Abstract
For quantitative real-time PCR (qRT-PCR) analysis, the key prerequisite that determines result accuracy is the selection of appropriate reference gene(s). Goji (Lycium barbarum L.) is a multi-branched shrub belonging to the Solanaceae family. To date, no systematic screening or evaluation of reference gene(s) in Goji has been performed. In this work, we identified 18 candidate reference genes from the transcriptomic sequencing data of 14 samples of Goji at different developmental stages and under drought stress condition. The expression stability of these candidate genes was rigorously analyzed using qRT-PCR and four different statistical algorithms: geNorm, BestKeeper, NormFinder and RefFinder. Two novel reference genes LbCML38 and LbRH52 showed the most stable expression, whereas the traditionally used reference genes such as LbGAPDH, LbHSP90 and LbTUB showed unstable expression in the tested samples. Expression of a target gene LbMYB1 was also tested and compared using optimal reference genes LbCML38 and LbRH52, mediocre reference gene LbActin7, and poor reference gene LbHSP90 as normalization standards, respectively. As expected, calculation of the target gene expression by normalization against LbCML38, LbActin7 or LbHSP90 showed significant differences. Our findings suggest that LbCML38 and LbRH52 can be used as reference genes for gene expression analysis in Goji.
Similar content being viewed by others
Introduction
Fluorescent quantitative real-time PCR (qRT-PCR) is a fast, accurate method for nucleic acid analysis. Unlike the standard reverse transcription polymerase chain reaction (RT-PCR), which detects the reaction product at the end, qRT-PCR detects and quantifies the amplified target nucleic acid in “real time” by measuring accumulated fluorescent signal during each cycle of polymerization. Therefore, qRT-PCR is more specific, sensitive and reproducible compared with standard RT-PCR1. However, reference gene is required for qRT-PCR to adjust the initial cDNA levels and transcriptional efficiency to offset the variation in nucleic acid purity and concentration during sample preparation, and to avoid the errors generated during sample treatment2. Previous studies demonstrated that very few reference genes were absolutely stable, but were only “relatively” stable under certain conditions in specific types of cells or tissues3,4. To date, some reference genes including those encoding actin, α- and β-tubulin, GAPDH, EF1α and ubiquitin have been identified. However, expression of these reference genes varied with different treatments and at different developmental stages of plants, which greatly affected the accuracy of target gene expression evaluation1,5,6. Thus, stable reference gene screening and evaluation are essential for functional studies of target genes.
As an importantly dietary and medicinal plant, Goji (Lycium barbarum L., 2n = 24) is cultivated in the northwest part of China for over 5 millennia due to its strong resistance to abiotic stresses as well as its economic value7. Its roots, leaves, and fruits contribute significant medicinal ingredients such as polysaccharide, betaine, carotene and anthocyanin, which function in improving immunity8, anti-oxidative stress9 and anti-tumor10 ability, scavenging free radicals11, as well as promoting sexual function12. Current researches on Goji are mainly limited in the isolation, extraction and development of active ingredients. Studies related to pharmaceutically active intermediate synthesis and molecular mechanisms underlying plant metabolism, development and stress resistance are still unavailable. Unlike plants from the same Solanaceae family such as tobacco, tomato, pepper and potato, the whole genome data of Goji are still not available. Previously, Liu and co-workers13 used actin as a reference gene to analyze the expression pattern of genes involved in carotene synthesis in Goji. However, the validity of results is questionable due to the lack of systematic and scientific screening of reference genes.
For plants lacking whole genome information, one of the standard approaches for reference gene identification is to clone gene homologous to the known housekeeping gene identified in other model plants. Alternatively, emerging chip or next-gen sequencing technology provides ample data, which can be used for reliable reference gene screening6,14,15,16. In Arabidopsis, a new approach using ATH1 chip was used to screen the super reference genes17. They found that a novel reference gene such as PP2A showed stable expression at different developmental stages and under different treatments than the classical reference gene such as actin. Macrae et al. used the spliceosome and proteasome genes from RNA-Seq data to normalize and calibrate target gene expression pattern in human cancer tissues18. Similarly, based on the RNA-Seq data of sika deer antlers, Liu et al. evaluated the stability of 16 standard reference genes and 5 expression-stable genes from the sequencing platforms of different tissues and treatments19.
In woody plants, screening of stably expressed reference genes using gene expression data from next-generation sequencing platform has been performed in plum16 (P. salicina cv. Lindl.) and wine grape20 (Vitis vinifera). But no such kind of research has been done in Goji. Here, the transcriptomic sequencing data from the leaves, flowers and fruits of Goji were systematically assessed, and 8 classical reference genes and 10 stably expressed transcripts with little variation among different tissues and treatments were selected as candidate reference genes. Their expression levels in different organs, at different development stages, and under drought stress condition were investigated by qRT-PCR. The stability of candidate gene expression was evaluated to select the best reference gene, and the newly selected reference gene was tested to normalize and analyze the expression level of a target gene LbMYB1 under different conditions. Our findings provide a foundation for the functional studies of genes in Goji.
Results
Sequencing data analyses
From the transcriptomic sequencing of 14 sample databases, a total of 8,091,979,192 raw reads were obtained, including751,495,092 clean reads and 67,634,558,280 clean nucleotides after impurity filtration. The average Q20 value was up to 97.6%. We also found 144,250 Unigenes with a total length of 172,036,673 nt after sequence assembly. The average length of Unigene was 1193 nt, and that of N50 was1885 nt.
Candidate reference gene selection
The mean values of raw fragments, coefficient of variance and annotation of the 18 selected candidate references were listed in Table 1. LbTUB displayed the maximal coefficient of variance (CV) value (1.005) of raw fragments. On the contrary, LbEIF4A showed the minimal CV value (0.079). The CV value variation among different reference genes suggests that these genes have different expression levels in different organs, at developmental stages and under different treatment conditions. Four classical reference genes LbCYC, LbEIF4A, LbEF1β and LbUBQ using our screening criteria (CV < 0.3, mean raw fragments > 500) were identified, suggesting the validity of our approach using raw fragment CV as preliminary screening criteria.
qRT-PCR analyses of reference genes
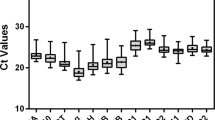
CT value reflects the abundance of reference gene expression. The higher the CT value, the lower the expression level, and vice versa. The CT values of the 18 reference genes ranged from 17.18 to 25.02 (Table 2). LbHIS3 showed the lowest CT value, whereas LbSKIP exhibited the highest CT value. Compared with LbActin7 and the newly identified reference genes LbRH52 and LbCML38, reference genes LbEF1α, LbHSP90 and LbHIS3 manifested a higher expression upon different treatments. In addition, the dispersion level (standard deviation, SD) of CT values is a schematic indicator of the stability of candidate reference gene expression in all tested samples. Among the 18 candidate reference genes, LbPP2A showed the lowest SD value. LbCML38 and LbRH52 also showed lower SD values (Table 2). In addition, PCR products of these candidate reference genes were checked on 1% agarose gel, and unique amplicons of expected length without distinct dimmers or non-specific products were observed (Supplementary Fig. 1).
Evaluation of reference gene expression stability
In order to screen the best reference gene or gene combination in different organs, at different developmental stages, and under different treatment conditions, geNorm, NormFinder, BestKeeper and RefFinder were employed to evaluate and rank their expression stability, as shown in Tables S1, S2 and S3.
geNorm analysis
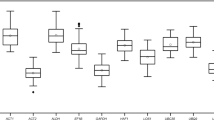
We used geNorm to compare and rank the M value of each candidate reference gene in terms of expression stability. The higher the M value, the lower the stability, and vice versa. The default cutoff value of geNorm software is 1.55. All candidate reference genes showed an M value lower than 1.5 (Fig. 1b). Based on the scores obtained from the 14 samples in different organs, at different developmental stages, and after different treatments, LbCML38 and LbRH52 were chosen as the best reference genes with an M value of 0.374. In leaves, LbCML38 and LbRH52 showed the best expression stability with an M value of 0.273 (Supplementary Table S3). In fruits and flowers, LbTBP demonstrated better stability with an M value of 0.249 and 0.142, respectively. Under drought stress condition, LbCYC and LbCML38 were the most stably expressed genes with an M value of 0.32 (Supplementary Table S2). However, commonly used reference genes such as LbCYC, LbActin7 and LbHIS3 showed mediocre expression stability. LbHSP90, LbTUB and LbGAPDH showed poor expression stability with the lowest ranking among these tested candidate reference genes (Supplementary Tables S2 and S3).
Sampling of materials and ranking of candidate reference genes.
(a) Samples representing different developmental stages of fruits for sequencing. (b) Comprehensive ranking of candidate reference genes based on their expression stability in all 14 samples calculated by geNorm. Stage 2 represents the flowering stage. Stages 3, 4 and 7 represent samples 18, 31 and 40 days after flowering, respectively. Scale bar = 1 cm. The y-axis represents the expression stability of gene based on normalization M. Mthreshold < 1.5 for solo calculation.
We also determined the optimal number of reference genes required under a particular condition by analyzing their paring difference value Vn/n+1. Typically, the threshold was set to 0.15 to select the best reference gene. When the paired value is lower than 0.15, additional (n + 1) reference genes are not necessary. After combining the analyses of all samples in different organs, at different developmental stages and after drought stress treatment together, V2/3 (0.139) was lower than the threshold of 0.15, indicating that the optimal number of reference genes needed was 2 (Fig. 2), and no need to introduce a third reference gene for calibration. The best combination of reference genes was LbCML38 and LbRH52. A combination of LbCYC and LbCML38 is optimal to analyze samples in different organs and at different developmental stages (Supplementary Tables S1, S2 and S3).
NormFinder analysis
Similar to geNorm, NormFinder evaluates reference genes by calculating their expression stability. Combined analyses of all samples showed that expression of LbCML38 and LbRH52 was the most stable with values of 0.16 and 0.254, respectively (Fig. 3). In different organs, LbRH52 was the most stably expressed reference gene in leaves (0.079), whereas LbActin7 was appropriate reference gene in fruits and flowers (Supplementary Table S3). Under drought stress condition, LbCML38 showed the highest expression stability with a value of 0.16 (Supplementary Table S2). Among the commonly used reference genes, NormFinder analyses confirmed the validity of LbCYC, LbHIS3 and LbEIF4A, with a value less than 0.5. On the contrary, LbHSP90, LbGAPDH and LbTUB showed poor expression stability (Fig. 3, Supplementary Table S3), and were not suitable to be used as reference genes.
BestKeeper analysis
Results from BestKeeper analysis is slightly different from that of geNorm and NormFinder analyses. When combining the analyses of all samples in different organs, at different developmental stages and under drought stress condition together, LbPP2A (SD = 1.13) was identified as the best reference gene (Fig. 4). LbPP2A was the most stably expressed gene in leaves (SD = 0.85). Whereas LbEF1β (SD = 0.77) and LbHSP90 (SD = 0.11) were respectively selected as the best reference gene in fruits and flowers (Supplementary Table S3). LbGAPDH (SD = 1.8) and LbTUB (SD = 2.03) were the worst candidate reference genes. Under drought stress condition, LbHIS3 (SD = 0.98) was the most stably expressed reference gene (Supplementary Table S2).
RefFinder analysis
Statistical analyses showed that the stability value ranged from 1.57 to 18 among the 18 selected candidate reference genes in the combined analyses of all samples in different organs, at different developmental stages and under drought stress condition evaluated by RefFinder (Fig. 5). LbCML38 and LbRH52 were identified as the most stably expressed two reference genes with an average value less than 2. LbGAPDH, LbHSP90 and LbTUB were the worst. In addition, RefFinder ranked LbCML38, LbRH52, LbHIS3 and LbCYC as the top four reference genes, which was consistent with the rankings obtained with geNorm and NormFinder. Collective evidence suggested that LbCML38 and LbRH52 were the best reference genes under the tested conditions. Individual factor analysis indicated that LbCML32 was the best reference gene under drought stress condition (Supplementary Table S2). LbRH52 and LbActin7 were identified as the best reference genes for target gene calibration in leaves, fruits and flowers (Supplementary Table S3).
In summary, these four software algorithms yielded various results in selecting the best reference genes in different organs, at different developmental stages, and under different treatment conditions in Goji. LbCML38 and LbRH52 showed relatively stable expression (Fig. 6), whereas LbTUB, LbHSP90 and LbGAPDH were not so stable (Fig. 7). Specifically, LbActin7 should be carefully used for calibrating gene expression in fruits and flowers. LbCML38, especially combination of LbCML38 and LbRH52 performed the best stability under most conditions.
Venn diagram showing the most unstable reference genes identified by the geNorm, NormFinder, BestKeeper and RefFinder algorithms.
The intersection part shows the most unstable genes in common, especifically, LbTUB, LbEF1α, LbGAPDH, LbRPL7A and LbHSP90. Mapping data were derived from Figs 1b, 3, 4 and 5.
Target gene expression analyses
To further re-evaluate the validity of selected reference genes, the expression level of LbMYB1 in 14 samples was normalized with LbCML38, LbRH52, LbActin7 and LbHSP90. As illustrated in Fig. 8, no significant difference between LbCML38 and LbRH52 was observed in most of the treatments when they were used for normalization of LbMYB1 (P < 0.01). However, compared with LbCML38, LbActin7 yielded a higher normalization value of LbMYB1 in leaves and lower values in fruits and flowers after some specific treatments (P < 0.01). LbMYB1 normalization against LbHSP90 resulted in higher values in most cases (P < 0.01). These results demonstrate that LbActin7 and LbHSP90 introduced errors when they were used as reference genes. Therefore, it is crucial to select appropriate reference gene(s) for evaluation of gene expression.
Expression profiles of LbMYB1 in 14 samples of L. barbarum determined by qRT-PCR using LbAcitn7, LbHSP90, LbCML38 and LbRH52 as reference genes.
Data were compared and analyzed with analyses of variance (ANOVA) and multiple comparisons using the statistical analysis software of SPSS 21 (P < 0.01). Error bars show the mean standard error based on triplicates. Letters and numbers in the abscissa axis stand for: C, control group; D, drought stress treatment group; L, leaf; G, Gouji berry; F, flower; 3, development stage 3 (18 days after flowering); 4, development stage 4 (31 days after flowering); 7, development stage 7 (40 days after flowering).
Discussion
The acreage of Goji in Ningxia and Qianghai provinces of China is growing rapidly due to its economical and medicinal value. However, the molecular mechanism of fruit development, pharmacologically active ingredient accumulation, and stress resistance of it is largely unknown. Screening and selection of stable reference genes for gene expression study in Goji will provide a foundation for elucidating the molecular mechanism. In the absence of systematic profiling of reference genes, we screened the reference genes suitable for samples in different organs, at developmental stages and under drought stress condition of Goji using transcriptomic sequence database for the first time.
Several studies on the screening of reference genes in Solanaceae family plants have been reported. In pepper, EF1α and UEP were found to be the most stably expressed genes in roots, stems, leaves and flowers under different treatment conditions (salicylic acid, gibberellic acid, cold, heat, salt, and drought)21. EF1α and APRT were the most stably expressed reference genes among 8 commonly used ones when potato plants were exposed to salt and drought stress, respectively22,23. In tomato (Solanum lycopersicum, cv. Suzanne), RPL2 and PP2Acs exhibited as stable expression as ACT and UBI under nitrogen deficiency, low temperature and different light conditions24. Gantasala and co-workers25 investigated 6 commonly used reference genes (18sRNA, APRT, GAPDH, CYP, Actin and RuBP) in egg plants (Solanum melongena) and found that 18sRNA, CYP and APRT had the best expression stability.
From the results discussed above, EF1α or Actin showed relative stable expression in some of the Solanaceae family plants. Liu et al. used Actin as a reference gene in fruits of two goji cultivars (Lycium barbarum L. and L. ruthenicum Murr.) for the profiling of genes involved in carotenoid biosynthesis and metabolism13. However, our study demonstrated that LbActin7 exhibited moderately stable expression in some of the treatments in flowers and fruits, but introduced significant errors when normalizing the target gene expression level in leaves and fruits at specific developmental stages (Fig. 8). These results suggest that special caution should be paid when using LbActin7 as a reference gene. Besides, commonly used LbGAPDH, LbHSP90, and especially LbTUB, were not suitable to be used as reference genes in Goji (Figs 6 and 7). This was further confirmed in the normalization of LbMYB1 gene expression (Fig. 8). We also tested the expression stability of established reference genes in Solanaceae family plants including LbRPL, LbPP2A and LbEF1α. Our data suggest that they are not suitable to be used as reference genes in all the tested samples of Goji (Figs 1b, 2, 3, 4, 5 and Supplementary Tables S1, S2 and S3). The expression stability of GAPDH, EF1α, Actin and other reference genes has been questioned in some reports6,26,27,28. The discrepancies between these results tested in Solanaceae materials and our data could be due to the different genetic backgrounds of plant species as well as the different treatment conditions. Therefore, selection of appropriate reference genes is critical in the genomic function study of Goji or a comparative study of different plant lines in the Solanaceae family.
DNA chips and next-generation sequencing provide a novel approach for reference gene screening of non-model organisms that lack whole genome information. Czechowski and co-workers firstly proposed to screen reference genes using Arabidopsis whole genome Affymetrix ATH1 chips17. González-Agüero et al. further summarized and refined the analytic process for reference gene screening from RNA-Seq data in grape20. A total number of 19 candidate reference genes were identified from 242 non-differentially expressed genes (NDE) using CV < 0.4 of the total read as the screening threshold. qRT-PCR results showed that VvAIG1 and VvTCPB were the most stably expressed reference genes in 14 grape lines, at 4 developmental stages, and under gibberellic acid treatment condition. Similarly, studies with oil-tea camellia29, Striga hermonthica30 and plums16 corroborated the approach of using RNA-Seq database to screen reference genes. In this study, we adopted even stricter screening threshold (CV < 0.3) to select 10 functional genes from 1272 raw fragments according to the analytic protocol proposed previously20, and three classical reference genes LbEIF4A, LbUBQ and LbEF1α were included with a CV value less than 0.1 (Table 1). They showed better stability than that of LbTUB, LbHSP90 and LbGAPDH as confirmed by qRT-PCR (Table 2), all of which had higher CV values of raw fragments and CT values.
Taken together, our studies confirmed the correlation between the transcriptional expression stability and qRT-PCR results, thus it is appropriate to select the transcripts with smaller CV values as candidate reference genes during preliminary screening with sequencing data. Collective evidence suggests that LbCML38 and LbRH52 could be used as the best reference genes for gene expression study in Goji.
Methods
Sample preparation and treatment
Ningxia goji (Lycium barbarum L.) Ningqi I was cultivated in field and collected. Plants grown under normal condition were used as control (control, C). Drought stress treatment was performed as described previously31 with minor modification: four-year-old adult plants were transplanted into pots filled with a mixture of fertilizer, sand and loam (1:1:3 v/v). The maximal water holding capacity of soil in field was measured and determined as 18%. Transplanted plants at vegetative growth stage with similar physiological state were selected for drought stress treatment. During the drought stress period, water holding capacity of soil in pots was maintained at 40% to 45% of the maximal field level. The water capacity of soil was measured at a fixed time point every day using soil moisture meter TDR300 (Spectrum, USA), and maintained at a specific level by artificial replenishment. Ten plants were individually cultivated in pots for each treatment. Sequencing samples including equal amounts of leaves (L), flowers (F) and fruits (G) from control and drought-treated plants were collected 18 (developmental stage 3), 31 (developmental stage 4) and 40 (developmental stage 7) days after flowering (Fig. 1a). Control leaves were collected from the same plants at developmental stage 3.
Total RNA isolation and sequencing database establishment
Total RNA was extracted using plant total RNA extraction kit (Tiangen Biotechnology, PRC). Genomic DNA was eliminated by treating each sample with RNase-free DNase I (TAKARA BIO INC., code 2270A) according to the instruction manual. The purity of total RNA extracted was checked using a NanoDrop 2000 spectrophotometer. Samples with an absorbance ratio at OD260/280 between 2.0 and 2.2 were used for further analyses. The concentration and quality of extracted RNA was determined using Agilent 2000 bioanalyzer (Agilent, USA). First-strand cDNA was synthesized from magnetic beads-enriched poly(A)-mRNA using random hexamers, followed by buffer addition to synthesize the complement strand. Synthesized cDNA was purified using Qiaquick PCR purification kit (Qiagen, USA) and eluted with EB elution buffer. Purified cDNA was mixed with poly(A) tail and sequencing adaptors. Appropriately sized cDNA fragments were selected by agarose electrophoresis and amplified by PCR. The library was sequenced on Illumina HiSeq2000 using de novo PE 100 sequencing strategy.
Sequence assembly and annotation of basic bioinformatics
The original imaging data obtained from sequencing were processed into raw reads by base calling, followed by filtration of noises and low quality data to obtain clean reads. The de novo assembler program Trinity was used to assemble the short reads into contigs, scaffolds and Unigene, respectively32. Unigene sequences were blasted against that of Nr, SwissProt, GO, KEGG and COG databases (E-value < 0.00001) to obtain homologous proteins with high sequence similarity to the reference protein. When Unigene sequences failed to match sequences in the databases, ESTScan software was used to predict the coding region and sequence orientation33. Unigene expression level was calculated using FPKM method (Fragments Per kb per Million fragments)34. The RNA-Seq data used in this study is available at the Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra) of the National Center for Biotechnology Information (NCBI) with an accession number SRP063577.
Reference gene screening and primer design
Two approaches were used to select reference genes: 1) Excavation of classical reference genes: 8 classical reference genes including CYP, TBP, GAPDH, TUB, EF1α, Actin7, HSP90 and HIS3 derived from tomato or Arabidopsis were used as template to screen homologous genes from prepared RNA-Seq database of L. barbarum; 2) Based on the method described by González-Agüero et al.20, 10 stably expressed Unigenes (LbCYC, LbEIF, LbPP2A, LbUBQ, LbCML38, LbLEA, LbEF1β, LbSKIP, LbRH52 and LbRPL7A) were selected out of 1272 transcripts (raw fragments > 500, and the coefficient of variation < 0.3). Primers were designed using Primer 3 software (http://frodo.wi.mit.edu/primer3/). Data information about the candidate genes and primers are listed in Tables 1 and 2.
qRT-PCR and data analysis
Synthesis of cDNA was performed with 0.5–1 μL total RNA (the final content of RNA in the reaction mixture was adjusted to 1 μg for all samples) according to the instruction manual of the cDNA synthesis System (TRANSGEN BIOTECH INC., code AU311–02) in a total volume of 20 μL. Quantitative RT-PCR analysis of the cDNA of 14 samples was performed using StepOne Real-time PCR Systems (Applied Biosystems, USA). A total amount of 20 μl of PCR reaction mix containing 10 μl of Power SYBR Green PCR Master Mix, 5 μl of diluted cDNA, 0.5 μl (10 pmol) each of forward and reverse primers, and ddH2O was prepared. The thermocycling condition was set as follows: initial denaturation at 95 °C for 10 mins, 40 cycles of denaturation at 95 °C for 15 seconds, annealing at 60–62 °C for 15 seconds, and extension at 72 °C for 25 seconds. Fluorescent signals were collected after each cycle. Each sample was run in triplicate along with a negative control. Amplified products were checked on 1% agarose gel.
The first-strand cDNA was synthesized after six rounds using a five-fold serial dilution. Each PCR reaction was conducted in triplicate using diluted cDNA as template. The CT values of samples were generated automatically after qRT-PCR. A standard curve was also generated and the melting curve was analyzed to determine the specificity of PCR products. The amplification efficiency (E) of each candidate reference gene was calculated with the slope of the standard curve according to the equation E = (10−1/slope − 1) × 100%17.
The stability of candidate reference genes was evaluated by geNorm, NormFinder, BestKeeper and RefFinder using obtained CT values of the samples. The average expression stability value (M) of each candidate reference gene was calculated by geNorm5. The higher the M value, the less stable the gene expression was, and vice versa. Meanwhile, paired difference analysis (Vn/n+1) of candidate reference gene normalization factor was used to determine the optimal number of required reference genes. NormFinder was used to determine the candidate reference gene stability using the combined variances within and between groups35. The lower the stability value, the more stable the reference gene expression was, and vice versa. BestKeeper was used to determine the candidate reference genes’ standard deviation (SD) and coefficient of variation (CV)36. Reference gene with the smallest CV ± SD value was considered the most stably expressed one. All reference genes with a SD value less than 1 were established as stably expressed genes. The smaller the SD value, the more stable the reference gene expression was, and vice versa. To calculate the geometric mean values and the ranking of each candidate reference gene, results respectively obtained with geNorm, NormFinder, BestKeeper and Delta CT were integrated using RefFinder (http://www.leonxie.com/referencegene.php). The lower the ranking index, the more stable the reference gene expression was, and vice versa.
Reference gene validation
For confirmation of selected reference gene validity, LbMYB1 which encodes a MYB transcription factor involved in drought stress response and flavonoid anabolism was selected as a target gene37. The expression levels of LbMYB1 in difference samples were normalized against those of the most stable reference genes LbCML38 and LbRH52, the moderate stable reference gene LbActin7, and the less stable reference gene LbHSP90, respectively. Data were compared and analyzed with analysis of variance (ANOVA) and multiple comparisons using the statistical analysis software of SPSS 21. Values of P < 0.01 were considered statistically significant difference.
Additional Information
How to cite this article: Gong, L. et al. LbCML38 and LbRH52, two reference genes derived from RNA-Seq data suitable for assessing gene expression in Lycium barbarum L. Sci. Rep. 6, 37031; doi: 10.1038/srep37031 (2016).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Huggett, J., Dheda, K., Bustin, S. & Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284 (2005).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Shivhare, R. & Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 6, 23036 (2016).
Guo, J., Ling, H., Wu, Q., Xu, L. & Que, Y. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Sci. Rep. 4, 7042 (2014).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.0031-research0034.0011 (2002).
Long, X. Y. et al. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 74, 307–311 (2010).
Amagase, H. & Farnsworth, N. R. A review of botanical characteristics, phytochemistry, clinical relevance in efficacy and safety of Lycium barbarum fruit (Goji). Food Res. Int. 44, 1702–1717 (2011).
Lin, F. Y. et al. Effects of Lycium barbarum extract on production and immunomodulatory activity of the extracellular polysaccharopeptides from submerged fermentation culture of Coriolus versicolor. Food Chem. 110, 446–453 (2008).
Ma, M., Liu, G., Yu, Z., Chen, G. & Zhang, X. Effect of the Lycium barbarum polysaccharides administration on blood lipid metabolism and oxidative stress of mice fed high-fat diet in vivo. Food Chem. 113, 872–877 (2009).
Chao, J. C. et al. Hot water-extracted Lycium barbarum and Rehmannia glutinosa inhibit proliferation and induce apoptosis of hepatocellular carcinoma cells. World J. Gastroenterol. 12, 4478–4484 (2006).
Qian, J. Y., Liu, D. & Huang, A. G. The efficiency of flavonoids in polar extracts of Lycium chinense Mill fruits as free radical scavenger. Food Chem. 87, 283–288 (2004).
Yu, M. S. et al. Neuroprotective effects of anti-aging oriental medicine Lycium barbarum against β-amyloid peptide neurotoxicity. Exp. Gerontol. 40, 716–727 (2005).
Liu, Y. et al. Comparative analysis of carotenoid accumulation in two goji (Lycium barbarum L. and L. ruthenicum Murr.) fruits. BMC Plant Biol. 14, 269–269 (2014).
Coker, J. S. & Davies, E. Selection of candidate housekeeping controls in tomato plants using EST data. Biotechniques 35, 748–740 (2003).
Libault, M. et al. Identifi cation of Four Soybean Reference Genes for Gene Expression Normalization. Plant Genome 1, 44–54 (2008).
Kim, H. Y. et al. RNA-Seq Analysis of Spatiotemporal Gene Expression Patterns During Fruit Development Revealed Reference Genes for Transcript Normalization in Plums. Plant Mol. Biol. Rep. 33, 1–16 (2015).
Tomasz, C., Mark, S., Thomas, A., Udvardi, M. K. & Wolf-Rüdiger, S. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 (2005).
Macrae, T. et al. RNA-Seq Reveals Spliceosome and Proteasome Genes as Most Consistent Transcripts in Human Cancer Cells. Plos One 8, e72884 (2013).
Liu, M. et al. Identification of novel reference genes using sika deer antler transcriptome expression data and their validation for quantitative gene expression analysis. Genes Genom. 36, 573–582 (2014).
González-Agüero, M. et al. Identification of two putative reference genes from grapevine suitable for gene expression analysis in berry and related tissues derived from RNA-Seq data. BMC Genomics 14, 499–501 (2013).
Wang, S. B. et al. Evaluation of appropriate reference genes for gene expression studies in pepper by quantitative real-time PCR. Mol. Breed. 30, 1393–1400 (2012).
Nicot, N., Hausman, J.-F., Hoffmann, L. & Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 56, 2907–2914 (2005).
Lopez-Pardo, R., Galarreta, J. I. R. D. & Ritter, E. Selection of housekeeping genes for qRT-PCR analysis in potato tubers under cold stress. Mol. Breed. 31, 39–45 (2013).
Løvdal, T. & Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 387, 238–242 (2009).
Gantasala, N. P. et al. Selection and validation of reference genes for quantitative gene expression studies by real-time PCR in eggplant (Solanum melongena L). BMC Research Notes 6, 1–11 (2013).
Wu, Z. J., Tian, C., Jiang, Q., Li, X. H. & Zhuang, J. Selection of suitable reference genes for qRT-PCR normalization during leaf development and hormonal stimuli in tea plant (Camellia sinensis). Sci. Rep. 6, 19748 (2016).
Tong, Z., Gao, Z., Fei, W., Zhou, J. & Zhen, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10, 1–13 (2009).
Galli, V., Messias, R. D. S., Silva, S. D. D. A. E. & Rombaldi, C. V. Selection of reliable reference genes for quantitative real-time polymerase chain reaction studies in maize grains. Plant Cell Rep. 32, 1869–1877 (2013).
Zhou, C. F. et al. Selection of reference genes for quantitative real-time PCR in six oil-tea camellia based on RNA-seq. Mol. Biol. 47, 836–851 (2013).
Fernández-Aparicio, M. et al. Application of qRT-PCR and RNA-Seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Mol. Biol. Rep. 40, 3395–3407 (2012).
Zhao, J. H. et al. Influence of Drought Stress on Plant Growth and Sugar Accumulation in Fruit of Lycium barbarum L. Plant Physiology Journal (in Chinese: Zhiwu shengli xuebao) 48, 1063–1068 (2012).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Iseli, C., Jongeneel, C. V. & Bucher, P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proceedings/… International Conference on Intelligent Systems for Molecular Biology Heidelberg, Germany. CA, USA: AAAI Press (1999).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Shin, D. et al. Expression of StMYB1R-1, a Novel Potato Single MYB-Like Domain Transcription Factor, Increases Drought Tolerance. Plant Physiol. 155, 421–432 (2011).
Acknowledgements
This work has been jointly supported by the following grants: the Natural Science Foundation of Ningxia University (ZR15037), National Natural Science Foundation of China (31371228, 31401444) and Open project of National Key Laboratory of Plant Molecular Genetics (52ZKF2014). We thank Mr. Jiehui Guanzhang (University of Toronto, Canada) for the critical reading and comments on this manuscript.
Author information
Authors and Affiliations
Contributions
H.X.Z. and L.G. initiated and designed the research. L.G. and Y.J.Y. performed the experiments. L.G., Y.J.Y., Y.C.C., J.S., and Y.X.S. analyzed the data. L.G. and H.X.Z. wrote the paper. All of the other authors reviewed and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gong, L., Yang, Y., Chen, Y. et al. LbCML38 and LbRH52, two reference genes derived from RNA-Seq data suitable for assessing gene expression in Lycium barbarum L.. Sci Rep 6, 37031 (2016). https://doi.org/10.1038/srep37031
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep37031
This article is cited by
-
Integrated metabolome and transcriptome analysis of differences in quality of ripe Lycium barbarum L. fruits harvested at different periods
BMC Plant Biology (2024)
-
Identification and validation of the reference genes in the echiuran worm Urechis unicinctus based on transcriptome data
BMC Genomics (2023)
-
Reference gene selection for qRT-PCR analyses of luffa (Luffa cylindrica) plants under abiotic stress conditions
Scientific Reports (2021)
-
Identification and validation of reference genes for qRT-PCR based studies in horse gram (Macrotyloma uniflorum)
Physiology and Molecular Biology of Plants (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.