Abstract
Confocal endomicroscopy is an emerging imaging technology that has recently been introduced into the clinic to instantaneously collect “optical biopsies” in vivo with histology-like quality. Here, we demonstrate a fast scanner located in the distal end of a side-viewing instrument using a compact lens assembly with numerical aperture of 0.5 to achieve a working distance of 100 μm and field-of-view of 300 × 400 μm2. The microelectromechanical systems (MEMS) mirror was designed based on the principle of parametric resonance and images at 5 frames per second. The instrument has a 4.2 mm outer diameter and 3 cm rigid length, and can pass through the biopsy channel of a medical endoscope. We achieved real time optical sections of NIR fluorescence with 0.87 μm lateral resolution, and were able to visualize in vivo binding of a Cy5.5-labeled peptide specific for EGFR to the cell surface of pre-cancerous colonocytes within the epithelium of dysplastic crypts in mouse colon. By performing targeted imaging with endomicroscopy, we can visualize molecular expression patterns in vivo that provide a biological basis for disease detection.
Similar content being viewed by others
Introduction
Many human diseases originate within the epithelium of hollow organs and ducts1. This thin layer of tissue has dimensions of only a few hundred microns2, a simple repetitive architecture3, and intense self-renewal kinetics4. Optical sectioning is being developed to visualize molecular expression patterns unique to biological processes that drive disease progression with sub-cellular resolution. For in vivo imaging, these instruments must be made sufficiently small in size and fast in speed. Confocal endomicroscopes have been developed for this purpose with front-viewing optics, and are now being used routinely in the clinic5,6. Front-view optics requires instrument placement perpendicular to the tissue surface. This can be difficult in areas where space to maneuver is limited, such as in colon, biliary tract, and pancreatic duct. Side-viewing optics may be more useful for these imaging applications.
Confocal endomicroscopy is an emerging imaging technology that is being developed to instantaneously collect “optical biopsies” in vivo with histology-like quality7. These instruments use the core of a single mode optical fiber that acts as a “pinhole” to reject light scattered by tissue. A flexible fiber is used to deliver and collect light through a medical endoscope. Current clinical endomicroscopes are limited in imaging performance by simple scanning mechanisms that are either too slow in speed resulting in motion artifacts or too bulky in dimension requiring a proximal location that reduces resolution. A fast compact scanner placed in the distal end of the endomicroscope can provide improved performance and flexibility for instrument control. We have previously developed a miniature scanner to perform high speed in vivo imaging8. This design uses parametric resonance where drive signals are applied at frequencies near 2ωo/n (ωo is the natural frequency of vibrational modes and n is an integer ≥1)9. These electrostatic devices were fabricated with compact dimensions using micro-electro-mechanical systems (MEMS) processes10.
In clinical confocal endomicroscopy, contrast has been generated using non-specific methods, such as reflectance11, endogenous fluorescence12, topically applied intra-vital dyes13, and intravenously injected fluorescein14. Targeted imaging agents have recently been developed that detect molecules that are specific to disease and have known biological function. These contrast agents require high resolution in vivo imaging to validate specific binding to the intended target. We have previously identified a peptide specific for domain 2 of EGFR15, a cell surface target that is overexpressed in up to 80% of human colonic adenomas16 and 97% of adenocarcinomas17. We have demonstrated targeted images from the mucosal surface of mouse colonic adenomas in vivo with topical administration using wide-field fluorescence endoscopy15. EGFR is also overexpressed in most epithelial-derived cancers, including breast18, esophagus19, head and neck20, lung21, and pancreas22, and is the target for a number of antibody and small molecule therapies, such as cetuximab and gefitinib23,24. Here, we aim to systemically administer a peptide specific for EGFR, and demonstrate use of a high resolution side-viewing endomicroscope to validate specific cell surface binding to dysplastic colonocytes in vivo.
Results
Side-viewing confocal endomicroscope
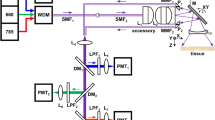
The schematic for a side-viewing endomicroscope is shown, Fig. 1A. A laser provides excitation at λex = 660 nm. We performed ray trace simulations to design the optics to achieve diffraction-limited resolution with a working distance of 100 μm, Fig. 2A–D. This imaging depth is representative of the thickness of the epithelium in mouse colon. Only lenses with outer diameter <3 mm were considered for use. Commercially available lenses were chosen to demonstrate proof-of-concept. The illumination beam is reflected by a dichroic mirror M1, focused by lens L1 into a 2 meter long single mode optical fiber (SMF), collimated by L2, and focused by L3. The beam is reflected 90° for side-viewing by the MEMS mirror M2 into a solid immersion lens L4 that provides contact with tissue and index matching. Near-infrared (NIR) fluorescence is collected by the same optics, transmits through the SMF optical fiber, passes through the dichroic M1, reflects off the static mirror M3, passes through band pass filter F1 (696–736 nm), focused by lens L5 into a 1 meter long multimode fiber (MMF), and transmits to the photomultiplier tube (PMT) detector. This design produced an effective numerical aperture (NA) of 0.5 with a field-of-view (FOV) of 300 × 400 μm2. Full instrument details are provided in Methods.
The packaging scheme for the mirror, optics, and fiber is shown, Fig. 3A. The assembled endomicroscope is shown passed through a 6 mm diameter biopsy channel of a medical endoscope, Fig. 3B. The rigid end of the endomicroscope was soaked in distilled water overnight to ensure an adequate seal. No leaks were observed after more than 20 in vivo uses in mouse colon.
Side-viewing endomicroscope.
(A) CAD drawing shows how instrument is packaged. (B) Dimensions allow for endomicroscope to pass through a 6 mm diameter biopsy channel of Olympus GIF XTQ160 endoscope. (C) Lateral resolution of 0.87 μm was measured from width of transition from 10% to 90% of maximum intensity from profile across knife-edge target. (D) Axial resolution of 13.2 μm was measured from full-width-at-half-maximum (FWHM) of intensity profile produced by translating a plane mirror in axial direction at a neutral position of 100 μm.
We plot the intensity profile along a knife-edge target to measure a lateral resolution of 0.87 μm, defined as the distance spanned by 10% to 90% of maximum intensity, Fig. 3C. We measured the signal collected while vertically translating a reflective surface, and measured a full-width-at-half-maximum (FWHM) of 13.2 μm for the axial resolution, Fig. 3D. We collected reflectance images from a standard resolution target to qualitatively estimate a lateral resolution <2 μm (group 7, element 6 in expanded view of box), Fig. 4A. A reflectance image from a target with 100 × 100 μm2 square grids showed a FOV of 300 × 400 μm2, Fig. 4B. Mild distortion in the image results from use of the scan mirror that rotates about separate axes, and can be corrected with a look-up table, if needed.
Image parameters.
(A) Reflectance image of standard resolution target (USAF 1951) qualitatively supports lateral resolution of <2 μm. Vertical and horizontal bars from group 7, element 6 are clearly seen in expanded view of box. (B) Reflectance image of grid with 100 × 100 μm2 squares shows a field-of-view of ∼300 × 400 μm2. Note: interference patterns seen in either image result from multiple reflections between the outer face of the distal lens L4 and the reflective surface of the resolution target. This condition does not occur when imaging tissue. The corners of the image appear darker due to curvature of the image plane when scanning. The focal spots in the corners are offset by 7 μm compared to the center. Images were collected at a working distance of ~100 μm.
MEMS scanner
We have previously designed, fabricated, and verified a compact 2D MEMS scanner with chip size of 3 × 3 mm2 and a 1.8 mm diameter circular reflector, Fig. 5A25. We reduced the diagonal dimension to 3.5 mm by cutting the four corners to match the reduced dimensions of the side-viewing endomicroscope. The reflector is mounted on a gimbal frame to minimize cross-talk between axes. Orthogonal sets of electrostatic comb-drive actuators are coupled to inner and outer torsional springs designed to achieve resonant frequencies near ~4 and ~1 kHz, respectively. A Lissajous scan pattern is created with a frame rate ≥5 Hz to cover 400 × 400 pixels. Aluminum was sputtered on the front-side silicon surface to achieve reflectivity >90% between 400 and 700 nm. We mounted the mirror onto a MEMS holder using copper pins to wire bond the electrical contacts, Fig. 5B. We measured the angular deflection of the MEMS scanner in either axis to characterize the frequency response to a sine-wave drive signal at 60 Vpp. For in vivo imaging, we used frequencies of 8210 and 1960 Hz to produce tilting frequencies at 4105 Hz and 985 Hz in the X and Y axes, respectively, Fig. 5C,D. We packaged the assembly in a 4.2 mm diameter stainless steel tube. The distance between lens L3 and the 2D resonant mirror M2 was adjusted to achieve a working distance of 100 μm. For in vivo imaging, the endomicroscope was mounted onto a motorized rotational and translational platform to control the position of the distal end.
MEMS scanner.
(A) An aluminum-coated reflector is mounted on a gimbal frame and is driven by orthogonal sets of electrostatic comb-drive actuators coupled to inner and outer torsional springs to produce rapid Lissajous scanning. (B) Mirror is secured by custom holder using copper pins as contacts to wire bond electrical connections. Frequency response shows mechanical scan angles for (C) inner and (D) outer axes with a drive voltage of 60 Vpp with either an upsweep (low-to-high) or downsweep (high-to-low) in frequency. The inner and outer axes were driven at 8210 Hz and 1960 Hz to achieve mechanical deflections of 5.5 and 3.5 degrees, respectively.
In vivo imaging of EGFR expression in mouse colonic adenoma
We performed in vivo imaging first with wide-field endoscopy using white light illumination in the colon of n = 6 CPC;Apc mice, Fig. 6A. These genetically-engineered animals develop adenomas spontaneously on the side wall of the colon that can be difficult to access with front-viewing optics26. We defined the distance between the endoscope tip and the anus and the clockwise location of the adenoma as landmarks to guide subsequent placement of the endomicroscope. We performed an intraperitoneal injection of Cy5.5-labeled peptide QRH* at a concentration of 300 μM in a volume of 600 μL and molecular weight = 1336.48. After ∼90 minutes, we observed peak fluorescence intensity from the adenoma, Fig. 6B. We then inserted the side-viewing endomicroscope, and used a motorized rotational and translational platform to position the distal tip onto the adenoma using the landmarks as a guide, Fig. S1. Optical sections were collected in vivo using a laser power of 1.5 mW. We observed strong binding of the EGFR peptide to the cell surface of dysplastic colonocytes (arrow), Fig. 6C, and minimal staining to normal colonic epithelium, Fig. 6D. A video (visualization) shows real time images being collected at 5 frames per sec while the side-viewing endomicroscope moves from an adenoma to normal colonic mucosa. We quantified each image by measuring the average fluorescence intensity from n = 5 regions of interest (ROI) with dimensions of 20 × 20 pixels that represented a range of intensities for dysplasia and adjacent normal. We found significantly greater mean fluorescence intensities in vivo for dysplasia compared with normal colonic mucosa, Fig. 6E.
Imaging of mouse colonic dysplasia.
(A) White light image of CPC;Apc mouse colon collected in vivo with wide-field endoscope shows location of spontaneous adenoma (arrow). (B) Maximum uptake of the NIR-labeled EGFR peptide QRH*-Cy5.5 is seen from the adenoma ~90 minutes after systemic administration. From the in vivo confocal images, (C) increased fluorescence intensity is seen from cell surface of dysplastic colonocytes (arrow) while (D) minimal staining is seen in normal crypts. Real time video (visualization) of side-viewing endomicroscope moving from adenoma to normal is shown. (E) A significantly greater mean fluorescence intensity was measured from dysplasia compared with that of adjacent normal, 107 ± 19 versus 62 ± 8 AU, from a group of n = 6 mice, P = 3.5 × 10−4 by paired two-way t-test.
Validation of EGFR expression in mouse colonic epithelium
After completion of endomicroscopy, we euthanized the animals and performed macroscopic imaging of excised colonic mucosa to validate selective uptake of the EGFR peptide in adenomas. Representative NIR fluorescence images were collected using a Cy5.5 filter set (λex = 675 nm excitation, λem = 720 nm emission, 0.05 sec exposure), and showed increased fluorescence intensities from adenomas compared with adjacent normal colonic mucosa, Fig. 7A. A ruler was placed next to the specimen to confirm the distance of the adenomas from the anus to register the images. Fluorescence intensities were measured from ROI (dotted boxes) around dysplasia (target) and adjacent normal (background). The target-to-background (T/B) ratio for each mouse was calculated by dividing the average intensity from a ROI of dysplasia with that from normal. We measured a T/B ratio of 3.47 ± 1.87 from n = 6 mice. Co-registration with white light images further confirmed the location of the adenomas, Fig. 7B.
Uptake of EGFR peptide in adenomas.
(A) Representative NIR fluorescence image collected ex vivo show selective uptake of QRH*-Cy5.5 in adenomas compared with adjacent normal colonic mucosa. Intensities were measured from the ROIs defined by the dotted rectangles. We calculated a mean T/B ratio of 3.47 ± 1.87 from the group of n = 6 mice. (B) Co-registered white light image of exposed mucosal surface confirms locations of adenomas.
We then validated specific binding of the EGFR peptide to pre-cancerous crypts using our endomicroscope on mouse colonic mucosa ex vivo and compared our images with that collected using a tabletop confocal microscope. Increased intensity (arrow) is seen at the cell surface of dysplastic colonocytes, Fig. 8A,B. We observed greatly reduced signal from the EGFR peptide for normal colon, Fig. 8D,E. Representative histology (H&E) is shown for dysplasia and normal, Fig. 8C,F.
Immunofluorescence of mouse colon ex vivo.
Strong fluorescence intensity is seen from binding of the EGFR peptide QRH*-Cy5.5 to the cell surface (arrow) of dysplastic colonocytes using the (A) side-viewing confocal endomicroscope and (B) a conventional tabletop confocal microscope. (C) Representative histology (H&E) for dysplasia. Minimal fluorescence intensity is seen for normal colonocytes using the (D) side-viewing confocal endomicroscope and (E) a conventional tabletop confocal microscope. (F) Representative histology (H&E) for normal colonic mucosa.
Panorama view
We rapidly created mosaic images of colonic mucosa to enlarge the FOV. We used the motorized platform to rapidly rotate and translate the endomicroscope along the colon lumen. ~1 hour after injection of Cy5.5, we collected a panorama image of normal colonic mucosa in vivo while translating the endomicroscope a distance of 1 mm and rotating the instrument by 90°, Fig. S2A. We simulated the in vivo anatomy of colon by warping the image in custom software, Fig. S2B.
Discussion
Here, we demonstrate a side-viewing confocal endomicroscope that uses a fast, compact, distal scanner to collect real time in vivo optical sections in colonic epithelium. Using MEMS technology, we were able to scale down the dimensions of the device for efficient packaging and perform rapid beam scanning. This capability provides significantly improved imaging performance in terms of speed and resolution compared with other endomicroscope designs currently used in the clinic. The instrument showed stable operation at 5 frames per second with minimal interference from mechanical vibrations while imaging. With further optimization, we can achieve frame rates as high as 10 per second. Using a Cy5.5 labeled peptide, we were able to collect bright optical sections of EGFR expression in mouse colonic epithelium at a depth of ∼100 μm. After completion of packaging, the scanner performed reliably, and was able to collect images continuously without any noticeable phase shifts at fixed drive frequencies. We fine tuned the phase at the beginning of each experiment. Position sensing of the MEMS device may be needed for use in more versatile environments.
We collected NIR fluorescence images with sub-micron lateral resolution. At an imaging depth of 100 μm, we were able to visualize binding of a Cy5.5-labeled peptide specific for EGFR to the cell surface of colonocytes within the epithelium of dysplastic crypts. The extra-cellular domain of EGFR in human and mouse has 97.5% homology, and is 100% conserved in domain 2 where the EGFR peptide binds27. We used the CPC;Apc mouse that has been genetically engineered to sporadically delete an APC gene26, which is mutated in >80% of human colorectal carcinomas28. Adenomas protrude spontaneously from the side wall in the distal colon where they are more readily accessible to imaging with a side-viewing rather than front-viewing endomicroscope. EGFR is overexpressed in the vast majority of human colonic adenomas16, thus may serve as an early target for detection of colorectal cancer. These results were validated ex vivo on macroscopic imaging and with immunofluorescence using a conventional tabletop confocal microscope with the EGFR peptide and a known EGFR antibody.
We used a peptide to visualize EGFR expression in colonic adenomas because of their small size and low molecular weight by comparison to antibodies. These properties result in better vascular permeability, enhanced tissue diffusion, and greater tumor penetration for improved targeting29. Also, peptides clear rapidly for reduced background and less biodistribution in non-target tissues30. Peptides have less potential for immunogenicity than antibodies, which allows for repetitive use31. We used Cy5.5, a NIR fluorophore, to label the peptide to maximize imaging depth, avoid hemoglobin absorption, reduce tissue scattering, and minimize autofluorescence background32. By performing endomicroscopy with targeted imaging agents, we can visualize molecular expression patterns in vivo that provide a biological basis for disease detection. By comparison with static biopsies from ex vivo specimens, real time in vivo imaging may provide a more robust approach to evaluate pathology of neoplastic lesions and has potential for use in point-of-care diagnostics. This approach may also identify patients most likely to respond to targeted therapies.
By comparison, the Optiscan ISC-1000® uses a tuning fork mechanism to mechanically scan a single mode optical fiber. In vivo images are acquired at a slow frame rate of 0.8 to 1.6 per second, and imaging can be limited by motion artifacts from gut peristalsis, respiratory motion, and heart beats33. The Mauna Kea Technologies Cellvizio® uses a flexible optical fiber bundle that ranges in diameter from 0.3 to 4.2 mm. The smaller probes can be used to image small caliber hollow organs, such as the biliary tract and pancreatic duct34. Lateral scanning is performed using galvo mirrors located at the proximal end. The image resolution is limited by the number of individual fibers in the bundle33. The fibers are hexagonally packed, and the space in between fibers constitutes a large fraction of area where no light transmits. The fiber cores are multi-mode, and not optimal for rejecting scattered light. Side-viewing endomicroscopy has also been demonstrated using a 50 mm long graded-index (GRIN) lens coupled to a prism that deflects the beam at 90° 35. Scanning is also performed at the proximal end using a bulky galvo mirror. Long rigid GRIN lenses incur chromatic aberrations that may need correction when imaging fluorescence, and are not compatible with flexible medical endoscopes. Side-viewing has also been used in a spectrally encoded confocal endomicroscope (SECM)36. This design uses a diffraction grating to spatially deflect different wavelengths of light delivered from a broad spectrum illumination source. Images are formed by mechanically rotating and translating the optics. This technique is limited to collection of reflectance, and has not been used to image molecular expression.
To our knowledge, we have demonstrated the first side-viewing confocal endomicroscope using a MEMS-based scanner. These devices can provide fast imaging speeds in a small footprint. Scanning in the distal tip of the endomicroscope provides greater flexibility for image formation. Other MEMS-based instruments include a 7 mm diameter front-viewing confocal endomicroscope that uses an electrothermal mirror. Lateral scanning is performed with ±26° of deflection, and a FOV of 180 × 180 mm2 is produced37. A separate electrothermal actuator consisting of 4 bimorphs provides ~400 μm of out-of-plance displacement. The lateral and axial resolutions were found to be 1 and 7 μm, respectively. Also, a 5.5 mm diameter front-viewing dual axes confocal endomicroscope was developed using a lateral scanning electrostatic MEMS mirror38. The instrument achieved a FOV of 362 × 212 μm2, and a micromotor was used to image at a depth of 140 μm. The lateral and axial resolution was 5 and 6.5 μm, respectively, and images were collected at 5 frames per sec. A handheld 12 mm diameter dual axes confocal microscope has been demonstrated using a lateral scanning electrostatic MEMS mirror with a resonance frequency of ~200 Hz39. Line scanning was performed to improve the imaging speed to 16 frames per sec. A custom, multi-element objective lens was to achieve a lateral and axial resolution of 1.1 and 2 μm, respectively, with a FOV of 300 × 300 μm2.
Our endomicroscope design has potential to be used as an adjunct to wide-field endoscopy by providing instantaneous “optical” biopsy. Targeted imaging can be used to stage early cancer, guide choice for therapy, and monitor therapeutic response. Furthermore, this instrument can quickly evaluate numerous tissue sites to minimize the number of physical biopsies collected, risk for bleeding, and procedure time. It may also be useful for detecting pre-malignant lesions that may otherwise be missed because of flat appearance, focal size, and patchy distribution. In the future, we can add the capability to perform depth scanning by using a MEMS device that performs out-of-plane displacement8. We found side-viewing to be effective for in situ imaging of adenomas located on the side wall of mouse colon that would otherwise be inaccessible with a front-viewing geometry. For the same reason, this instrument can potentially be useful in the clinic to provide real-time pathology and assess important clinical conditions, such as flat colonic adenomas, indeterminant biliary strictures, and pancreatic intraepithelial (PanIN) lesions.
Methods
Side-viewing confocal endomicroscope
We performed ray-trace simulations (ZEMAX, ver 13) to design optics for the side-viewing endomicroscope. Our design criteria included diffraction-limited resolution on axis with working distance of 100 μm. We used a solid-state diode laser (660-S, Toptica Photonics) to deliver fluorescence excitation at λex = 660 nm. The beam is reflected by a dichroic mirror M1 (FF685-Di02–25 × 36, Semrock) and focused by lens L1 (f = 11 mm, C220TMD-B, Thorlabs) into a 2 meter long single mode optical fiber (SMF, SM600, Thorlabs). The illumination is collimated by lens L2 (f = 12 mm, 45–262, Edmund Optics) to a beam diameter of 3 mm and focused by lens L3 (f = 6 mm, 45–089, Edmund Optics). The focused beam is reflected 90° by the 2D resonant MEMS mirror M2 into a solid immersion lens L4 with (f = 1 mm, OD 1.5 mm, 65–261, Edmund Optics). For L2, L3, and L4, only commercially available lenses with outer diameter <3 mm were considered for endoscope compatibility.
NIR fluorescence (dashed red arrows, Fig. 1) is collected by the same optics and travels in the opposite direction along the same path, is descanned by M2, and focused into the same single mode fiber (SMF). After exiting the fiber, fluorescence is collimated by L1, passes through the dichroic M1, reflected by M3, and passes through a band pass filter F1 (FF01-716/40-25, Semrock) that transmits from 696 to 736 nm with >93% transmission efficiency, and is focused by lens L5 (f = 6.24 mm, C110TMD-B, Thorlabs) into a 1 meter long multimode fiber (MMF, FT400UMT, Thorlabs). The multimode fiber transmits fluorescence to a photomultiplier tube (PMT) detector (H7422–40, Hamamatsu). The PMT signal is amplified by a high-speed current amplifier (59–178, Edmund Optics). A high-speed multi-function data acquisition board (National Instruments, PCI-6115) is used to digitize the amplified PMT signal. The same board was used to generate control signals to drive the 2D MEMS scanner. The data acquisition board was controlled using custom software (LabVIEW, National Instruments).
MEMS scanner
A compact scanner was designed and fabricated using micro-electro-mechanical systems (MEMS) processes. The reflector has diagonal dimensions of 3.5 mm, and is mounted on a gimbal frame to minimize vibrational cross-talk between orthogonal axes. The mirror is rotated about either axis by drive signals applied to panels of electrostatic comb-drive actuators coupled to inner and outer torsional springs. Custom software was developed in LabView to drive the MEMS scanner and reconstruct the image by remapping the time series signal in a 2D matrix.
At the center of the FOV of the endomicroscope, we measured a lateral resolution of ∼0.90 μm. To satisfy the Nyquist sampling criteria, we must sample with spacing <0.45 μm between pixels. We used different scanning parameters for in vivo versus ex vivo images. For in vivo imaging, we used tilting frequencies of 4105 and 985 Hz to create a Lissajous scan pattern with 400 × 400 pixels at 5 frames per sec to cover a FOV of 300 × 300 μm2. The pixel spacing is 0.75 μm, and we are under sampling a little to maintain a higher frame rate to incur less motion artifact. For ex vivo imaging, we used frequencies of 4105 and 976 Hz to create a scan pattern with 1000 × 1000 pixels at 1 frame per sec to cover a FOV of 300 × 400 μm2. The pixel spacing is ≤0.4 μm, and meets the Nyquist sampling criteria.
We used computer-aided design (CAD) software (Solidworks, ver 2015) to design the housing to package the optics and scanner. We used an stainless steel tube with an outer diameter (OD) of 3.4 mm and an inner diameter (ID) of 3 mm to center and align the lenses L2 and L3, and a second stainless steel tube with OD 4.2 mm and ID 3.4 mm to enclose the MEMS scanner and lens L4. We fabricated the MEMS holder with OD 3.8 mm using M3 Crystal, an acrylic based resin with high strength and stiffness. We used a high precision 3D printer (ProJet 3500HD MAX, 3DSystems) with 16 μm resolution. A total of 3 copper pins were inserted for wire bonding. The distance between L3 and M2 was adjusted to achieve a working distance of 100 μm. The position of M2 was adjusted using a 6-axis precision manipulator to ensure that the reflected beam was at normal incidence to L4. After assembly, the probe was sealed using epoxy (G14250, Thorlabs).
We verified the dimension of the packaged endomicroscope by passing it through the 6 mm diameter instrument channel of a medical endoscope (Olympus GIF XTQ160). Prior to use, the distal tip of the endomicroscope was soaked in distilled water overnight to ensure an adequate seal. For in vivo imaging, the endomicroscope was mounted onto a rotational and translational platform driven by a motorized linear stage (CONEX-LTA-CC, Newport) and a stepper motor (324, Adafruit) to precisely control the distal end of the endomicroscope in the mouse colon.
In vivo imaging of EGFR expression in mouse colonic adenoma
All experimental procedures were performed in accordance with relevant guidelines and regulations of the University of Michigan, and the mouse imaging studies and all experimental procedures were conducted with approval by the University Committee on the Use and Care of Animals (UCUCA). The mice were housed in pathogen-free conditions and supplied water ad libitum under controlled conditions of humidity (50 ± 10%), light (12/12 hour light/dark cycle) and temperature (25 °C). We have previously developed a peptide that is specific for domain 2 of EGFR15. We synthesized the monomeric linear peptide with sequence QRHKPRE, hereafter QRH*, with linker GGGSK using standard Fmoc-mediated solid-phase synthesis40. QRH* was labeled with a water soluble NIR fluorophore using sulfo-Cy5.5- N-hydroxysuccinimide ester (Lumiprobe LLC). After lyophilization, the peptides were stored at −80 °C.
We used a CPC;Apc mouse that spontaneously develops adenomas in the distal colon where they are accessible by the side-viewing endomicroscope. During imaging, 12-week-old CPC;Apc mice were anesthetized with 2% isoflurane (Fluriso; MWI Veterinary Supply Co.). We first used a wide-field small animal endoscope (27030BA, Karl Storz Veterinary Endoscopy) with white light illumination to examine the colon for presence of grossly visible adenomas. We waited ∼90 minutes after intraperitoneal injection of QRH*-Cy5.5 to achieve maximum target uptake. The distal tip of the endomicroscope was placed into the colon of anesthetized mice. The insertion depth and angle were controlled using the translational stage and the rotational fixture in order to image normal colonic mucosa and adenomas at the same locations as first detected on the wide-field endoscope.
Live video streams were collected ~100 μm below the mucosa surface at 5 frames/sec using a laser power of 1.5 mW. Individual frames with minimal motion artifacts were identified. After completion of imaging, the mice were euthanized, and the colon was harvested for ex vivo imaging. The optical window of probe was positioned directly en face over the exposed mucosal surface of resected colon to optimize contact. Images were collected with the same laser power as that used in vivo. Previously identified landmarks were used to confirm the locations of the adenomas. The colon was fixed in 10% buffered formalin and paraffin embedded for routine histology (H&E). Images and tissues were collected and analyzed in a total of n = 6 mice. One representative image was selected from either dysplasia or normal for each mouse. We quantified each image by measuring the average fluorescence intensity from n = 5 regions of interest (ROI) with dimensions of 20 × 20 pixels that represented a range of intensities for dysplasia and adjacent normal.
Validation of EGFR expression in mouse colonic epithelium
After completion of imaging, the animals were euthanized, and the colon was excised, flushed with PBS, and divided longitudinally for macroscopic imaging (IVIS 200, Caliper Life Sciences), to validate specific uptake by the EGFR peptide. NIR fluorescence images were collected using a Cy5.5 filter with λex = 675 nm excitation and 720 nm emission with 0.05 sec exposure. A ruler was placed next to the specimen to determine the distance from the anus for registration with the endoscopy and histology images. Whole mount tissue was imaged with a conventional tabletop confocal microscope (Leica TCS SP5 Microsystems) using Cy5.5 and DAPI filters. The colon was then fixed in 10% buffered formalin and processed for routine histology (H&E).
Panorama view
The side-viewing endomicroscope can be rapidly rotated and translated in the lumen of the colon to enlarge the dimensions of the images collected using the motorized platform. We first injected Cy5.5, and waited approximately 1 hour for the dye to circulate. We collected images in a video stream of normal colonic mucosa in vivo while translating the endomicroscope a distance of 1 mm and rotating the instrument by 90°. We stitched together individual images to form a mosaic and warped the composite image to simulated the in vivo anatomy of colon using custom software (Matlab, Mathworks Inc).
Additional Information
How to cite this article: Duan, X. et al. Visualizing epithelial expression of EGFR in vivo with a distal scanning side-viewing confocal endomicroscope. Sci. Rep. 6, 37315; doi: 10.1038/srep37315 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Torre, L. A., Bray, F., Siegel, R. L., Ferlay, J., Lortet-Tieulent, J. et al. Global cancer statistics, 2012. CA Cancer J Clin. 65, 87–108 (2015).
Wang, T. D., Van Dam, J., Crawford, J. M., Preisinger, E. A., Wang, Y. et al. Fluorescence endoscopic imaging of human colonic adenomas. Gastroenterology. 111, 1182–1191 (1996).
Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell. 154, 274–284 (2013).
Barker, N., Ridgway, R. A., van Es, J. H., van de Wetering, M., Begthel, H. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 457, 608–611 (2009).
Kiesslich, R., Burg, J., Vieth, M., Gnaendiger, J., Enders, M. et al. Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo. Gastroenterology. 127, 706–713 (2004).
Wang, T. D., Friedland, S., Sahbaie, P., Soetikno, R., Hsiung, P. L. et al. Functional imaging of colonic mucosa with a fibered confocal microscope for real-time in vivo pathology. Clin Gastroenterol Hepatol. 5, 1300–1305 (2007).
Wang, T. D. Confocal Microendoscopy – From the Bench to the Bedside. Gastrointestinal Endoscopy. 62, 696–697 (2005).
Li, H., Qiu, Z., Duan, X., Oldham, K. R., Kurabayashi, K. et al. A Monolithically-Integrated 3D MEMS Scanner for Switchable Real-time Vertical/Horizontal Cross-sectional Imaging in Dual Axes Confocal Endomicroscope. Optics Express. 24, 2145–2155 (2016).
Turner, K. L., Miller, S. A., Hartwell, P. G., MacDonald, N. C., Strogatz, S. H. et al. Five parametric resonances in a microelectromechanical system. Nature. 396, 149–152 (1998).
Lee, K., Krisnamoorthy, K., Yu, K. & Solgaard, O. Single-crystalline silicon micromirrors actuated by self-aligned vertical electrostatic comb drives with piston-motion and rotational capabilities. Sensors and Actuators A: Physical. 114, 423–428 (2004).
Maitland, K. C., Gillenwater, A. M., Williams, M. D., El-Naggar, A. K., Descour, M. R. et al. In vivo imaging of oral neoplasia using a miniaturized fiber optic confocal reflectance microscope. Oral Oncol. 44, 1059–1066 (2008).
Thiberville, L., Moreno-Swirc, S., Vercauteren, T., Peltier, E., Cavé, C. et al. In vivo imaging of the bronchial wall microstructure using fibered confocal fluorescence microscopy. Am J Respir Crit Care Med. 175, 22–31 (2007).
Thiberville, L. & Salaün, M. Bronchoscopic advances: on the way to the cells. Respiration. 79, 441–449 (2010).
Wallace, M. B., Meining, A., Canto, M. I., Fockens, P., Miehlke, S. et al. The safety of intravenous fluorescein for confocal laser endomicroscopy in the gastrointestinal tract. Aliment Pharmacol Ther. 31, 548–552 (2010).
Zhou, J., Joshi, B. P., Duan, X., Pant, A., Qiu, Z. et al. EGFR Overexpressed in Colonic Neoplasia Can be Detected on Wide-Field Endoscopic Imaging. Clin Transl Gastroenterol. 6, e101 (2015).
Porebska, I., Harlozińska, A. & Bojarowski, T. Expression of the tyrosine kinase activity growth factor receptors (EGFR, ERB B2, ERB B3) in colorectal adenocarcinomas and adenomas. Tumour Biol. 21, 105–115 (2000).
Spano, J. P., Lagorce, C., Atlan, D., Milano, G., Domont, J. et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 16, 102–108 (2005).
Bhargava, R. I., Gerald, W. L., Li, A. R., Pan, Q., Lal, P. et al. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 18, 1027–1033 (2005).
Hanawa, M., Suzuki, S., Dobashi, Y., Yamane, T., Kono, K. et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer. 118, 1173–1180 (2006).
Reuter, C. W., Morgan, M. A. & Eckardt, A. Targeting EGF-receptor-signalling in squamous cell carcinomas of the head and neck. Br J Cancer. 96, 408–416 (2007).
Hirsch, F. R., Varella-Garcia, M., Bunn, P. A., Di Maria, M. V., Veve, R. et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 21, 3798–3807 (2003).
Jimeno, A., Tan, A. C., Coffa, J., Rajeshkumar, N. V., Kulesza, P. et al. Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res. 68, 2841–2849 (2008).
Van Cutsem, E., Köhne, C. H., Hitre, E., Zaluski, J., Chang Chien, C. R. et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 360, 1408–1417 (2009).
Paez, J. G., Jänne, P. A., Lee, J. C., Tracy, S., Greulich, H. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 304, 1497–1500 (2004).
Duan, X., Li, H., Qiu, Z., Joshi, B. P., Pant, A. et al. MEMS-based multiphoton endomicroscope for repetitive imaging of mouse colon. Biomed Opt Express. 6, 3074–3083 (2015).
Hinoi, T., Akyol, A., Theisen, B. K., Ferguson, D. O., Greenson, J. K. et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res. 67, 9721–9730 (2007).
Choi, Y. S., Yoon, S., Kim, K. L., Yoo, J., Song, P. et al. Computational design of binding proteins to EGFR domain II. PLoS One. 9, e92513 (2014).
Rowan, A. J., Lamlum, H., Ilyas, M., Wheeler, J., Straub, J. et al. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci. 97, 3352–3357 (2000).
Lee, S., Xie, J. & Chen, X. Peptides and peptide hormones for molecular imaging and disease diagnosis. Chem Rev. 110, 3087–3111 (2010).
Thurber, G. M., Schmidt, M. M. & Wittrup, K. D. Antibody tumor penetration: transport opposed by systemic and antigen-mediated clearance. Adv Drug Deliv Rev. 60, 1421–1434 (2008).
Wu, C. H., Kuo, Y. H., Hong, R. L. & Wu, H. C. α-Enolase-binding peptide enhances drug delivery efficiency and therapeutic efficacy against colorectal cancer. Sci Transl Med. 7, 290ra91 (2015).
Hilderbrand, S. A. & Weissleder, R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol 14, 71–79 (2010).
Wang, T. D. In-Vivo Microscopy in Translational Multi-Modality Optical Imaging. (ed. Intes, X., Azar, F. ) 19–32 (Artech House, 2008).
Slivka, A., Gan, I., Jamidar, P., Costamagna, G., Cesaro, P. et al. Validation of the diagnostic accuracy of probe-based confocal laser endomicroscopy for the characterization of indeterminate biliary strictures: results of a prospective multicenter international study. Gastrointest Endosc. 81, 282–290 (2015).
Kim, P., Chung, E., Yamashita, H., Hung, K. E., Mizoguchi, A. et al. In vivo wide-area cellular imaging by side-view endomicroscopy. Nat Methods. 7, 303–305 (2010).
Kang, D., Carruth, R. W., Kim, M., Schlachter, S. C., Shishkov, M. et al. Endoscopic probe optics for spectrally encoded confocal microscopy. Biomed Opt Express. 4, 1925–1936 (2013).
Liu, L., Wang, E., Zhang, X., Liang, W., Li, X. et al. MEMS-based 3D confocal scanning microendoscope using MEMS scanners for both lateral and axial scan, Sensors Actuators A Phys. 215, 89–95 (2014).
Piyawattanametha, W., Ra, H., Qiu, Z., Friedland, S., Liu, J. T. C. et al. In vivo near-infrared dual-axis confocal microendoscopy in the human lower gastrointestinal tract. J. Biomed. Opt. 17, 21102 (2012).
Yin, C., Glaser, A. K., Leigh, S. Y., Chen, Y., Wei, L. et al. Miniature in vivo MEMS-based line-scanned dual-axis confocal microscope for point-of-care pathology. Biomed. Opt. Express. 7, 251 (2016).
Fields, G. B. & Noble, R. L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res. 35, 161–214 (1990).
Acknowledgements
Funding was provided in part by National Institutes of Health R01 CA200007 (TDW), R01 EB020644 (KRO), and R01 CA193377 (TDW) and by gift funds from Mary L. Petrovich (TDW). We thank C Qiu for technical support.
Author information
Authors and Affiliations
Contributions
X.D., K.R.O., and T.D.W. conceived and designed the experiments; X.D. and H.L. developed the instrument; X.D. and Q.Z. performed the experiments; X.D. and T.D.W. analyzed the data; J.Z. contributed materials/analysis tools; X.D. and T.D.W. wrote the paper.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Duan, X., Li, H., Zhou, J. et al. Visualizing epithelial expression of EGFR in vivo with distal scanning side-viewing confocal endomicroscope. Sci Rep 6, 37315 (2016). https://doi.org/10.1038/srep37315
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37315
This article is cited by
-
Wide-field endoscope accessory for multiplexed fluorescence imaging
Scientific Reports (2023)
-
Cell-to-cell variation in gene expression and the aging process
GeroScience (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.