Abstract
DNA methylation is an important epigenetic mark that regulates gene expression. Dnmt1 plays an important role in maintaining DNA methylation patterns on daughter DNA strands. Studies have shed light into the functional role of Dnmt1 regulation in the hematopoietic and epidermal systems. Here we show that Dnmt1 is required for myogenesis. Loss of Dnmt1 results in reduced expression of myogenic genes and defects in myogenic differentiation. We have utilized a conditional knockout mouse approach to examine the functional consequences of Dnmt1 depletion specifically in the developing muscle. These mice were born runted, with smaller body weights, and reduced ability to form myotubes in vitro. We show that expression of Id-1, a negative regulator of myogenesis, is enhanced in Dnmt1-deficient cultures, leading to enhanced transdifferentiation of myoblasts toward the osteogenic lineage. Thus, these studies demonstrate that Dnmt1 influences cellular identity and determines lineage fidelity.
Similar content being viewed by others
Introduction
Epigenetic modification of the chromatin plays crucial roles in maintaining cellular states, genomic stability, ensuring proper gene transcription and DNA repair1,2. Methylation of cytosine residues in DNA is carried out by a class of enzymes, the DNA methyltransferases (Dnmts), that tightly regulate the initiation and the maintenance of these methyl marks3,4. Dnmt3a and Dnmt3b are responsible for establishing the de novo patterns of methylation during embryogenesis, while Dnmt1 is responsible for the propagation of methylation patterns. Errors in this enzymatic machinery have profound effects on development and disease5.
All three Dnmts are required for embryonic development, with previous studies reporting Dnmt1−/− and Dnmt3b−/− mice to be embryonically lethal6,7. While Dnmt3a−/− mice survive to full term, they die around 4 weeks of age7. Surprisingly, embryonic stem cells (ESCs) can be derived without Dnmts and maintain their stem cell characteristics8. Studies have also highlighted critical roles for DNA methylation in adult stem cells. Loss of Dnmt1 in neuronal progenitors leads to global genomic hypomethylation and neonatal death9, while Dnmt1−/− fibroblasts undergo growth arrest and widespread apoptosis10. In the hematopoietic system, Dnmt3a and Dnmt3b deficiency leads to defects in self-renewal of hematopoietic stem cells, but had no effect on cellular differentiation11. However, the loss of Dnmt1 resulted in self-renewal and differentiation abnormalities in the same cell type12. Studies in the mammalian epidermis reported a premature differentiation of epidermal progenitor cells and tissue loss in the absence of Dnmt1, further emphasizing a role of DNA methylation in maintaining the undifferentiated progenitor cell state13.
In skeletal muscle, the importance of DNA methylation in the control of myogenesis was shown in the regulation of master myogenic transcriptional factor, MyoD. MyoD is selectively expressed in skeletal muscle cells and its expression in non-muscle cells is suppressed by DNA methylation14. Demethylating agents such as 5-azacytidine can induce MyoD transcription and lead to myogenic conversion of non-muscle cells such as in the CH310T1/2 and NIH3T3 cell lines15,16,17. These studies suggest that epigenetic marking through DNA methylation plays a significant role in determining cell fate during muscle development. Limited data exists on the function of Dnmts during myogenesis. It has been reported that DNMT1 mRNA expression in human myoblasts decreased with differentiation, and this coincided with increases in myogenic differentiation gene expressions13. While these results are consistent with the notion that DNA methylation in self-renewal of skeletal muscle stem cells exists, more detailed analyses are needed to formally establish this.
In the present study, we have analysed the effect of Dnmt1 depletion during murine myogenesis. We demonstrate the Dnmt1 is essential for proper myogenic differentiation and cell fate transition. Absence of Dnmt1 in myoblasts lead to reduced myogenic gene expressions and defects in myoblast fusion that results in the formation of smaller myotubes. Mice with muscle-specific deletion of Dnmt1 are runted and display smaller body weight. We show that loss of Dnmt1 results in hypomethylation of the Id-1 promoter, a negative regulator of myogenesis, leading to increased propensity of myoblasts transdifferentiate into the osteogenic lineage. These studies will lead to a better understanding of the epigenetic mechanisms that regulate muscle development, having implications in interventions aimed at combating musculoskeletal disorders such as muscular dystrophy.
Results
Loss of Dnmt1 lead to the formation of immature myotubes
We first determined the expression pattern of Dnmt1 during differentiation using the well-established myogenic cell line, C2C12 (Supplementary Fig. 1). We show that Dnmt1 transcript level was downregulated during myogenic differentiation (Fig. 1A). To study the functional significance of Dnmt1 in myogenesis, we generated a stable Dnmt1 knockdown C2C12 cell line where the myoblasts were transduced with lentiviral shRNA to target Dnmt1 and selected using puromycin (shDnmt1). Efficient reduction of Dnmt1 expression was observed in the transduced C2C12 cells (Supplementary Fig. 2A). Furthermore, knockdown of Dnmt1 was highly specific and did not affect Dnmt3a or Dnmt3b expression (Supplementary Fig. 2B). Myf5 and MyoD are required for myogenesis and are expressed in both myoblasts and myotubes. Myogenin (Myog) is an early marker of myoblasts entering the differentiation pathway18,19, and myosin heavy chain 2 (Myh2) is highly expressed when myoblasts differentiate and fuse to form myotubes20. We detected no difference in Myf5 transcript levels in the Dnmt1 depleted C2C12 cultures, while MyoD, Myog and Myh2 expression were significantly reduced (Fig. 1B). The ability of shDnmt1 myoblasts to undergo myogenic differentiation was assessed 7 days after differentiation media was added to confluent myoblast cultures. Consistent with the qPCR data, we observed decreased number of Myog- and Myh2-expressing cells in the knockdown cells which translated to significant reduction in the number of myotubes formed in the shDnmt1 cultures (Fig. 1C). We found this difference was not due to differences in cellular proliferation between the control and knockdown cultures as the number of cells were similar between groups (Supplementary Fig. 3), but rather a defect in the ability of the shDnmt1 cells to fuse (Fig. 1D,E). These data indicated that knockdown of Dnmt1 prevented differentiation of C2C12 myoblasts into myotubes.
Dnmt1 knockdown adversely affects myotube formation.
(A) Dnmt1 expression following differentiation of C2C12 cells. *P < 0.05 over D0. (B) qPCR analysis for myogenic genes in the shCtrl compared to shDnmt1 myoblasts. *P < 0.05 over shCtrl cultures. (C) Myog and Myh2 immunofluorescence in Dnmt1 knockdown upon induction of myogenic differentiation. Quantitation of the staining results showing number of Myog+ and Myh2+ cells in the knockdown cultures compared to shCtrl are shown in the bar graph. Scale bar = 50 μm. (D) Fusion indexes were calculated after 7 days of differentiation. *P < 0.05 over shCtrl cells. (E) Analysis of the number of nuclei calculated in Myh2-stained myotubes. *P < 0.05 over shCtrl myoblasts.
Dnmt1 loss coincides with a loss of myogenic gene expression in myoblasts
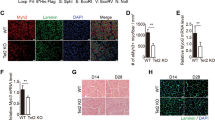
To further establish a role for Dnmt1 in myogenic differentiation, we conditionally deleted Dnmt1 expression in murine myoblasts by crossing the Dnmt1fl/fl mice12 with a transgenic line expressing Cre recombinase under the regulation of the human skeletal alpha-actinin 1 promoter (Acta1-cre)21. The Acta1 promoter is transcribed specifically in skeletal muscle at post-coitum day 9.5 (E9.5) coinciding with the earliest stages of skeletal muscle development and differentiation22. Acta1-cre+: Dnmt1fl/fl mice were viable and born in Mendelian ratios, however the conditional knockout mice were born runted with smaller body weight compared to their littermate controls (Fig. 2A). This smaller body composition persisted to adulthood (Fig. 2B).
Loss of Dnmt1 results in attenuated myogenesis.
(A) Appearance of the Acta1-cre+: Dnmt1f/f mice and littermate controls (Acta1-cre+: Dnmt1+/+) during postnatal development. Asterisks indicate Acta1-cre+: Dnmt1f/f mice. (B) Weight measurements of mice during development. *P < 0.05 over WT and Het mice of same age. (C) Myotube formation in the littermate control and the Acta1-cre+: Dnmt1f/f mice. Number of myotubes per field were counted and graphed on the right. Five individual fields over three independent cultures were analysed. *P < 0.05 over Acta1-cre+: Dnmt1+/+ littermate controls. Scale bar = 100 μm. (D) Acta1-cre+: Dnmt1f/f myoblasts were transduced with retroviruses expressing EGFP or overexpression (OE) of Dnmt1 for 48 hours, and cultured in differentiation media for 5 days. Scale bar = 100 μm. Number of myotubes per field (five individual fields over three independent experiments) counted in the EGPF and Dnmt1 O/E cultures are shown in the bar graph.
We are able to isolate myoblasts with >98% purity based on gene expression and staining for myogenic markers (Supplementary Fig. 4). We verified by qPCR that Dnmt1 was efficiently deleted in the myoblasts isolated from the Acta1-cre+: Dnmt1fl/fl mice, and had no effect on Dnmt3a and Dnmt3b expression (Supplementary Fig. 5). Consistent with our Dnmt1 depleted C2C12 cells, we observed attenuated ability of myoblasts from the Acta1-cre+: Dnmt1fl/fl mice to form myotubes in vitro (Fig. 2C). This coincided with reduced expression levels of myogenic genes in the knockout mice compared to littermate controls (Acta1-cre+: Dnmt1+/+) at baseline (Supplementary Fig. 5). To confirm these observations were indeed caused by an absence of Dnmt1 in myoblasts, myoblasts isolated from the Acta1-cre+: Dnmt1fl/fl mice were transduced with a Dnmt1 over-expressing retrovirus, and myogenic gene expression then re-analyzed. Dnmt1 levels following transduction were restored to levels similar to that of the littermate controls. Similarly, expression for Myf5, MyoD, Myog and Myh2 were also increased in Acta1-cre+: Dnmt1fl/fl myoblasts with Dnmt1 overexpression (Supplementary Fig. 6). However, this restoration of Dnmt1 was not able to increase myotube formation in the Acta1-cre+: Dnmt1fl/fl myoblasts (Fig. 2D). Together, these data confirm an important role of Dnmt1 in regulating myogenesis.
Loss of Dnmt1 leads to increased activation of Inhibitor of DNA binding
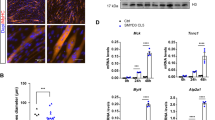
In addition to the basic helix-loop-helix (bHLH) myogenic factors (MyoD, Myf5 and Myog) that are known to have critical roles in orchestrating the muscle phenotype, the myocyte enhancer factor 2 (Mef2) family of genes are also required for the regulation of myogenic gene expression23. In line with the function of Mef2c during muscle maturation, we show that Mef2c expression is markedly increased with C2C12 differentiation (Fig. 3A). However, Mef2c expression in Dnmt1 depleted cells was almost 4-fold lower compared to scrambled control at baseline, and remained low throughout the duration of the differentiation (more than 7-fold lower than shCtrl cultures over the differentiation time course) (Fig. 3A). The inhibitor of DNA binding family of HLH proteins (Id1-4) are thought to affect the balance between cell growth and differentiation by negatively regulating the function of bHLH transcription factors24. Id-1 is a negative regulator of MyoD25, and its overexpression impairs the ability of myoblasts to differentiate into myotubes24,26. Similar to previous reports, Id-1 expression decreases with myogenic differentiation (Fig. 3B). However, we observed elevated Id-1 baseline expression in the shDnmt1 myoblasts compared to shCtrl cells, as well as increasing levels of Id-1 in the shDnmt1 cells as the cells underwent differentiation (Fig. 3B). The presence of lower expression of Mef2c together with higher levels of Id-1 gene expression following differentiation could in part explain the observation of an arrest in cellular differentiation and the failure to form myotubes as seen in the Dnmt1-deficient cells.
Absence of Dnmt1 leads to hypomethylation of the Id-1 promoter.
(A) qPCR analysis of Mefc expression in shCtrl and shDnmt1 myoblasts over 7 days of differentiation. *P < 0.05 over D0 shCtrl; #P < 0.05 over D0 shDnmt1. (B) qPCR analysis of Id-1 expression in shCtrl and shDnmt1 myoblasts over 7 days of differentiation. *P < 0.05 over D0 shCtrl; #P < 0.05 over D0 shDnmt1. (C,D) ChIP-qPCR was performed with H3K4me3 or H3K27me3 antibodies on chromatin obtained from day 0 myoblasts (C) or day 7 myotubes (D). The precipitated DNA was amplified by qPCR using specific primers targeting a region of Id-1 promoter *P < 0.05 over shCtrl.
Dnmt1 is known to promote DNA methylation. Therefore we sought to evaluate the chromatin status of Id-1 loci in Dnmt1-deficient myoblasts. Chromatin immunoprecipitation (ChIP) assays were performed in shCtrl and shDnmt1 C2C12 cells with antibodies directed against H3K4me3 (active) and H3K27me3 (inactive) chromatin. ChIP-qPCR analysis showed similar levels of H3K4me3 and H3K27me3 between the shCtrl and shDnmt1 myoblasts at the Id-1 promoter prior to differentiation (Fig. 3C). Following induction of myogenic differentiation, significant enrichment for the H3K27me3 mark at the Id-1 promoter in the shCtrl cells was observed (Fig. 3D), consistent with decreased expression of Id-1 levels in shCtrl myoblasts following differentiation as seen by qPCR. In stark contrast, the Id-1 promoter remained in the euchromatic state in the shDnmt1 myoblasts 7 days after initiation of myogenic differentiation as demonstrated by the extensive enrichment for the H3K4me3 mark (Fig. 3D). These results are consistent with the differentiation defects observed with Dnmt1 knockdown in C2C12 myoblasts. Therefore, these results demonstrate that Dnmt1 in myoblasts is required for methylation of Id-1, and that a diminished level of Dnmt1 prevents transcriptional repression of Id-1, leading to dysfunctional myogenesis.
Deficiency of muscle-specific Dnmt1 leads to increased osteogenesis
Muscle cells have the potential to transdifferentiate into the osteogenic lineage in the presence of bone morphogenetic proteins (BMPs)27. BMP induce the expression of Id-1, which negatively regulates myogenesis25. To test whether the increased expression of Id-1 as a result of Dnmt1 deficiency can affect the ability of myoblasts to undergo osteogenic differentiation, shCtrl and shDnmt1 myoblasts were cultured in media in the absence or presence of BMP-4. In agreement with previously published reports, addition of BMP-4 decreased the expression of myogenic genes in shCtrl myoblasts, and this reduction was further exacerbated in shDnmt1 myoblasts in the presence of BMP-4 (Fig. 4A). Concomitant with this decrease in myogenic genes was an increase in expression of early (Runx2, alkaline phosphatase, Alp, and osterix, Osx) and late (bone sialoprotein, Bsp; and osteocalcin; Ocn) osteogenic markers (Fig. 4B). Induction of osteogenic gene expression was greater in the shDnmt1 cells compared to shCtrl cultures as demonstrated by ALP staining and activity, and the ability to form mineralized nodules as assessed by Alizarin Red S (Fig. 4C). In agreement with the osteogenic differentiation data, a more transcriptionally active Ocn promoter was evident in shDnmt1 cells compared to shCtrl myoblasts at both baseline and following osteogenic differentiation (Fig. 4D). Taken together, these results demonstrate that Dnmt1 safeguards myoblasts against transdifferentiation into alternative lineages such as the osteogenic lineage.
Dnmt1 knockout myoblasts undergo enhanced osteogenic differentiation.
(A) shCtrl and shDnmt1 C2C12 myoblasts were grown in osteogenic media in the absence or presence of BMP-4 to stimulate osteogenic differentiation, and expression for myogenic markers MyoD and Myog was assessed by qPCR. *P < 0.05 over shCtrl - BMP-4; #P < 0.05 over shDnmt1 - BMP-4. (B) qPCR analysis of early (Runx2, ALP, Osx) and late (Bsp, Ocn) osteogenic genes in shCtrl and shDnmt1 myoblasts without and with BMP-4. *P < 0.05 over shCtrl - BMP-4; #P < 0.05 over shDnmt1 - BMP-4. (C) ALP staining performed 4 days following osteogenic differentiation, and Alizarin Red S staining performed 7 days following differentiation, in the shCtrl and shDnmt1 cells. ALP activity in shCtrl and shDnmt1 cells after 4 days of BMP-4 treatment is shown in the bar graph. *P < 0.05 over shCtrl - BMP-4. (E) ChIP-qPCR was performed with H3K4me3 or H3K27me3 antibodies on chromatin obtained from day 0 myoblasts or day 7 myotubes. The precipitated DNA was amplified by qPCR using specific primers targeting a region in the Ocn promoter. *P < 0.05 over shCtrl.
Discussion
It was hypothesised more than 30 years ago in two independent seminal papers by Riggs28 and Holliday and Pugh29 that DNA methylation could alter gene expression by influencing the binding affinities of transcription factors or other proteins to DNA. DNA methylation has been well studied in embryos and during development, but only recently has the role of Dnmts been examined in somatic cells. Concentrating on the muscular system, we found the abrogation of Dnmt1 reduced myogenic gene expression and differentiation capacities in myogenic cells (Figs 1 and 2). Previous studies have raised the possibility that de novo methylation carried out by Dnmt3a and Dnmt3b may also contribute to maintenance methylation in the absence of Dnmt130,31. However, we did not find any compensatory up-regulation of these de novo methyltransferases when Dnmt1 was depleted in our cells (Fig. 3, Supplementary Figs 1 and 2), suggesting that the observed effects were specific to the enzymatic actions of Dnmt1. Interestingly, when we overexpressed Dnmt1 back into Dnmt1-deficient myoblasts, we were able to increase myogenic gene expression. However, we were unable to rescue their inability to form multinucleated myotubes. These data suggest that in addition to affecting myogenic gene expression, some other critical factors during myogenesis are controlled by Dnmt1.
During development, mesenchymal cells can either undergo myogenic, adipogenic, osteogenic or chondrogenic differentiation. Stable modifications made to the methylation pattern of DNA activate lineage-specific genes and prevent the transcription of genes from other lineages. Treatment of mouse C3H10T1/2 fibroblasts with 5-azacytidine32, or the overexpression of antisense RNA against Dnmt133 induced a myogenic program in this cell type that do not normally undergo myogenesis. This effect on lineage specificity associated with the absence of Dnmt1 is consistent with our data where Dnmt1 deficiency in C2C12 myoblasts alters their cellular identity and led to enhanced differentiation into the osteogenic lineage (Fig. 4). To confirm an effect of Dnmt1 in regulating cell fate, the Id genes were examined for their role in transdifferentiation34,35. We further show that the Id-1 promoter is hypomethylated in Dnmt1 knockdown cells.
Errors in DNA methylation has been linked to a number of human diseases1. Aberrant methylations of tumor suppressor genes or oncogenes are frequently linked to the metastatic potential of many tumour types. Mutations in DNMT3B have been linked to human ICF syndrome36,37, and abnormalities in genomic methylation patterns of DNMT3L has been linked to infertility38. Mutations in DNMT1 have also recently been implicated in neurodegenerative diseases39, while hypomethylation of the A161 allele is associated with the pathogenesis of facioscapulohumeral muscular dystrophy40,41. We have shown here that Dnmt1 plays a functional role in myogenesis, and that the Acta1-cre+: Dnmt1fl/fl mice display a runted, dystrophic-like phenotype. Altogether, these results indicate that Dnmt1 is necessary to maintain the correct level of myogenic differentiation, and to prevent promiscuous transdifferentiation into alternate lineages. It provides a new direction for the study of myogenesis and will be interesting in future studies to determine whether Dnmt1 loss translates to premature aging of the tissue or muscular dystrophy.
Materials and Methods
Mouse lines
The Dnmt1fl/fl mice were generously provided by Dr. Stuart Orkin (Children’s Hospital Boston, Harvard Stem Cell Institute, Boston, MA, USA)12 and the Acta1-Cre transgenic mice were purchased from the Jackson Laboratory, with the strain originally published by Miniou and colleagues21. All mice were maintained on a predominantly C57BL/6J background. Acta1-cre+: Dnmt1fl/fl mice were generated by breeding mice heterozygous for the transgenes; Acta1-cre+: Dnmt1+/+ littermates were used as controls. Genotyping was performed on genomic DNA extracted from tail samples and PCR performed using previously published primers and PCR conditions. All experiments were approved by and carried out in accordance with the guideline of Yale University’s Institutional Animal Care and Use Committees.
Cell culture
C2C12 myoblasts were cultured in DMEM supplemented with 20% FBS. Primary myoblasts were isolated from 8–12 week old mice as previously described27. Briefly, hindlimb muscles were enzymatically digested with 0.25% pronase at 37 °C for 1 hour and digestion terminated with the addition of 10% horse serum. Cells were cultured in DMEM containing 20% FBS and antibiotics. Myogenic differentiation was induced by culturing cells in 2% horse serum. Osteogenic differentiation was induced by culturing cells in osteogenic media27 and treated with 0 or 50 ng/ml BMP-4 (120-05, Peprotech). MTS assays were performed according to manufacturer’s protocol (Promega).
Lentiviral production and shRNA knockdown
Dnmt1 shRNA (shDnmt1) and scrambled controls (shCtrl) lentiviruses were generated using standard protocols. Briefly, plasmids containing the control or Dnmt1 constructs were transfected into HEK293 cells using Fugene 6, and the viral supernatant collected and concentrated by ultracentrifugation. Cells were transduced for 48 hours and selected using puromycin (2 μg/ml). Surviving cells were then sequentially passaged to establish stable cell lines, and lines where more than 70% of Dnmt1 was knockdown was used for subsequent studies.
Semi-quantitative and quantitative reverse transcriptase-PCR analysis
Total RNA were extracted from cells using the RNeasy mini kit (Qiagen) and quantified by Nanodrop. cDNA was prepared using the first strand cDNA synthesis kit (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed with SYBR green PCR master mix (Biorad) using the Biorad C1000 thermal cycler. Samples were run in triplicate and normalised to β-actin. Primer sequences used are listed in Table 1.
Histochemical staining and immunocytochemistry staining
Cellular viability was determined using the CellTitre 96 Aqueous One Solution Cell Proliferation Assay kit (Promega) according to the manufacturer’s instructions. Alkaline phosphatase activity and staining was detected as previously described42. Calcium deposits were assessed by Alizarin Red S staining27.
For immunofluorescence staining, cells were fixed with 3.7% paraformaldehyde, permeablized with 0.1% Triton X followed by washing and blocking with 10% FBS. Cells were then incubated with the following primary antibodies overnight at 4 °C: Myh2 (1:100) and Myog (1:50), all from DSHB. Samples were incubated with Alexa 488- and 555-conjugated goat anti-mouse IgG (Invitrogen, 1:250) and DAPI stained.
Western blots and chromatin immunoprecipitation (ChIP)
Cell lysates were used for immunoblotting and ran on a 12.5% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% skim milk and incubated in primary antibodies overnight. Secondary antibodies were incubated for 30 min. Primary antibodies used include β-actin (Santa Cruz, sc-1616), Dnmt1 (Abcam, #13537), Dnmt3a (Abcam, #13888), Dnmt3b (Abcam, #13604). Rabbit anti-mouse-horseradish peroxidise (Sigma, A0168) or goat anti-mouse-horseradish peroxidise (Fisher, 62–6520) secondary antibodies were used.
Chromatin immunoprecipitation (ChIP) was performed as previously described43. Briefly, 1 × 107 cells were crosslinked and used for each immunoprecipitation. DNA was sheared to 200–750 bp by sonication. Protein G Dynabeads (Invitrogen) were used to immunoprecipitate the antibody-antigen complexes and antibodies against H3K4me3 and H3K27me3 were used. H3 and IgG were also included as positive and negative controls, respectively. Following cross-link reversal and proteinase K treatment, immunoprecipated DNA was extracted with phenol-chloroform, ethanol precipitated and eluted. Recovered DNA was purified with PCR Purification Kit (Qiagen) and analysed by quantitative PCR. Primers spanning the promoter regions of Ocn, and Id1, were used to detect amplification of input and immunoprecipitated DNA. Primer sequences are listed in Table 2. All analysis was performed relative to % input.
Statistical analysis
Statistical analyses were performed with unpaired Student’s t test, with P < 0.05 considered significant.
Additional Information
How to cite this article: Liu, R. et al. Dnmt1 regulates the myogenic lineage specification of muscle stem cells. Sci. Rep. 6, 35355; doi: 10.1038/srep35355 (2016).
References
Robertson, K. D. DNA methylation and human disease. Nature reviews. Genetics 6, 597–610 (2005).
Bird, A. DNA methylation patterns and epigenetic memory. Genes & development 16, 6–21 (2002).
Bestor, T., Laudano, A., Mattaliano, R. & Ingram, V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. Journal of molecular biology 203, 971–983 (1988).
Bestor, T. H. The DNA methyltransferases of mammals. Human molecular genetics 9, 2395–2402 (2000).
Shames, D. S., Minna, J. D. & Gazdar, A. F. DNA methylation in health, disease, and cancer. Current molecular medicine 7, 85–102 (2007).
Li, E., Bestor, T. H. & Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926 (1992).
Okano, M., Bell, D. W., Haber, D. A. & Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999).
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes to cells: devoted to molecular & cellular mechanisms 11, 805–814 (2006).
Fan, G. et al. DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals. The Journal of neuroscience: the official journal of the Society for Neuroscience 21, 788–797 (2001).
Jackson-Grusby, L. et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature genetics 27, 31–39 (2001).
Tadokoro, Y., Ema, H., Okano, M., Li, E. & Nakauchi, H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. The Journal of experimental medicine 204, 715–722, 10.1084/jem.20060750 (2007).
Trowbridge, J. J., Snow, J. W., Kim, J. & Orkin, S. H. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell stem cell 5, 442–449, 10.1016/j.stem.2009.08.016 (2009).
Sen, G. L., Reuter, J. A., Webster, D. E., Zhu, L. & Khavari, P. A. DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature 463, 563–567, 10.1038/nature08683 (2010).
Tapscott, S. J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development (Cambridge, England) 132, 2685–2695, 10.1242/dev.01874 (2005).
Weintraub, H. et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proceedings of the National Academy of Sciences of the United States of America 86, 5434–5438 (1989).
Jones, P. A. & Taylor, S. M. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20, 85–93 (1980).
Konieczny, S. F. & Emerson, C. P. Jr. 5-Azacytidine induction of stable mesodermal stem cell lineages from 10T1/2 cells: evidence for regulatory genes controlling determination. Cell 38, 791–800 (1984).
Wright, W. E., Sassoon, D. A. & Lin, V. K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 56, 607–617 (1989).
Edmondson, D. G. & Olson, E. N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes & development 3, 628–640 (1989).
Perry, R. L. & Rudnick, M. A. Molecular mechanisms regulating myogenic determination and differentiation. Frontiers in bioscience: a journal and virtual library 5, D750–D767 (2000).
Miniou, P. et al. Gene targeting restricted to mouse striated muscle lineage. Nucleic acids research 27, e27 (1999).
Sassoon, D. A., Garner, I. & Buckingham, M. Transcripts of alpha-cardiac and alpha-skeletal actins are early markers for myogenesis in the mouse embryo. Development (Cambridge, England) 104, 155–164 (1988).
Olson, E. N. & Klein, W. H. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes & development 8, 1–8 (1994).
Jen, Y., Weintraub, H. & Benezra, R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes & development 6, 1466–1479 (1992).
Katagiri, T. et al. Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes to cells: devoted to molecular & cellular mechanisms 7, 949–960 (2002).
Gundersen, K. & Merlie, J. P. Id-1 as a possible transcriptional mediator of muscle disuse atrophy. Proceedings of the National Academy of Sciences of the United States of America 91, 3647–3651 (1994).
Liu, R. et al. Myoblast sensitivity and fibroblast insensitivity to osteogenic conversion by BMP-2 correlates with the expression of Bmpr-1a. BMC musculoskeletal disorders 10, 51, 10.1186/1471–2474–10–51 (2009).
Riggs, A. D. X inactivation, differentiation, and DNA methylation. Cytogenetics and cell genetics 14, 9–25 (1975).
Holliday, R. & Pugh, J. E. DNA modification mechanisms and gene activity during development. Science (New York, N.Y.) 187, 226–232 (1975).
Riggs, A. D. & Xiong, Z. Methylation and epigenetic fidelity. Proceedings of the National Academy of Sciences of the United States of America 101, 4–5, 10.1073/pnas.0307781100 (2004).
Jones, P. A. & Liang, G. Rethinking how DNA methylation patterns are maintained. Nature reviews. Genetics 10, 805–811, 10.1038/nrg2651 (2009).
Taylor, S. M. & Jones, P. A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell 17, 771–779 (1979).
Szyf, M., Rouleau, J., Theberge, J. & Bozovic, V. Induction of myogenic differentiation by an expression vector encoding the DNA methyltransferase cDNA sequence in the antisense orientation. The Journal of biological chemistry 267, 12831–12836 (1992).
Kowanetz, M., Valcourt, U., Bergstrom, R., Heldin, C. H. & Moustakas, A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Molecular and cellular biology 24, 4241–4254 (2004).
Zebedee, Z. & Hara, E. Id proteins in cell cycle control and cellular senescence. Oncogene 20, 8317–8325, 10.1038/sj.onc.1205092 (2001).
Hansen, R. S. et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proceedings of the National Academy of Sciences of the United States of America 96, 14412–14417 (1999).
Xu, G. L. et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402, 187–191, 10.1038/46052 (1999).
Kobayashi, H. et al. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. European journal of human genetics: EJHG 17, 1582–1591, 10.1038/ejhg.2009.68 (2009).
Klein, C. J. et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nature genetics 43, 595–600, 10.1038/ng.830 (2011).
Padberg, G. W. & van Engelen, B. G. Facioscapulohumeral muscular dystrophy. Current opinion in neurology 22, 539–542, 10.1097/WCO.0b013e328330a572 (2009).
van Overveld, P. G. et al. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Annals of neurology 58, 569–576, 10.1002/ana.20625 (2005).
Katagiri, T. et al. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. The Journal of cell biology 127, 1755–1766 (1994).
Lee, T. I., Johnstone, S. E. & Young, R. A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nature protocols 1, 729–748, 10.1038/nprot.2006.98 (2006).
Acknowledgements
We thank Dr. Yifei Liu for expert advice on ChIP assays. R.L. was the recipient of the Sir Keith Murdoch Fellowship from the American Australian Association. I.-H.P. was supported in part by the Charles Hood Foundation, NIH (GM0099130-01, GM111667-01), CSCRF (12-SCB-YALE-11, 13-SCB-YALE-06), KRIBB/KRCF (NAP-09-3) and CTSA Grant UL1 RR025750 from the National Center for Advancing Translational Science (NCATS), a component of the National Institutes of Health (NIH), and NIH roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Author information
Authors and Affiliations
Contributions
R.L. performed and designed the experiments, and wrote the manuscript. K.-Y.K. and Y.-W.J. performed experiments. I.-H.P. supervised the project, discussed the results and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Liu, R., Kim, KY., Jung, YW. et al. Dnmt1 regulates the myogenic lineage specification of muscle stem cells. Sci Rep 6, 35355 (2016). https://doi.org/10.1038/srep35355
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep35355
This article is cited by
-
Disturbance of calcium homeostasis and myogenesis caused by TET2 deletion in muscle stem cells
Cell Death Discovery (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.