Abstract
A novel nitrate- and selenite reducing bacterium strain ZYKT was isolated from a rice paddy soil in Dehong, Yunnan, China. Strain ZYKT is a facultative anaerobe and grows in up to 150, 000 ppm O2. The comparative genomics analysis of strain ZYKT implies that it shares more orthologues with B. subtilis subsp. subtilis NCIB 3610T (ANIm values, 85.4–86.7%) than with B. azotoformans NBRC 15712T (ANIm values, 84.4–84.7%), although B. azotoformans NBRC 15712T (96.3% 16S rRNA gene sequence similarity) is the closest Bacillus species according to 16S rRNA gene comparison. The major cellular fatty acids of strain ZYKT were iso-C14:0 (17.8%), iso-C15:0 (17.8%), and C16:0 (32.0%). The polar lipid profile consisted of phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol and an unidentified aminophospholipid. Based on physiological, biochemical and genotypic properties, the strain was considered to represent a novel species of the genus Bacillus, for which the name Bacillus oryziterrae sp. nov. is proposed. The type strain is ZYKT (=DSM 26460T =CGMCC 1.5179T). Strain ZYKT can reduce nitrate to nitrite and ammonium and possesses metabolic genes for nitrate reduction including nar, nap and nrf. Biogenic selenium nanoparticles of strain ZYKT show a narrow size distribution and agree with the gaussian distribution. These selenium nanoparticles show significant dose-dependent inhibition of the lung cancer cell line H157, which suggests potential for application in cancer therapy.
Similar content being viewed by others
Introduction
Rice is widely cultivated and feeds more than half of the world’s population1. Paddy soils typically undergo wet and dry cycles during rice cultivation2 and diverse bacterial groups are active and play key roles in biogeochemical cycling, thus influencing the levels of trace elements, such as Se, in rice3,4. Members of the genus Bacillus are numerically dominant proteolytic bacteria in organic-matter amended paddy soils at different stages of rice development (e.g., vegetative growth, maximal tilling, and harvest stage)5. Bacillus spp. are crucial in both organic-nitrogen and inorganic-nitrogen transformation because of their high extracellular protease and nitrate reduction ability6. In paddy soils, Bacillus spp. may play an important role in the biological cycling of C and N, also because of their high resistance to a wet-dry rotation environment through endospore formation. Although dissimilatory nitrate reduction to ammonium (DNRA) in paddy soils has been well studied, Bacillus spp. responsible for this process were not considered until recently7.
Selenium is a key constituent of selenoproteins and is often lacking in rice-based diets where soils are Se-deficient and traditional rice diets do not provide Se levels adequate to maintain health1,8. Se availability in soils is of great importance in how this influences Se content in crops and consequently human Se status. Se concentrations in rice are strongly controlled by bioavailable Se in paddy soils, and water soluble and exchangeable Se is generally considered to be directly taken up by rice9,10. Hence, the cycling of Se in paddy soil is of considerable interest since the microbially mediated oxidation-reduction reactions of Se affects its bioavailability, mobility and solubility11,12,13. However little is known about the bacterial species involved and how they influence Se transformations. Bacterial reduction of selenite to elemental Se can result in selenium nanoparticles (SeNPs) precipitation and accumulation by binding to high-affinity proteins, including alkyl hydroperoxide reductase, peroxiredoxin, NADH peroxidase, and ribosomal protein14,15,16,17. Most SeNPs-bound proteins in bacteria are non-inducible selenite reduction-related oxidoreductases17. Artificial SeNPs have attracted increasing attention in the past decade because of their anticancer activities and low toxicity18,19. SeNPs were found to suppress the growth of prostate LNCaP cancer cells through Akt/Mdm2/AR controlled apoptosis20. Polyethylene glycol (PEG) is the most popular polymer for protein conjugation, and PEGylation has become a leading approach for overcoming most of the limits of the therapeutic application. PEG nanosized ultrasmall SeNPs overcome drug resistance in hepatocellular carcinoma HepG2 cells through induction of mitochondrial dysfunction21. Several synthetic techniques exist to produce SeNPs, however a major limitation is that they do not yield the narrow size distribution important for industrial applications. Biogenic SeNPs are more uniform in size than those from chemical processing and thus have better application potential.
We previously reported on the genome sequence of Bacillus sp. strain ZYKT22. Here, we provide additional details on its characteristics and phylogenetic position, indicating that the strain represents a new species of the genus Bacillus. Strain ZYKT is a facultative anaerobic bacterium with the potential for dissimilatory nitrate reduction to ammonium, selenite reduction and consequent SeNPs production. Our analysis identifies the encoding genes that are responsible for DNRA in strain ZYKT. A proposed model is then established for the mechanisms of DNRA. Selenium transformation occurs through assembly of elemental selenium with intracellular high-affinity proteins. These biogenic SeNPs can significantly inhibit the non-small lung cancer cell line H157.
Results
Characterization of strain ZYKT
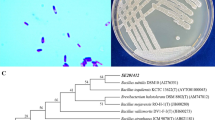
Here we provide a detailed taxonomic characterization indicating that stain ZYKT represents a new species in the genus Bacillus. Phylogenetic relationships were inferred using the maximum-likelihood (ML) method (Supplementary Fig. S1), reconstructed using a selection of well-known Bacillus species, including Bacillus azotoformans NBRC 15712T with 96.3% 16S rRNA gene sequence similarity, Bacillus cohnii DSM 6307T with 95.2% 16S rRNA gene sequence similarity and Bacillus subtilis subsp. subtilis NCIB 3610T with 93.0% gene sequence similarity, and additional species that are known for their ability to transform selenium22,23. Of these, Bacillus selenitireducens ATCC 700615T24 was the closest known selenite reducer and shared 90.0% 16S rRNA gene sequence similarity with strain ZYKT22.
Strain ZYKT is a Gram-positive, spore-forming, motile, flagellated, bacilliform (0.2–0.3 μm wide and 1.5–2.0 μm long) rod (Supplementary Fig. S2)22. Strain ZYKT is a facultative anaerobic bacterium and can grow in the presence of up to 15% O2 on agar plates. Strain ZYKT grew between 21 and 45 °C, with an optimum temperature for growth at 30–40 °C. Growth at pH 6.0–7.6 was observed, with an optimum pH value of at about 7.0. Strain ZYKT grew in the presence of NaCl concentrations ranging from 0% to 1.1% with an optimum near to 0% NaCl (Table 1)22. Strain ZYKT was positive for nitrate and selenite reduction, and acid production from D-glucose and maltose. Strain ZYKT was negative for acid production from D-mannose, D-mannitol and D-xylose, and hydrolysis of casein, starch, gelatin and aesculin, and can use L-histidine and tween-20 as carbon source. The DNA base composition was 36.1 mol% G+C, which is lower than compared to the phylogenetically most closely related Bacillus species (Table 1). This mol% G+C determined by the Tm technique matched that from genome analysis22. A detailed comparison of the characteristics of strain ZYKT with B. azotoformans, B. cohnii and B. subtilis is presented in Table 1.
The fatty acid composition of strain ZYKT, B. azotoformans and B. cohnii are shown in Table 2. The major cellular fatty acids of strain ZYKT were iso-C14:0 (17.8%), iso-C15:0 (17.8%), anteiso-C15:0 (10.4%), iso-C16:0 (10.5%) and C16:0 (32.0%), clearly separating it form B. azotoformans and B. cohnii. The polar lipid profile of strain ZYKT contains PE, phosphatidylethanolamine; DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; UPAL, and an unidentified aminophospholipid (Fig. S3a). The pattern of thin layer chromatogram of strain ZYKT and Bacillus azotoformans were distinctly different in terms of UPL, unknown phospholipid and UL, unknown lipid (Fig. S3b).
General genomic features and examples of gene family expansions
The strain ZYKT genome shows large differences from the previously published genomes of B. azotoformans and B. subtilis subsp. subtilis NCIB 3610T (Fig. S4a–c)22,27,28. Interestingly however, strain ZYKT shared more orthologues with B. subtilis subsp. subtilis NCIB 3610T than with B. azotoformans, although B. azotoformans is the closest Bacillus species according to 16S rRNA gene comparison. The ANIm values between strains ZYKT and B. azotoformans, B. subtilis subsp. subtilis NCIB 3610T were 84.4–84.7% and 85.4–86.7, respectively (Supplementary Tables S1 and S2). These values were below standard ANI criteria for species identity (95–96%).
Selenite, nitrate reduction, and prediction metabolic pathway of dissimilatory nitrate reduction to ammonium
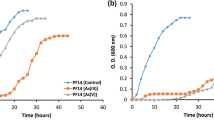
Strain ZYKT reduced SeO32− (up to 80% of 1 mM over 15 days) with production of a red precipitate after 15 days of incubation (Fig. 1A). Figure 1B shows the reduction of NO3− to nitrite and ammonium over 15 days of incubation.
Genes encoding for proteins Bacillus ZYK-3GL000484, Bacillus ZYK-3GL000748, Bacillus ZYK-3GL000884 were identified in the genome of strain ZYKT that may be responsible for the metabolism of nitrogen. The predicted protein sequence of NarG (Bacillus ZYK-3GL000484) in strain ZYKT was 83% similar to NarG found in B. licheniformis 9945A. The respiratory nitrate reductase NarG subunit (1228 amino acids) forms a heterotrimetric structure with subunit NarH and NarI22,29 (Fig. 2). NarI has been shown to anchor the other subunits to the cytoplasmic membrane29. Thus, the presence of a NarI homolog in strain ZYKT suggests that NarGH is oriented towards the cytoplasmic side of the membrane22,30 (Fig. 3). The respiratory nitrate reductase subunit NarJ plays an integral role in assembly of the NarGHI membrane complex22,31, and is predicted to be involved in the transport of nitrate into the cell. A transporter encoded by narL is predicted to transport nitrate into the cell or reduce nitrate into nitrite.
Nap operons have been identified in many prokaryotes, and Nap is found in the soluble periplasmic fraction32. In strain ZYKT, the predicted protein sequence of NapA (Bacillus ZYK-3GL000748) (850 amino acids long) was 83% similar to NapA found in B. azotoformans ATCC 29788T. The catalytic subunit encoded by napA is responsible for the reduction of nitrate to nitrite. Although the function of NapD is not understood, it was speculated to be necessary in maturation of NapA33. napG and napH are predicted to encode for a membrane-bound menaquinol complex22,33 (Figs 2 and 3).
Strain ZYKT is capable of reducing nitrite to ammonium (Fig. 1B). We identified nrfA, which encodes for a membrane-localized protein, which is probably responsible for nitrite reduction in periplasm22. The predicted protein sequence of NrfA (Bacillus ZYK-3GL000884) (477 amino acids long) was 82% similar to that found in Bacillus azotoformans ATCC 29788T (Fig. 3).
Identification of SeNPs High-Affinity Proteins
We separated SeNPs high-affinity proteins from strain ZYKT by SDS-PAGE (Fig. S5). Nano-LC-LTQ results showed a qualitative characterization of high-affinity proteins associated with self-assembled SeNPs in ZYKT cells (Fig. S6). Proteins with imino groups tend to bind with elemental selenium through the formation of a Se-NH-bond, which leads to highly stable SeNPs structures34,35. Hence, proteins play a critical role in the nucleation and self-assembly of SeNPs15,16. The protein types with the highest number of peptides matched (46% of total) are hypothetical proteins because of insufficient information in the database (Table S3). About 27% of the high-affinity proteins were identified as energy metabolic enzymes, 12% as replication and transcription factors and 9% as amino acid metabolic enzymes.
Inhibition of cancer cells H157 by SeNPs
Biogenic SeNPs of strain ZYKT show a narrow size distribution and in line with the gaussian distribution (Fig. 4A–C). Dose-dependent effects of SeNPs on viability of H157 cells show that SeNPs are significantly toxic to H157 cells, only less than 40% cells survive expose to 0.3 μg/μl wet weight SeNPs (Fig. 4D). The results were superior to the effect of chemical generated SeNPs on HepG2 cells which the survival ratio is the same but with nearly 2.0 μg/μl SeNPs35.
Representative SEM images of SeNPs from strain ZYKT (A), The scale is 0.5 μm. Energy dispersive X-ray spectroscopy (EDX) spectrum of the electron-dense SeNPs (B). Corresponding size distribution histograms (C). Dose-dependent inhibitions of SeNPs on the viability of cancer cell line H157 following 24 h exposure (D).
Discussion
In the absence of oxygen, many Bacillus species can respire on nitrate, but the molecular and genetic basis remains poorly characterized. The comparative genomic analysis of strain ZYKT imply that it shares more orthologues with B. subtilis subsp. subtilis NCIB 3610T than with B. azotoformans, although B. azotoformans is the closest Bacillus species according to 16S rRNA gene comparison. Three putative genes that encode for Nar, Nap and Nrf reductases were assigned through sequence comparison. The reductases were closely related to those of B. azotoformans and B. licheniformis. Their locations on either cytoplasmic or periplasmic side were also predicted. Strain ZYKT carries multiple copies of key denitrification genes, encodes both membrane-bound and periplasmic nitrate reductases, and the key genes for nitrite reduction. Modularity and redundancy in nitrate reduction pathways may be a general feature of nitrate-reducing members of Bacillus. Unlike Gram-negative bacteria, Gram-positive microorganisms typically have a very limited space in their periplasm36,37. In this respect, most DNRA enzymes were consistently associated with membrane fractions in ZYKT. Considering the wide-spread occurrence of Bacillus species in paddy soil5, the in situ activity of Bacillus–mediated DNRA warrants further research.
We could not identify a selenite reductase gene because of the lack of reference sequences. Selenite reduction in ZYKT may be performed by Nrf as suggested for Thauera selenatis and Clostridium sp. BXM11,38. Proteomic analysis of SeNPs high-affinity proteins shows abundant proteins with diverse cellular functions. As a general microbial detoxification reaction to oxyanions, selenite can be reduced to elemental Se by thiol groups of proteins/peptides such as peroxiredoxins and NADH peroxidase through “Painter-type” reaction17,35. In this study, no peroxiredoxins or NADH peroxidase was found in those high-affinity proteins, here the mechanism for selenite reduction is not clear. The energy metabolic enzymes may also relate to selenite reduction located in periplasm. The hypothetical proteins might be produced for intracellular binding with SeNPs and subsequent export of SeNPs. Further study is needed on the association of the protein binding with nanoparticles, and hypothetical proteins should be identified as well.
Microbial redox transformations of metals can result in the precipitation of metal nanoparticles across a wide range of environmental conditions39. The aggregation state of these particles may have a strong impact on metal mobility. Our results suggest that aggregation induced by elemental selenium high-affinity proteins plays an important role in limiting elemental selenium dispersal and thus influence the fate of selenium in paddy soils.
Here we demonstrate that the SeNPs generated by ZYKT show a stronger inhibition effect on lung cancer cells than HepG2 cells and human breast carcinoma cells MDA-MB-231 treated by chemically generated SeNPs35,40. This protein derived control of biogenic SeNPs could have implications for industrial-scale production. SeNPs have also been tested in vivo with less toxicity compared with other seleno-compounds41. Further investigation on the underlying molecular mechanisms revealed that depletion of mitochondrial membrane potential and generation of superoxide anions contributed to SeNPs-induced apoptotic R-HepG2 cell death41. The size of SeNPs plays an important role in their biologic activity, and nanoparticle surface-induced oxidative stress has been confirmed as side effect, especially <100 nm, which means the smaller the better42. Nanoparticles of various sizes and chemical compositions preferentially mobilize to mitochondria, giving the possibility of ROS production and therefore interfering with antioxidant defenses21. Future study should be focus on reducing average diameter of biogenic SeNPs to improve their application in cancer therapy.
Description of Bacillus oryziterrae sp. nov
Bacillus oryziterrae (oryziterrae: o.ry.zi.ter’rae. L. n. oryza, rice; L. n. terra, earth, soil; N. L. gen. n. oryziterrae, of the rice soil or field, referring to the source of the type strain). Cells are Gram-positive, spore-forming, motile, flagellated, bacilliform (0.2–0.3 μm wide and 1.5–2.0 μm long) rod. Facultative anaerobic. Grows optimally at pH 7.0 (range 6.0–7.6), 30 °C (range 21–45 °C) without NaCl (range 0–1.1%). Positive for nitrate and selenite reduction, and acid production from D-glucose and maltose. Negative for acid production from D-mannose, D-mannitol and D-xylose, and hydrolysis of casein, starch, gelatin and aesculin, can use citrate, L-histidine and tween-20 as carbon sources. The major cellular fatty acids are iso-C14:0 (17.8%), iso-C15:0 (17.8%), anteiso-C15:0 (10.4%), iso-C16:0 (10.5%) and C16:0 (32.0%). Polar lipid profile of strain contains PE, phosphatidylethanolamine; DPG, diphosphatidylglycerol; PG, phosphatidylglycerol; UPAL, unidentified aminophospholipid. The genomic DNA G+C content is 36.1 mol%.
The type strain is ZYKT (=DSM 26460T =CGMCC 1.5179T), isolated from a rice paddy soil sample, collected from Dehong, Yunnan, China.
Methods
Enrichment and isolation
A soil slurry for the enrichment of anaerobic bacteria was established from a rice paddy soil sample, collected from Dehong, Yunnan (24°41′29″N, 97°58′37″E), China. The vessels with soil slurry were fully filled, sealed with a stopper and kept at 4 °C until used for culture setup. To determine soil characteristics the soil was air-dried, ground with a mortar and crushed to pass through a 2.0 mm sieve. Soil organic carbon (2.3%) was determined by potassium dichromate oxidation titration, and total Fe (28.4 g kg−1) was determined by inductively coupled plasma optical emission spectroscopy (ICP-OES). Plant-available sulfur (14.4 mg kg−1) was extracted by Ca(H2PO4)2 (0.01 mol l−1) and determined by turbidimetry. Soil-available Fe (36.5 mg kg−1) was extracted by DTPA-CaCl2-TEA and determined by ICP-OES. Soil-available Mn (17.1 mg kg−1) was extracted by ammonium acetate (1.0 mol l−1) and determined by ICP-OES.
An anoxic freshwater medium was used for cultivation22. To establish anoxic cultures, fresh soil sample (5.0 g) was mixed with 100 ml anoxic water, then 5.0 ml of soil suspension was added to a serum bottle containing 50 ml medium and incubated at 30 °C in the dark. Anaerobic culturing techniques were used throughout the enrichment43. During incubation, the enrichment culture was briefly shaken once a day. After 7 days growth, new cultures were inoculated (10% v/v), and after 15 transfers, a stable culture was obtained. For the isolation of bacteria, the method was the dilution to extinction technique. The final dilution culture was purified by streaking on agar plates containing sodium selenite in anoxic jars under an atmosphere of 15% O2 and 6% CO2. The single colony that produced red precipitate was chosen, because selenite reduction products, SeNPs is in red color.
Characterization of strain ZYKT
The cell morphology and spore formation of strain ZYKT was assessed by phase-contrast light microscopy (Nikon E-2100). The 16S rRNA gene sequence (1,544 nt) of strain ZYKT was used to identify related taxa using the BLAST program22,44 and EzTaxon server 2.1 (http://eztaxon-e.ezbiocloud.net). The 16S rRNA gene sequences of related taxa were obtained from GenBank. In total, 299 Bacillus sequences were retrieved from the database and CLUSTAL 2.019,45 used to align the sequences. A phylogenetic tree was constructed using the maximum likelihood method in MEGA 5.05 with bootstrap analysis based on 1000 replications22,46,47.
Experiments comparing strain ZYKT22 to other Bacillus species were conducted in duplicate to test pH, temperature and NaCl ranges for growth in anoxic freshwater medium. Strains ZYKT, Bacillus azotoformans ATCC 29788T, Bacillus cohnii DSM 6307T and Bacillus subtilis subsp. subtilis NCIB 3610T were sub-cultured at least once under the same experimental conditions. For pH studies, the medium was adjusted with anaerobic stock solutions of either HCl or NaOH to give the desired pH of 6.0, 6.2, 6.6, 7.0, 7.3, 7.6, or 7.8. Temperature range for growth was determined at 21, 25, 30, 37, 40, and 45 °C. NaCl requirement/tolerance was tested at concentrations of 0, 0.2, 0.4, 0.6, 0.8, and 1.1% (w/v). The reduction of nitrate and selenite was tested at a concentration of 1.0 mM and monitored by ion chromatography (GP50 gradient pump, a column oven LC25, an electrochemical detector ED50) with a Dionex Ionpac AS14 column (4.6 × 3100 mm). Catalase activity was tested by adding 3% H2O2 to culture plates. The oxidase reaction was performed on filter paper moistened with a 1% (w/v) aqueous solution of N, N, N′, N′-tetramethyl-p-henylenediamine. Carbon source utilization was tested with sodium citrate, D-cellobiose, L-arginine, L-proline, L-serine, L-histidine, and Tween-20 in the medium. Hydrolysis of casein, starch, gelatin and aesculin was determined according to the modified methods described by Lanyi48 and Smibert & Krieg49. Acid production from D-mannose, D-glucose, maltose, D-mannitol and D-xylose was tested as described50.
The bacterial culture grown at 30 °C for 7 days was used for lipid extraction according to a procedure51 employing a single-phase organic solvent system comprised of sodium hydroxide, methanol, distilled water, concentrated hydrochloric acid, n-hexane, methyl tertiary butyl ether. After extraction, 3 ml sodium hydroxide and a few drops of saturated sodium chloride solution were added to the organic-phase, which resulted in a two-phase system. The lipids confined to the upper phase were collected. The lipids were identified by the Sherlock Microbial Identification System (MIDI) using an Agilent 6890 series gas chromatograph (GC-FID) equipped with a 25 m UItra-2 column (0.22-mm internal diameter, 0.33 μm film thickness). Polar lipids of strain ZYKT and reference strain Bacillus azotoformans ATCC 29788T were extracted and analyzed by two-dimensional TLC52.
Genome synteny comparisons
Pairwise alignments for dot plot representations were performed on the six-frame amino acid translation of the genome sequences of strain ZYKT (ANOK00000000) Bacillus azotoformans ATCC 29788T (AJLR00000000) and Bacillus subtilis subsp. subtilis NCIB 3610T (ABQL01000000) using the Promer program in the MUMmer 3.23 package53. For all analyses default parameters were applied; that is, exact matches longer than six amino acids were identified, adjacent exact matches were joined if separated by a gap no longer than 30 amino acids, the resulting clusters were further processed if the total length of their matches was longer than 20 amino acids and then aligned using a BLOSUM62 amino acid substitution matrix. The average nucleotide identity (ANI) estimate values between two genomes were calculated using JSpecies (V1.2.1).
Selenite and nitrate reduction analysis
To examine the reduction of selenite and nitrate strain ZYKT was cultivated anaerobically with either selenite (1.0 mmol l−1) or NaNO3 (1.0 mmol l−1) in 100 ml serum bottles in the dark. Each batch of experiments were established in triplicate, inoculated with 1.0 ml (OD600 0.16) of an exponential phase culture, and incubated at 30 °C, respectively. Samples were taken from the medium after 0, 6, 9, 12, and 15 days of incubation and analyzed for nitrate, nitrite and ammonium. To determine nitrate and nitrite, 0.5 ml of the sample was filtered (0.22 μm) in order to remove particulates that could interfere with ion chromatography. The ion chromatography system consisted of a GP50 gradient pump (Thermo Fisher Scientific Inc. Sunnyvale, California, USA), a column oven LC25, an electrochemical detector ED50 and a Dionex Ionpac AS14 column (4.6 × 3100 mm). The operating condition was operated as described11, with an eluent of 3.5 mmol l−1 Na2CO3 and 1.0 mmol l−1 NaHCO3 at a flow rate of 1.2 ml min−1. The measurement of ammonium was performed by the indophenol-blue colorimetric method and the concentration of ammonium monitored by spectrophotometry (Pgeneral T6, Beijing, China). The measurement of selenite reduction was performed by ion chromatography, and the selenite reduction product, elemental selenium particles, was identified by field emission scanning electron microscopy (FESEM, HITACHI SU8020).
Phylogenetic analysis of nitrate reductases
The protein sequence of putative nitrate reductases Nar, Nap and Nrf, as well as selected putative homologues of the catalytic subunits retrieved via BLASTP searches45 of non-redundant databases at NCBI, were aligned using CLUSTALW54 and analyzed in MEGA 5.0546.
Proteome analysis of SeNPs high-affinity proteins: protein identification, quantification, database construction, and bioinformatic data analysis
SeNPs were extracted from ZYKT culture grown with 1.0 mM selenite for 15 days. SeNPs were treated in loading buffer to dissolve high-affinity proteins, and 20 μl suspension was loaded onto sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The samples were then loaded onto an Agilent C18 trap, followed by nano-LC-ESI-MS/MS analysis55. The peptides were sequentially eluted from the HPLC column with a gradient of 0–90% of buffer B (acetonitrile: water: acetic acid, 80: 19.9: 0.1) in buffer A (acetonitrile: water: acetic acid, 5: 94.9: 0.1) at a flow rate of approximately 0.5 ml min−1 (after split) using surveyor pumps. The eluted peptides were sprayed directly from the tip of the capillary column to the LTQ mass spectrometer (Thermo Finnigan, San Jose, CA) for mass spectrometry analysis. The LTQ mass spectrometer was operated in the data-dependent mode in which first the initial MS scan recorded the mass to charge (m/z) ratios of ions over the mass range from 300–1800 Da. The five most abundant ions were automatically selected for subsequent collision-activated dissociation.
All MS/MS data were searched against the predictive protein sequence according to the genomic data of strain ZYKT (ANOK00000000) and the NCBI database using the SEQUEST (v.28). All searches were performed using a precursor mass tolerance of 3.0 Da calculated using average isotopic masses. Variable modification was set for methionine with the addition of 15.999 Da to represent methionine oxidation, static modification was set for cysteine with the addition of 57.052 Da to represent cysteine carboxyamidation; a fragment ion mass tolerance of 1.0 Da was used. Enzyme cleavage specificity was set to trypsin and no more than two missed cleavages were allowed. The SEQUEST outputs were analyzed using commercial software Thermo Electron BioWorks (Rev.3.3.1 sp1). The filter settings for peptides were as follows:- distinct peptide, Xcorr ≥ 1.9 (z = 1), 2.7 (z = 2), 3.50 (z = 3), Sp ≥ 500, Rsp ≤ 5, at least two distinct peptides per protein.
Cancer cell viability measurements
To test for the toxicity of SeNPs on cancer cells, a common and fatal cancer, non–small lung cancer cell line H157 was chosen. Cell line H157 bought from the Cell Culture Center of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences School of Basic Medicine Peking Union Medical College. H157 cells were grown in RPMI medium with 10% FBS and 1% PEST at 37 °C in a 5% CO2 incubator55. Viability was measured by means of the MTT viability kit in 96-well cell culture plates (Nunc). Cells were counted in a Burke’s chamber, and 10,000 cells per well (200,000 cells/ml) were plated and pre-incubated for 24 h to allow adhesion. After pre-incubation, the medium was discarded, and treated with a series of dosages of SeNPs (0.003, 0.006, 0.012, 0.075, 0.15, and 0.3 μg μl−1 wet weight, in 50 μl RPMI 1640) for 24 h in five replicates. Determination of cells viability was performed according to the operating instructions.
Nucleotide sequence accession numbers
The GenBank accession number for the 16S rRNA gene sequence of strain ZYKT is JX103165. A draft genome is available under accession number ANOK00000000.
Additional Information
How to cite this article: Bao, P. et al. Characterization and Potential Applications of a Selenium Nanoparticle Producing and Nitrate Reducing Bacterium Bacillus oryziterrae sp. nov. Sci. Rep. 6, 34054; doi: 10.1038/srep34054 (2016).
References
Williams, P. N. et al. Selenium characterization in the global rice supply chain. Environ Sci Technol 43, 6024–6030 (2009).
Wassmann, R. et al. Characterization of methane emissions from rice fields in Asia. II. Differences among irrigated, rainfed, and deepwater rice. Nutr Cycl Agroecosyst 58, 13–22 (2000).
Liesack, W., Schnell, S. & Revsbech, N. P. Microbiology of flooded rice paddies. FEMS Microbiol Rev 24, 625–645 (2000).
Weber, S., Stubner, S. & Conrad, R. Bacterial populations colonizing and degrading rice straw in anoxic paddy soil. Appl Environ Microbiol 67, 1318–1327 (2001).
Watanabe, K. & Hayano, K. Distribution and identification of proteolytic Bacillus spp. in paddy field soil under rice cultivation. Can J Microbiol 39, 674–680 (1993).
Watanabe, K. & Hayano, K. Seasonal variation of soil protease activites and their relation to proteolytic bacteria and Bacillus spp. in paddy field soil. Soil Biol Biochem 27, 197–203 (1995).
Ishii, S., Ikeda, S., Minamisawa, K. & Senoo, K. Nitrogen cycling in rice paddy environments: past achievements and future challenges. Microbes Environ 26, 282–292 (2011).
Sun, G. X., Liu, X., Williams, P. N. & Zhu, Y. G. Distribution and translocation of selenium from soil to grain and its speciation in paddy rice (Oryza sativa L.). Environ Sci Technol 44, 6706–6711 (2010).
Cao, Z. H., Wang, X. C., Yao, D. H., Zhang, X. L. & Wong, M. H. Selenium geochemistry of paddy soils in Yangtze River Delta. Environ Int 26, 335–339 (2001).
Zhu, Y. G., Pilon-Smits, E. A., Zhao, F., Williams, P. N. & Meharg, A. A. Selenium in higher plants: understanding mechanisms for biofortification and phytoremediation. Trends Plant Sci 14, 436–442 (2009).
Bao, P., Huang, H., Hu, Z. Y., Häggblom, M. M. & Zhu, Y. G. Impact of temperature, CO2 fixation and nitrate reduction on selenium reduction, by a paddy soil Clostridium strain. J Appl Microbiol 114, 703–712 (2013).
Doran, J. W. Microorganisms and the biological cycling of selenium. In Marshall, K. L. (Ed.) Advances in Microbial Ecology Vol. 6, Plenum Press pp. 1–32 (1982).
Stolz, J. F., Basu, P., Santini, J. M. & Oremland, R. S. Arsenic and Selenium in Microbial Metabolism. Annu Rev Microbiol 60, 107–130 (2006).
Oremland, R. S. et al. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl Environ Microbiol 70, 52–60 (2004).
Debieux, C. M. et al. A bacterial process for selenium nanosphere assembly. Proc Natl Acad Sci USA 108, 13480–13485 (2011).
Dobias, J., Suvorova, E. I. & Bernier-Latmani, R. Role of proteins in controlling selenium nanoparticle size. Nanotechnology 22, 195605 (2011).
Lenz, M., Kolvenbach, B., Gygax, B., Moes, S. & Corvini, P. F. X. Shedding light on selenium biomineralization: Proteins associated with bionanominerals. Appl Environ Microbiol 77, 4676–4680 (2011).
Zhang, J., Wang, X. & Xu, T. Elemental selenium at nano size (Nano-Se) as a potential chemopreventive agent with reduced risk of selenium toxicity: comparison with se-methylselenocysteine in mice. Toxicol Sci 101, 22–31 (2008).
Ahmad, M. S., Yasser, M. M., Sholkamy, E. N., Ali, A. M. & Mehanni, M. M. Anticancer activity of biostabilized selenium nanorods synthesized by Streptomyces bikiniensis strain Ess_amA-1. Int J Nanomed 10, 3389–3401 (2015).
Kong, L. et al. The suppression of prostate LNCaP cancer cells growth by selenium nanoparticles through Akt/MDM2/AR controlled apoptosis. Biomaterials 32, 6515–6522 (2011).
Zheng, S. et al. PEG-nanolized ultrasmall selenium nanoparticles overcome drug resistance in hepatocellular carcinoma HepG2 cells through induction of mitochondria dysfunction. Int J Nanomed 7, 3939–3949 (2012).
Bao, P., Su, J. Q., Hu, Z. Y., Häggblom, M. M. & Zhu, Y. G. Genome sequence of the facultative anaerobic bacterium Bacillus sp. strain ZYK, a selenite and nitrate reducer from paddy soil. Stand Genomic Sci 9, 646–654 (2014).
Pichinoty, F., Durand, M., Job, C., Mandel, M. & Garcia, J. L. Etude morphologique, physiologique et taxonomique de Bacillus azotoformnns . Can J Microbiol 24, 608–617 (1978).
Switzer Blum, J., Burns, B. A., Buzzelli, J., Stolz, J. F. & Oremland, R. S. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloal-kaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch Microbiol 171, 19–30 (1998).
Logan, N. A. et al. Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59, 2114–2121 (2009).
Tindall, B. J., Rosselló-Móra, R., Busse, H. J., Ludwig, W. & Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 60, 249–266 (2010).
Srivatsan, A. et al. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet 4, 8 (2008).
Heylen, K. & Keltjens, J. Redundancy and modularity in membrane-associated dissimilatory nitrate reduction in Bacillus . Front Microbiol 3, 371 (2012).
Blasco, F., Guigliarelli, B., Magalon, A., Giordano, G. & Rothery, R. A. The coordination and function of the redox centers of the membrane-bound nitrate reductases. Cell Mol Life Sci 58, 179–193 (2001).
Martinez-Espinosa, R. M. et al. Look on the positive side! The orientation, identification and bioenergetics of ‘Archaeal’ membrane-bound nitrate reductases. FEMS Microbiol Lett 276, 129–139 (2007).
Dubourdieu, M. & DeMoss, J. A. The narJ gene product is required for biogenesis of respiratory nitrate reductase in Escherichia coli . J Bacteriol 174, 867–872 (1992).
Richardson, D. J., Berks, B. C., Russell, D. A., Spiro, S. & Taylor, C. J. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci 58, 165–178 (2001).
Rauschenbach, I., Yee, N., Häggblom, M. M. & Bini, E. Energy metabolism and multiple respiratory pathways revealed by genome sequencing of Desulfurospirillum indicum strain S5. Environ Microbiol 13, 1611–1621 (2011).
Zhang, W. et al. Biosynthesis and structural characteristics of selenium nanoparticles by Pseudomonas alcaliphila. Colloid Surf. B-Biointerfaces 88, 196–201 (2011).
Zhang, J. S., Gao, X. Y., Zhang, L. D. & Bao, Y. P. Biological effects of a nano red elemental selenium. Biofactors 15, 27–38 (2001).
Merchante, R., Pooley, H. M. & Karamata, D. A. Periplasm In Bacillus subtilis. J Bacteriol 177, 6176–6183 (1995).
Matias, V. R. & Beveridge, T. J. Lipoteichoic acid is a major component of the Bacillus subtilis periplasm. J Bacteriol 190, 7414–7418 (2008).
DeMoll-Decker, H. & Macy, J. M. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch Microbiol 160, 241–247 (1993).
Moreau, J. W. et al. Extracellular proteins limit the dispersal of biogenic nanoparticles. Science 316, 1600–1603 (2007).
Luo, H., Wang, F., Bai, Y., Chen, T. & Zheng, W. Selenium nanoparticles inhibit the growth of HeLa and MDA-MB-231 cells through induction of S phase arrest. Colloids Surf B Biointerfaces 94, 304–308 (2012).
Nel, A. E. et al. Understanding biophysicochemical interactions at the nano–bio interface. Nat Mater 8, 543–557 (2009).
Li, N. et al. Ultrafine particulate pollutants induce oxidative stress and mitochondrial damage. Environ Health Perspect 111, 455–460 (2003).
Widdel, F. & Bak, F. Gram-negative mesophilic sulfate reducing bacteria. In The Prokaryotes Balows, A., Trüper, H. G., Dworkin, M., Harder, W. & Schleifer, K. H. New York, Springer, pp 3352–3378 (1992).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J Mol Biol 215, 403–410 (1990).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22, 4673–4680 (1994).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24, 1596–1599 (2007).
Felsenstein, J. Confidence-limits on phylogenies-an approach using the bootstrap. Evolution 39, 783–791 (1985).
Lanyi, B. Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19, 1–67 (1987).
Smibert, R. M. & Krieg, N. R. Phenotypic characterization. In Methods for General and Molecular Bacteriology Edited by Gerhardt, P. Washington, DC: American Society for Microbiology pp 607–654 (1994).
Leifson, E. Determination of carbohydrate metabolism of marine bacteria. J Bacteriol 85, 1183–1184 (1963).
White, D. C., Davis, W. M., Nickels, J. S. & King, J. D. & Bobbie, R. J. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oceologia 40, 51–62 (1979).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2, 233–241 (1984).
Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol 5, R12 (2004).
Chenna, R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31, 3497–3500 (2003).
Bao, P. et al. Selenite-induced toxicity in cancer cells is mediated by metabolic generation of endogenous selenium nanoparticles. J Proteome Res 14, 1127–1136 (2015).
Acknowledgements
We thank Professor J. P. Euzéby for help with Latin. This research was financially supported by the National Natural Science Foundation of China (No. 41571240). We thank for the support of a visiting professorship to Max M. Häggblom, Chinese Academy of Sciences, Institute for Urban Environment.
Author information
Authors and Affiliations
Contributions
Y.-G.Z. and P.B. designed the study. P.B., K.-Q.X., H.-J.W., H.X. and Y.J. conducted the experiments. P.B., K.-Q.X. and P.-P.X. analyzed the data. P.B., M.M.H. and Y.-G.Z. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Bao, P., Xiao, KQ., Wang, HJ. et al. Characterization and Potential Applications of a Selenium Nanoparticle Producing and Nitrate Reducing Bacterium Bacillus oryziterrae sp. nov.. Sci Rep 6, 34054 (2016). https://doi.org/10.1038/srep34054
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep34054
This article is cited by
-
From contaminant to commodity: a critical review of selenium usage, treatment, and recovery
Reviews in Environmental Science and Bio/Technology (2024)
-
A newly isolated Bacillus amyloliquefaciens SRB04 for the synthesis of selenium nanoparticles with potential antibacterial properties
International Microbiology (2021)
-
Biological Removal of Selenate and Selenite from Wastewater: Options for Selenium Recovery as Nanoparticles
Current Pollution Reports (2020)
-
Metal-tolerant thermophiles: metals as electron donors and acceptors, toxicity, tolerance and industrial applications
Environmental Science and Pollution Research (2018)
-
Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.