which motivated us to perform expression profile analyses of GmPEPC genes under various abiotic stress and hormones treatments. The results showed that Al, cold, salt or PEG stress could alter the expression level of soybean PEPC genes and soybean PEPC activity (Figs 7 and 8). GmPEPC6, GmPEPC8 and GmPEPC9 were significantly induced by Al, cold, salt or osmotic stress, suggesting that these three GmPEPC genes are important for soybean response to abiotic stresses (Fig. 7). It has been reported that the exudation of PEPC-derived organic acids functions to chelate metals in the rhizosphere to alleviate heavy metal toxicity by preventing their uptake into the cell2. Overexpression of a C4 PEPC isoenzyme in rice led to increased oxalate exudation and Al tolerance55. In our study, eight out of ten GmPEPC genes in soybean roots were responsive to Al stress (Fig. 7B). On the other hand, although the up-regulation of PEPC genes has been well documented in plants2, we still found that a few GmPEPC genes (such as GmPEPC2, GmPEPC4 and GmPEPC10) showed down-regulating or non-responsive tendency during the abiotic and phytohormone treatments. The down-regulation or non-responsiveness of GmPEPC2, GmPEPC4 and GmPEPC10 might be due to the low transcript abundance of GmPEPC2 and GmPEPC10 in all tested soybean tissues (Fig. 6) and few (only two) cis-elements responsive to hormones or stresses in the promoter region of GmPEPC4 (Fig. 5), which indicates their less importance in soybean response to these hormone and stress treatments. The possible biological meaning of this phenomenon could be that different GmPEPC genes have different regulation mechanisms and diverged functions. In addition, the PEPC activity in soybean leaves increased after salt or PEG treatment, and PEPC activity in soybean roots was upregulated in response to Al or cold stress (Fig. 8). These results suggested that PEPC might be involved in soybean response to abiotic stresses.
In vascular plants, PEPCs are strictly regulated enzymes by a variety of mechanisms due to their irreversible nature of the enzymatic reactions. PEPC is activated by its positive effector, glucose 6-phosphate, and inhibited by its negative effectors, malate, aspartate, and glutamate, as well as phosphorylation catalyzed by a specific Ca2+-independent serine/threonine kinase known as PPCK (PEPC protein kinase)2, and dephosphorylation by a PP2A (protein phosphatase 2 A)1,2,56. The control of reversible phosphorylation catalyzed by PPCK is an important mechanism that regulates the activity of PEPC57. In this reaction, phosphorylation catalyzed by PPCK changes the sensitivity of PEPC to its allosteric effectors2. Previously, four PPCK genes (GmPPCK1, Glyma.03G251400; GmPPCK2, Glyma.20G222600; GmPPCK3, Glyma.10G166600; GmPPCK4, Glyma.10G186000) have been characterized in soybean58. In order to investigate the relationship between GmPEPC activity and GmPPCK expression, we examined the expression patterns of GmPPCK genes in soybean leaves and roots in response to abiotic stress and phytohormone applications (Supplementary Figure S5). The results showed that the expression patterns of four GmPPCK genes were not same. For example, under ABA treatment, only GmPPCK1 was upregulated in soybean leaves, suggesting different GmPPCK genes have different regulation mechanisms in specific tissues and environments. And the changes in GmPEPC activities (Fig. 8) were not consistent with the changes in the transcript abundance of GmPPCK genes (Supplementary Figure S5) under these phytohormone and abiotic stress treatment, which revealed the complexity of the regulation mechanisms of PEPC activity. Indeed, degradation of both PEPC and PPCK by the polyubiquitin–proteasome pathway has been reported59,60. Recently, monoubiquitination of PEPC during sorghum seed development and germination were also documented61. Additionally, the dephosphorylation of PEPC is catalyzed by PP2A1,2,56. In summary, our results suggest a complex regulation mechanism of GmPEPC activity.
In conclusion, we conducted a genome-wide survey of the PEPC family in soybean. In silico analysis of the soybean genome database identified 10 PEPC genes, supported by conserved domain and multiple sequence alignments. Phylogenetic analyses of 75 PEPC genes from 13 speices indicated that these PEPCs could be divided into two subfamilies. This classification was further supported by gene structure and motif analyses, with each group sharing common features of exon-intron and protein motifs. The GmPEPC genes were expressed in soybean roots, stems, leaves, flowers and developing seeds. The presence of important cis-regulatory elements related to various stresses in the promoter regions of GmPEPC genes indicates their putative functions in soybean response to stresses. Their transcript abundance and enzyme activities in soybean leaves and roots were altered by Al, cold, salt or PEG stress, as well as the exogenous application of ABA, GA, JA, and SA, implying that soybean PEPCs may participate in soybean response to abiotic stresses. Taken together, this work would provide a foundation for future functional investigation of the PEPC family in soybean.
Methods
Plant material and stress treatments
The seeds of soybean [G. max (L.) Merr.] variety ‘Kefeng No. 1’ (Al-tolerant and drought-tolerant cultivar)62,63 were obtained from the National Center for Soybean Improvement (Nanjing Agricultural University, Nanjing), and were sterilized in 3% (v/v) sodium hypochlorite for 10 min and rinsed thoroughly with deionized water. Seeds were placed on wet paper towels for 4 d in a plant growth chamber (Dongnan Equipment, Ningbo, China) with a 16-h-light/8-h-dark cycle at 28 °C/25 °C and a light intensity of 150 μmol· m−2·s−1. The chamber had a relative humidity of 70%. The seedlings were then transferred to plastic boxes (1 L) filled with half-strength Hoagland nutrient solution for hydroponic culture64 and incubated in the same growth chamber. The nutrient solution was changed every 3 days. For tissue-specific expression analysis, young leaves, stems, and roots were collected from 4-week-old seedlings at V2 stage; blooming flowers were sampled from plants at R2 stage, whereas developing seeds were collected since the beginning of R3 to R7 stages, at five days intervals65. For different abiotic stress and hormone treatment, 14-day-old seedlings were subjected to 25 μM AlCl3 (pH 4.3), 20% (w/v) polyethylene glycol (PEG) 6000, 200 mM NaCl, 4 °C cold treatment, 100 μM abscisic acid (ABA), 100 μM 1-aminocyclopropane-1-carboxylic acid (ACC), 100 μM jasmonic acid (JA), and 100 μM gibberellins (GA), respectively. Meanwhile, control plants were treated with half-strength Hoagland nutrient solution or 0.5 mM CaCl2 (pH = 4.3) solution containing either 0 μ M AlCl3 (control). Leaves and roots from all treatments were harvested separately at 0, 3, 6, 12, 24 and 48 h after treatment, then immediately frozen in liquid nitrogen and stored at −80 °C until use. Each sample was the mixture of three seedlings and each treatment was repeated three times.
Identification and sequence analysis of PEPC genes
We searched the PEPC genes from thirteen plant genomes and their sequences and corresponding annotations were downloaded from Phytozome database (http://phytozome.jgi.doe.gov/pz/portal.html). We used the following steps to identify the PEPC genes. First, the amino acid consensus sequences of four Arabidopsis PEPC were used as queries to conduct BLASTP searches in the soybean Proteome (http://phytozome.jgi.doe.gov/pz/portal.html). The protein domain and motif analysis was performed using PFAM (http://pfam.sanger.ac.uk/), SMART (http://smart.emblheidelberg.de/) and MEME (http://meme.nbcr.net/meme/).
Multiple alignments of the sequences were performed using ClustalW with the default options66 in MEGA Version 6.067. Phylogenetic trees were constructed based on the neighbor-joining (NJ) method with a Kimura 2-parameter model using MEGA v6.0. The stability of the internal nodes was assessed with a bootstrap analysis of 1,000 replicates.
Gene structure analysis
The gene structures were predicted using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). Multiple expectation maximization for motif elicitation (MEME68) was employed to identify and analyze the conserved motifs of PEPC sequences in this study. Only the motifs with P values < 10−6 and no overlap with each other were reported.
Analysis of cis-acting elements in GmPEPC promoter regions
The 1500 bp upstream sequences of the start codon were used to analyze the cis-elements in GmPEPC promoter regions using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PLACE database69.
Quantitative real-time PCR
Total RNA samples were extracted using RNeasy Plant Mini Kits with on-column DNase (RNase free DNase set) treatment (Qiagen, Dusseldorf, Germany). The RNA concentration and integrity were checked by spectrophotometry and gel electrophoresis. A total of 0.5 μg RNA per sample was reverse transcribed into cDNA with the PrimeScript™ II reverse transcription kit (TaKaRa, Dalian, China). The cDNAs were diluted 1:10 with nuclease-free water prior to the qRT-PCR analyses. Soybean GmRP15 gene was used as the internal standard, and all gene-specific primers were designed using Primer-BLAST (http:// www.ncbi.nlm.nih.gov/tools/primer-blast/) and are listed in Supplementary Table S1. Quantitative real-time PCR (qRT-PCR) analysis was carried out with the IQ5 light cycler (Bio-Rad, Hercules, USA). Each PCR mixture contained 10 μL SYBR Premix Ex Taq II (TaKaRa), 0.5 μM gene-specific primers and 50–100 ng cDNA in a final volume of 20 μL. For tissue expression pattern analysis, the gene expression levels in different tissues were calculated using the 2−ΔCt method70. The relative gene expression levels in response to various abiotic and phytohormone treatments were calculated using the 2−ΔΔCt method71, using the control plants (0 h) as the reference. The expression heatmaps with clustering of genes were constructed by MeV 4.9 software with Euclidean distance metric using the average linkage method72.
PEPC activity measurement
Approximately 0.5 g soybean leaves or roots were homogenized with 1.5 mL of extraction buffer containing 100 mM Tris–HCl (pH 7.8), 1 mM EDTA, 1 mM dithiothreitol (DTT), 5 mM MgCl2, 1% (w/v) polyvinylpyrrolidone (PVP), and 1% (v/v) protease inhibitor cocktail (Sigma, USA). Then the extract was centrifuged at 12000× rpm at 4 °C for 15 min. PEPC activity was measured at 30 °C by an enzyme-coupled spectrophotometric method73 with slight modifications. The standard assay mixture (a total volume of 1.0 ml) contained 100 mM HEPES-NaOH (pH 8.0), 0.2 mM NADH, 1 mM NaHCO3, 6 mM PEP, 10 mM MgCl2, and 12 units of malate dehydrogenase (MDH) from porcine heart mitochondria (Roche Diagnostics, Germany). PEPC activity was recorded by monitoring NADH oxidation at 340 nm in a spectrophotometer (Beckman DU200, Germany) at 25 °C. Assays were initiated by the addition of protein extracts. Protein amounts were determined by the method of Bradford74.
Statistical analysis
Statistical analyses were performed with the SPSS version 17.0 software (SPSS, Chicago, IL, USA) for Windows. Data are presented as mean values ± SD of three independent experiments. Differences between treatments were analyzed using Student’s t test.
Additional Information
How to cite this article: Wang, N. et al. Genome-wide Analysis of Phosphoenolpyruvate Carboxylase Gene Family and Their Response to Abiotic Stresses in Soybean. Sci. Rep. 6, 38448; doi: 10.1038/srep38448 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Izui, K., Matsumura, H., Furumoto, T. & Kai, Y. Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu. Rev. Plant Biol. 55, 69–84 (2004).
O’Leary, B., Park, J. & Plaxton, W. C. The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 436, 15–34 (2011).
O’Leary, B. et al. Tissue-specific expression and post-translational modifications of plant- and bacterial-type phosphoenolpyruvate carboxylase isozymes of the castor oil plant, Ricinus communis L. J. Exp. Bot. 62, 5485–5495 (2011).
Cousins, A. B. et al. The role of phosphoenolpyruvate carboxylase during C4 photosynthetic isotope exchange and stomatal conductance. Plant Physiol. 145, 1006–1017 (2007).
Nakagawa, T. et al. Characterization and expression analysis of genes encoding phosphoenolpyruvate carboxylase and phosphoenolpyruvate carboxylase kinase of Lotus japonicas, a model legume. Mol. Plant Microbe Interact. 16, 281–288 (2003).
Doubnerová Hýsková, V., Miedzińska, L., Dobrá, J., Vankova, R. & Ryšlavá., H. Phosphoenolpyruvate carboxylase, NADP-malic enzyme, and pyruvate, phosphate dikinase are involved in the acclimation of Nicotiana tabacum L. to drought stress. J. Plant Physiol. 171, 19–25 (2014).
Cheng, G., Wang, L. & Lan, H. Cloning of PEPC-1 from a C4 halophyte Suaeda aralocaspica without Kranz anatomy and its recombinant enzymatic activity in responses to abiotic stresses. Enzyme Microb. Tech. 83, 57–67 (2016).
García-Mauriño, S., Monreal, J. A., Alvarez, R., Vidal, J. & Echevarríaa, C. Characterization of salt stress-enhanced phosphoenolpyruvate carboxylase kinase activity in leaves of Sorghum vulgare: independence from osmotic stress, involvement of ion toxicity and significance of dark phosphorylation. Planta 216, 648–655 (2003).
Carmo-Silva, A. E., Bernardes da Silva, A., Keys, A. J., Parry, M. A. J. & Arrabaca, M. C. The activities of PEP carboxylase and the C4 acid decarboxylases are little changed by drought stress in three C4 grasses of different subtypes. Photosynth. Res. 97, 223–233 (2008).
González, M. C., Sánchez, R. & Cejudo, F. J. Abiotic stresses affecting water balance induce phosphoenolpyruvate carboxylase expression in roots of wheat seedlings. Planta 216, 985–992 (2003).
Qin, N. et al. Drought tolerance and proteomics studies of transgenic wheat containing the maize C4 phosphoenolpyruvate carboxylase (PEPC) gene. Protoplasma 253, 1503–1512 (2016).
Chen, M., Tang, Y., Zhang, J., Yang, M. & Xu, Y. RNA interference-based suppression of phosphoenolpyruvate carboxylase results in susceptibility of rapeseed to osmotic stress. J. Integr. Plant Biol. 52, 585–592 (2010).
Sánchez R. & Cejudo, F. J. Identification and expression analysis of a gene encoding a bacterial-type phosphoenolpyruvate carboxylase from Arabidopsis and rice. Plant Physiol. 132, 949–957 (2003).
Svensson, P., Bläsing, O. E. & Westhoff, P. Evolution of C4 phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys. 414, 180–188 (2003).
Murmu, J. & Plaxton, W. C. Phosphoenolpyruvate carboxylase protein kinase from developing castor oil seeds: partial purification, characterization, and reversible control by photosynthate supply. Planta 226, 1299–1310 (2007).
Rontein, D., Dieuaide-Noubhani, M., Dufourc, E. J., Raymond, P. & Rolin, D. The metabolic architecture of plant cells. Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J. Biol. Chem. 277, 43948–43960 (2002).
Sima, B. D. & Desjardins, Y. Sucrose supply enhances phosphoenolpyruvate carboxylase phosphorylation level in in vitro Solanum tuberosum. Plant Cell Tiss. Org. Cult. 67, 235–242 (2001).
Duff, S. & Chollet, R. In vivo regulation of wheat-leaf phosphoenolpyruvate carboxylase by reversible phosphorylation. Plant Physiol. 107, 775–782 (1995).
Engelmann, S., Blasing, O. E., Gowik, U., Svensson, P. & Westhoff, P. Molecular evolution of C4 phosphoenolpyruvate carboxylase in the genus Flaveria–a gradual increase from C3 to C4 characteristics. Planta 217, 717–725 (2003).
Christin, P. A., Salamin, N., Savolainen, V., Duvall, M. R. & Besnard, G. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr. Biol. 17, 1241–1247 (2007).
Besnard, G. et al. Phylogenomics of C4 photosynthesis in sedges (Cyperaceae): multiple appearances and genetic convergence. Mol. Biol. Evol. 26, 1909–1919 (2009).
Vazquez-Tello, A. et al. Sequence of a soybean (Glycine max L.) phosphoenolpyruvate carboxylase cDNA. Plant Physiol. 103, 1025–1026 (1993).
Hata, S., Izui, K. & Kouchi, H. Expression of a soybean nodule-enhanced phosphoenolpyruvate carboxylase gene that shows striking similarity to another gene for a house-keeping isoform. Plant J. 13, 267–273 (1998).
Sugimoto, T. et al. cDNA sequence and expression of a phosphoenolpyruvate carboxylase gene from soybean. Plant Mol. Biol. 20, 743–747 (1992).
Sullivan, S., Jenkins, G. I. & Nimmo, H. G. Roots, cycles and leaves. Expression of the phosphoenolpyruvate carboxylase kinase gene family in soybean. Plant Physiol. 135, 2078–2087 (2004).
Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183 (2010).
Fukuda, Y. Interaction of tobacco nuclear protein with an elicitor-responsive element in the promoter of a basic class I chitinase gene. Plant Mol. Biol. 34, 81–87 (1997).
Kunitomo, H., Higuchi, T., Iino, Y. & Yamamoto, M. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11+ gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell 11, 3205–3217 (2000).
Eulgem, T., Rushton, P. J., Schmelzer, E., Hahlbrock, K. & Somssich, I. E. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699 (1999).
Gausing, K. & Jensen, C. B. Two ubiquitin-long-tail fusion genes arranged as closely spaced direct repeats in barley. Gene 94, 165–171 (1990).
Siebertz, B., Logemann, J., Willmitzer, L. & Schell, J. cis-analysis of the wound-inducible promoter WUN1 in transgenic tobacco plants and histochemical localization of its expression. Plant Cell 1, 961–968 (1989).
Mosser, D. D., Theodorakis, N. G. & Morimoto, R. I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol. Cell Biol. 8, 4736–4744 (1988).
Dunn, M. A., White, A. J., Vural, S. & Hughes, M. A. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol. Biol. 38, 551–564 (1998).
Nakagoshi, H., Nagase, T., Ueno, Y. & Ishii, S. Transcriptional trans-repression by the c-myb proto-oncogene product. Nucleic Acids. Res. 17, 7315–7324 (1989).
Hamel, L. P. et al. Activation of stress-responsive mitogen-activated protein kinase pathways in hybrid poplar (Populus trichocarpa x Populus deltoides). Tree Physiol. 25, 277–288 (2005).
Hagen, G. & Guilfoyle, T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385 (2002).
Sakai, T., Takahashi, Y. & Nagata, T. Analysis of the promoter of the auxin-inducible gene, parC, of tobacco. Plant Cell Physiol. 37, 906–913 (1996).
Itzhaki, H., Maxson, J. M. & Woodson, W. R. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA 91, 8925–8929 (1994).
Ohme-Takagi, M. & Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182 (1995).
Skriver, K., Olsen, F. L., Rogers, J. C. & Mundy, J. Cis-acting DNA elements responsive to gibberellin and its antagonist abscisic acid. Proc. Natl. Acad. Sci. USA 88, 7266–7270 (1991).
Beaudoin, N. & Rothstein, S. J. Developmental regulation of two tomato lipoxygenase promoters in transgenic tobacco and tomato. Plant Mol. Biol. 33, 835–846 (1997).
Schindler, U., Beckmann, H. & Cashmore, A. R. TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4, 1309–1319 (1992).
Goldsbrough, A. P., Albrecht, H. & Stratford, R. Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes. Plant J. 3, 563–571 (1993).
Vicente-Carbajosa, J., Moose, S. P., Parsons, R. L. & Schmidt, R. J. A maize zinc-finger binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 94, 7685–7690 (1997).
Kuhlemeier, C. et al. Localization and conditional redundancy of regulatory elements in rbcS-3A, a pea gene encoding the small subunit of ribulose-bisphosphate carboxylase. Proc. Natl. Acad. Sci. USA 85, 4662–4666 (1998).
Terzaghi, W. B. & Cashmore, A. R. Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474 (1995).
Giuliano, G. et al. An evolutionary conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85, 7089–7093 (1988).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Christin, P. A. & Besnard, G. Two independent C4 origins in Aristidoideae (Poaceae) revealed by the recruitment of distinct phosphoenolpyruvate carboxylase genes. Am. J. Bot. 96, 2234–2239 (2009).
Christin, P. A. et al. Complex evolutionary transitions and the significance of C3–C4 intermediate forms of photosynthesis in Molluginaceae. Evolution 65, 643–660 (2011).
Gregory, A. L. et al. In vivo regulatory phosphorylation of the phosphoenolpyruvate carboxylase AtPPC1 in phosphate-starved Arabidopsis thaliana. Biochem. J. 420, 57–65 (2009).
Xu, W., Zhou, Y. & Chollet, R. Identification and expression of a soybean nodule-enhanced PEP-carboxylase kinase gene (NE-PpcK) that shows striking up-/down-regulation in vivo. Plant J. 34, 441–452 (2003).
Bandyopadhyay, A. et al. Enhanced photosynthesis rate in genetically engineered indica rice expressing pepc gene cloned from maize. Plant Sci. 172, 1204–1209 (2007).
Sánchez, R., Flores, A. & Cejudo, F. J. Arabidopsis phosphoenolpyruvate carboxylase genes encode immunologically unrelated polypeptides and are differentially expressed in response to drought and salt stress. Planta 223, 901–909 (2006).
Begum, H. H., Osaki, M., Watanabe, T. & Shinano, T. Mechanisms of aluminum tolerance in phosphoenolpyruvate carboxylase transgenic rice. J. Plant Nutr. 32, 84–96 (2009).
Nimmo, H. G. The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci. 5, 75–80 (2000).
Monreal, J. A. et al. Factors involved in the rise of phosphoenolpyruvate carboxylase-kinase activity caused by salinity in sorghum leaves. Planta 237, 1401–1413 (2013).
Xu, W., Sato, S. J., Clemente, T. E. & Chollet, R. The PEP-carboxylase kinase gene family in Glycine max (GmPpcK1-4): an in-depth molecular analysis with nodulated, non-transgenic and transgenic plants. Plant J. 49, 910–923 (2007).
Agetsuma, M., Furumoto, T., Yanagisawa, S. & Izui, K. The ubiquitin–proteasome pathway is involved in rapid degradation of phosphoenolpyruvate carboxylase kinase for C4 photosynthesis. Plant Cell Physiol. 46, 389–398 (2005).
Schulz, M., Klockenbring, T., Hunte, C. & Schnabl, H. Involvement of ubiquitin in phosphoenolpyruvate carboxylase degradation. Bot. Acta 106, 143–145 (1993).
Ruiz-Ballesta, I. et al. New insights into the post-translational modification of multiple phosphoenolpyruvate carboxylase isoenzymes by phosphorylation and monoubiquitination during sorghum seed development and germination. J. Exp. Bot. 67, 3523–3536 (2016).
Liu, Y., Gai, J. Y., Lü, H. N., Wang, Y. J. & Chen, S. Y. Identification of drought tolerant germplasm and inheritance and QTL mapping of related root traits in soybean (Glycine max (L.) Merr.). Yi Chuan Xue Bao. 32, 855–863 (2005).
Qi, B. et al. Mapping quantitative trait loci associated with aluminum toxin tolerance in NJRIKY recombinant inbred line population of soybean (Glycine max). J. Integr. Plant Biol. 50, 1089–1095 (2008).
Song, H. et al. Functional properties and expression quantitative trait loci for phosphate transporter GmPT1 in soybean. Plant Cell Environ. 37, 462–467 (2014).
Chen, P., Zhang, X. L., Zhao, T. J., Li, Y. & Gai, J. Y. Genome-wide identification and characterization of RBR ubiquitin ligase genes in soybean. PLoS One 9, e87282 (2014).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Higo, K., Ugawa, Y., Iwamoto, M. & Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300 (1999).
Wang, S. B. et al. Evaluation of appropriate reference genes for gene expression studies in pepper by quantitative real-time PCR. Mol. Breeding 30, 1393–1400 (2012).
Nordgård, O., Kvaløy, J. T., Farmen, R. K. & Heikkilä, R. Error propagation in relative real-time reverse transcription polymerase chain reaction quantification models: the balance between accuracy and precision. Anal. Biochem. 356, 182–193 (2006).
Saeed, A. I. et al. TM4 microarray software suite. Methods Enzymol. 411, 134–193 (2006).
Nisi, P. D. & Zocchi, G. Phosphoenolpyruvate carboxylase in cucumber (Cucumis sativus L.) roots under iron deficiency: activity and kinetic characterization. J. Exp. Bot. 51, 1903–1909 (2000).
Bradford, M. M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31371645), the National High-tech R & D Program of China (2013AA102602), the Fundamental Research Funds for the Central Universities, the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT13073), the Program for New Century Excellent Talents in University (NCET-12-0891), the MOE 111 Project (B08025), the Program for MOA Innovative Research Team, the Program for High-level Innovative and Entrepreneurial Talents in Jiangsu Province, and the Jiangsu Higher Education PAPD Program. We thank the staff at Jiangpu Experimental Station for their assistance with soybean seed production.
Author information
Authors and Affiliations
Contributions
N.W., X.Z., Y.C., T.W., S.Y. performed the experiments, N.W. carried out the analyses and drafted the manuscript, Y.L. and J.G. conceived the study, N.W. and Y.L. revised the manuscript. All authors reviewed and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, N., Zhong, X., Cong, Y. et al. Genome-wide Analysis of Phosphoenolpyruvate Carboxylase Gene Family and Their Response to Abiotic Stresses in Soybean. Sci Rep 6, 38448 (2016). https://doi.org/10.1038/srep38448
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep38448
This article is cited by
-
Comparative Analysis of Root Transcriptome Profiles of Sesame (Sesamum indicum L.) in Response to Osmotic Stress
Journal of Plant Growth Regulation (2021)
-
Transcriptional analyses of two soybean cultivars under salt stress
Molecular Biology Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.