Abstract
With the advance of next generation sequencing techniques, whole genome sequencing (WGS) is expected to become the optimal method for molecular subtyping of bacterial isolates. To use WGS as a general subtyping method for disease outbreak investigation and surveillance, the layout of WGS-based typing must be comparable among laboratories. Whole genome multilocus sequence typing (wgMLST) is an approach that achieves this requirement. To apply wgMLST as a standard subtyping approach, a pan-genome allele database (PGAdb) for the population of a bacterial organism must first be established. We present a free web service tool, PGAdb-builder (http://wgmlstdb.imst.nsysu.edu.tw), for the construction of bacterial PGAdb. The effectiveness of PGAdb-builder was tested by constructing a pan-genome allele database for Salmonella enterica serovar Typhimurium, with the database being applied to create a wgMLST tree for a panel of epidemiologically well-characterized S. Typhimurium isolates. The performance of the wgMLST-based approach was as high as that of the SNP-based approach in Leekitcharoenphon’s study used for discerning among epidemiologically related and non-related isolates.
Similar content being viewed by others
Introduction
Molecular subtyping of bacterial isolates has been fundamental for epidemiologic study of infectious diseases. Subtyping methods used for disease outbreak investigation and surveillance across regions and countries must be standardized so that the results can be compared across laboratories. For example, pulsed-field gel electrophoresis (PFGE) is a good example; it has been standardized and successfully implemented as a common subtyping tool in the foodborne disease surveillance network—PulseNet1. Although PFGE is highly discriminatory to most bacterial organisms, it is labor- and time-consuming and sometimes insufficient in discerning among strains of highly clonal organisms. A multilocus variable-number tandem repeat analysis (MLVA) exhibits a much higher level of discrimination than PFGE in discerning among very closely related strains; however, MLVA is very organism-specific, and comparing its results across laboratories is difficult2,3. With the advance of next-generation sequencing (NGS) techniques, whole genome sequencing (WGS) has become a practical and powerful subtyping tool for disease outbreak detection4,5.
To use WGS as a standard subtyping tool for disease surveillance and the investigation of common outbreaks across regions or countries, the layout of fingerprints (genotypes) generated from WGS data must be comparable among laboratories. Currently, NGS platforms generally produce millions of short sequences (reads) for a bacterial strain. The millions of reads can be further assembled into longer sequences (contigs) and annotated using various assemblers6,7,8. A number of algorithms and approaches have been developed for analyzing WGS data9,10,11,12,13,14. Single nucleotide polymorphism (SNP) is an approach frequently used to analyze WGS data for evolutionary study and disease outbreak investigation15,16,17. To apply the SNP approach, a reference genome sequence is required for selecting SNPs from WGS data of strains. When different reference sequences are used, different SNP sets are generally yielded, making the SNP profiles incomparable across laboratories. Whole genome multilocus sequence typing (wgMLST)14,18, an extended concept of the traditional MLST19, is considered an ideal approach to sort out WGS data and generate genetic layouts that are portable and comparable among laboratories. To use wgMLST as a standard subtyping tool, a pan-genome allele database (PGAdb) for the population of a bacterial organism must first be established. In a PGAdb, genes (loci) and their sequence variants (alleles) are designated using a standardized numbering system. An allelic sequence consists of a series of digital numbers and can be portable and comparable across laboratories.
We present a web service tool, PGAdb-builder that can be used for the construction of bacterial pan-genome allele databases. In this paper, we demonstrate the function of the PGAdb-builder by constructing a S. Typhimurium PGAdb and generating a wgMLST tree for a panel of epidemiologically well-characterized S. Typhimurium isolates, which were sequenced previously by the DTU Food20.
Methods and Implementation
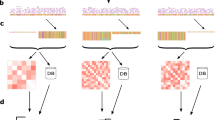
The flowchart for the proposed PGAdb-builder is illustrated in Fig. 1. The PGAdb-builder server comprises two functional modules: Build_PGAdb for creating a PGAdb database and Build_wgMLSTtree for constructing a wgMLST tree from uploaded genome contigs and formulating genetic relatedness trees by using the PGAdb for generating allelic sequences. The details of the Build_PGAdb and Build_wgMLSTtree modules are described herein.
Build_PGAdb
The Build_PGAdb module executes the annotation of uploaded genome contigs by using the Prokka pipeline21, a rapid bacterial genome annotation tool. Subsequently, the output gff file created in the annotation process is processed to place proteins into orthologous clusters by using the Roary pipeline22, a tool that can rapidly process a large-scale collection of genomes. In this module, paralogous genes are excluded from a pan-genome allele dataset. Each orthologous cluster consists of a protein family with 95% (adjustable between 90% and 99%) sequence identity. Each protein family is defined as a locus (gene). The orthologous proteins in each cluster are converted to nucleotide sequences through inference to the ffn file created in the annotation process to establish a pan-genome allele dataset. In this step, sequences in a locus with one or more mismatched nucleotides between each other are defined as different alleles. The loci of a pan-genome allele dataset are then encoded with a prefix string of three alphabetic letters followed by an eight digits serial number (e.g., SAL00000001, SAL00000002…) and the alleles in each locus are simply assigned by a series of integers beginning from 1 to n (e.g. 1, 2, 3, … n).
Build_wgMLSTtree
The Build_wgMLSTtree module compares the uploaded genome contigs of strains by using a PGAdb database and constructs genetic relatedness trees (wgMLST trees). To create a wgMLST tree, the uploaded genome contigs is compared with the built PGAdb using BLASTN23. If an allele is present in a locus, the predefined allele number is assigned; however, if an allele is absent, “0” is assigned. After the allele finding process is finished, an “allelic sequence” for an uploaded genome is created. A dendrogram with bootstrap values, which is calculated by the ETE tool kit24, is then constructed from allelic sequences with the PHYLIP program25 through use of UPGMA clustering algorithm.
Implementation
The PGAdb-builder server is created through an integration of the Build_PGAdb and Build_wgMLSTtree modules in PHP scripts. The web page was constructed using HTML, javascript, and PHP. The server runs on a Linux cluster with 2.40 GHz Intel Xeon processors comprising 24 cores. Dendrograms labeled with bootstrap values made using Build_wgMLSTtree module are output in the webpage and in a downloadable Newick and a pdf format.
Webserver
Input format
The two modules of PGAdb-builder accepts genome contigs in the FASTA format (Fig. 2A). When the default parameter was usded (protein sequence identity = 95%), Build_PGAdb required approcimately 19 hours to construct a database in a test using 487 S. Typhimurium genomes (487ST_set). Build_PGAdb creates a Database ID after the process finished. Build_wgMLSTtree required 4.5 hours to construct a wgMLST tree (with pan-genome scheme) for 34 S. Typhimurium genomes by using the PGAdb when the default parameters (alignment coverage ≥ 90%; alignment identity ≥ 90%) were set. Users are encouraged to provide e-mail addresses through which to receive a notification for when a job finishes.
Output format
The output of Build_PGAdb comprises (A) a summary of settings; (B) a pie chart illustrating the numbers (percentages) of loci for the core genome, dispensable genome, and unique genes in the PGAdb; (C) a checkbox menu for the selection of the user-defined scheme; and (D) buttons, to perform “Go To wgMLSTtree” and “Download User Defined Scheme.” The file of the user-defined scheme can be used as the input for the module of Build_wgMLSTtree from the “Upload User Defined Scheme” option. Through this mechanism, users can exchange their pan-genome database by sharing their scheme files. The output of Build_wgMLSTtree includes (A) a summary of settings, (B) a genetic relatedness tree constructed using the scheme, which is selected by users, and (C) a summary of output files to download. Examples of Build_PGAdb and Build_wgMLSTtree outputs are shown in Fig. 2B,C, respectively.
Example Analysis
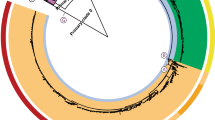
We tested the ability of the Build_PGAdb module to construct a PGAdb by using 487 Salmonella Typhimurium (487ST_set) strains of genome contigs (Table S1), which were downloaded from the National Center for Biotechnology Information (NCBI) Genome database (https://www.ncbi.nlm.nih.gov/genome). The operation required approximately 19 hours on a Linux server with 2.40 GHz Intel Xeon processors comprising 24 cores. The S. Typhimurium PGAdb contained 27,011 loci, of which 12.5% (3,375 loci) belonged to the core genome, 44% (11,905 loci) belonged to the dispensable genome, and 43.5% (11,731 loci) belonged to the unique genes. In this step, we defined the core genome as having genes present in 95% of the tested genomes, a dispensable genome as having genes present in two or more but less than 95% of the genomes, and unique genes as being present only in a single genome. The PGAdb from the 487ST_set was then used to construct a wgMLST tree for 34 epidemiologically well-characterized S. Typhimurium isolates by using the Build_wgMLSTtree module. The allelic sequences for the 34 isolates were formed on the basis of the 27,011 loci for the core genome. As illustrated in Fig. 3, the genetic relationships among the 34 isolates constructed using the wgMLST-based approach were highly concordant with the relationships of the isolates determined using the SNP-based method, as shown in a previous study20.
Dendrogram (genetic relatedness tree) for 34 epidemiologically well-characterized S.
Typhimurium isolates sequenced by DTU Food20. Isolates for 6 foodborne disease outbreaks are marked.
Conclusion
The proposed online tool PGAdb-builder, comprising two modules, Build_PGAdb and Build_wgMLSTtree, was established to enable users to use WGS data to construct bacterial pan-genome allele databases and to apply the databases to create genetic relatedness trees for bacterial strains. A strong advantage of the PGAdb-builder server is that the built PGAdb with the user-defined scheme can be reused through uploading the downloaded “User defined scheme file” (UDS file), which records the database ID and the defined scheme. Through this mechanism, users can exchange their PGAdbs by only sharing the UDS files. This PGAdb-builder would be a useful online tool for the construction of bacterial pan-genome allele databases and construction of genetic relatedness tree.
Additional Information
How to cite this article: Liu, Y.-Y. et al. PGAdb-builder: A web service tool for creating pan-genome allele database for molecular fine typing. Sci. Rep. 6, 36213; doi: 10.1038/srep36213 (2016).
References
Swaminathan, B., Barrett, T. J., Hunter, S. B., Tauxe, R. V. & Force, C. D. C. P. T. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis 7, 382–389, doi: 10.3201/eid0703.010303 (2001).
Chiou, C. S. Multilocus variable-number tandem repeat analysis as a molecular tool for subtyping and phylogenetic analysis of bacterial pathogens. Expert Review of Molecular Diagnostics 10, 5–7, doi: 10.1586/Erm.09.76 (2010).
Chiou, C. S. et al. A simple approach to obtain comparable Shigella sonnei MLVA results across laboratories. International Journal of Medical Microbiology 303, 678–684, doi: 10.1016/j.ijmm.2013.09.008 (2013).
Jonathan, S. B., Michael, G. & Adam, M. Whole-genome sequencing detection of ongoing contamination at a restaurant, Rhode Island, USA, 2014. Emerg Infect Dis 22, 1474, doi: 10.3201/eid2208.151917 (2016).
Jackson, B. R. et al. Implementation of nationwide real-time whole-genome sequencing to enhance Listeriosis outbreak detection and investigation. Clin. Infect. Dis. 63, 380–386, doi: 10.1093/cid/ciw242 (2016).
Zerbino, D. R. & Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Research 18, 821–829, doi: 10.1101/gr.074492.107 (2008).
Bankevich, A. et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. Journal of Computational Biology 19, 455–477, doi: 10.1089/cmb.2012.0021 (2012).
Luo, R. B. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, doi: Artn 1810.1186/2047-217x-1-18 (2012).
Leekitcharoenphon, P. et al. snpTree-a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics 13 Suppl 7, S6, doi: 10.1186/1471-2164-13-S7-S6 (2012).
Taylor, A. J. et al. Characterization of foodborne outbreaks of Salmonella enterica serovar Enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J. Clin. Microbiol. 53, 3334–3340, doi: 10.1128/JCM.01280-15 (2015).
de Been, M. et al. A core genome MLST scheme for high-resolution typing of Enterococcus faecium. J Clin Microbiol, doi: 10.1128/JCM.01946-15 (2015).
Cheng, J., Cao, F. & Liu, Z. AGP: a multimethods web server for alignment-free genome phylogeny. Mol Biol Evol 30, 1032–1037, doi: 10.1093/molbev/mst021 (2013).
Snipen, L. & Ussery, D. W. Standard operating procedure for computing pangenome trees. Stand Genomic Sci 2, 135–141, doi: 10.4056/sigs.38923 (2010).
Jolley, K. A. & Maiden, M. C. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC bioinformatics 11, 595, doi: 10.1186/1471-2105-11-595 (2010).
Pang, S. et al. Genetic relationships of phage types and single nucleotide polymorphism typing of Salmonella enterica Serovar Typhimurium. J Clin Microbiol 50, 727–734, doi: 10.1128/JCM.01284-11 (2012).
Schork, N. J., Fallin, D. & Lanchbury, J. S. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet 58, 250–264 (2000).
Collins, A., Lonjou, C. & Morton, N. E. Genetic epidemiology of single-nucleotide polymorphisms. Proceedings of the National Academy of Sciences of the United States of America 96, 15173–15177, doi: 10.1073/pnas.96.26.15173 (1999).
Maiden, M. C. et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 11, 728–736, doi: 10.1038/nrmicro3093 (2013).
Aanensen, D. M. & Spratt, B. G. The multilocus sequence typing network: mlst.net. Nucleic Acids Res 33, W728–W733, doi: 10.1093/nar/gki415 (2005).
Leekitcharoenphon, P., Nielsen, E. M., Kaas, R. S., Lund, O. & Aarestrup, F. M. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PloS one 9, e87991, doi: 10.1371/journal.pone.0087991 (2014).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069, doi: 10.1093/bioinformatics/btu153 (2014).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693, doi: 10.1093/bioinformatics/btv421 (2015).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410, doi: 10.1016/S0022-2836(05)80360-2 (1990).
Huerta-Cepas, J., Dopazo, J. & Gabaldon, T. ETE: a python Environment for Tree Exploration. BMC bioinformatics 11, 24, doi: 10.1186/1471-2105-11-24 (2010).
Felsenstein, J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17, 368–376 (1981).
Acknowledgements
We thank the researchers in DTU Food, Denmark for using their 34 S. Typhimurium genomes to assess the usefulness of the PGAdb-builder in fine typing of bacterial strains for disease outbreak investigation. This study was mainly supported by grant from ‘Academic Summit Program’ of Ministry of Science and Technology (MOST-105-2221-E-110-079), ‘Medical Science and Technology Center of Aiming for the Top University Program’ of National Sun Yat-sen University and Ministry of Education, Taiwan, and also a grant (MOHW105-CDC-C-315-123301) from Centers for Disease Control, the Ministry of Health and Welfare, Taiwan.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: Y.-Y.L., C.-S.C. and C.-C.C. Performed the experiments and analyzed the data: Y.-Y.L. and C.-C.C. Contributed materials/analysis tools and wrote the paper: Y.-Y.L., C.-S.C. and C.-C.C.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Liu, YY., Chiou, CS. & Chen, CC. PGAdb-builder: A web service tool for creating pan-genome allele database for molecular fine typing. Sci Rep 6, 36213 (2016). https://doi.org/10.1038/srep36213
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep36213
This article is cited by
-
Phylogenetic analysis revealed that Salmonella Typhimurium ST313 isolated from humans and food in Brazil presented a high genomic similarity
Brazilian Journal of Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.