Abstract
SET (Su(var), E(z), and Trithorax) domain-containing proteins play an important role in plant development and stress responses through modifying lysine methylation status of histone. Gossypium raimondii may be the putative contributor of the D-subgenome of economical crops allotetraploid G. hirsutum and G. barbadense and therefore can potentially provide resistance genes. In this study, we identified 52 SET domain-containing genes from G. raimondii genome. Based on conserved sequences, these genes are grouped into seven classes and are predicted to catalyze the methylation of different substrates: GrKMT1 for H3K9me, GrKMT2 and GrKMT7 for H3K4me, GrKMT3 for H3K36me, GrKMT6 for H3K27me, but GrRBCMT and GrS-ET for nonhistones substrate-specific methylation. Seven pairs of GrKMT and GrRBCMT homologous genes are found to be duplicated, possibly one originating from tandem duplication and five from a large scale or whole genome duplication event. The gene structure, domain organization and expression patterns analyses suggest that these genes’ functions are diversified. A few of GrKMTs and GrRBCMTs, especially for GrKMT1A;1a, GrKMT3;3 and GrKMT6B;1 were affected by high temperature (HT) stress, demonstrating dramatically changed expression patterns. The characterization of SET domain-containing genes in G. raimondii provides useful clues for further revealing epigenetic regulation under HT and function diversification during evolution.
Similar content being viewed by others

Introduction
Epigenetics is the study of inheritable genetic changes without a change in DNA sequence1. Molecular mechanisms of epigenetic regulation mainly consist of DNA methylation, chromatin/histone modifications and small non-coding RNAs etc2. Being one of most important epigenetic modifications, histone modification occurs primarily on lysines and arginines, including phosphorylation, ubiquitination, acetylation, methylation and others3. Among these covalent modifications, histone methylation and demethylation are catalyzed by Histone Lysine Methyltransferases (KMTs) and Histone Lysine Demethylases (KDMs), respectively.
KMTs commonly include an evolutionarily conserved SET (Su(var), E(z), and Trithorax) domain, which carries enzyme catalytic activity for catalyzing mono-, di-, or tri- methylation on lysine4. The SET domain typically constitutes a knot-like structure formed by about 130–150 amino acids, which contributes to enzymatic activity of lysine methylation5. To date, a number of SET domain-containing proteins have been discovered and analyzed in the released genomic sequences of model plants. Baumbusch et al. early reported that Arabidopsis thaliana had at least 29 active genes encoding SET domain-containing proteins6, and Springer et al. found 32 Arabidopsis SET proteins, which were divided into five classes and 19 orthology groups7, and then Ng et al. detected 7 classes, 46 Arabidopsis SET proteins8. Based on different substrate specificities, Huang et al. have recently proposed a new and rational nomenclature, in which plant SET domain-containing proteins were grouped into six distinct classes: KMT1 for H3K9, KMT2 for H3K4, KMT3 for H3K36, KMT6 for H3K27 and KMT7 for H3K4, while S-ETs contain an interrupted SET domain and are likely involved in the methylation of nonhistone proteins9. Besides the above major KMT classes, rubisco methyltransferase (RBCMT) family proteins are also identified as specific methyltransferases for nonhistone substrate in plants and consist of large subunit Rubisco methyltransferase (LSMT) and small subunit Rubisco methyltransferase (SSMT)8,10.
It was shown that SET domain-containing proteins regulated plant developmental processes such as floral organogenesis, seed development11 and plant senescence12. More recent studies demonstrated that SET domain-containing proteins were also involved in plant defense in response to different environmental stresses. In euchromatin, methylation of histone H3K4, H3K36 and H3K27me3 were shown to be associated with gene regulations including transcriptional activation and gene silencing13. For example, histone modifications (e.g. enrichment in H3K4me3) on the H3 N-tail activated drought stress-responsive genes14. By establishing the trimethylation pattern of H3K4me3 residues of the nucleosomes, ATX1/SDG27 (Arabidopsis Homolog of Trithorax) regulates the SA/JA signaling pathway for plant defense against bacterial pathogens by activating the expression of the WRKY70, which was a critical transcription factor15. By regulating H3K36 methylation of histone proteins in JA (jasmonic acid) and/or ethylene13 and brassinosteroids signaling pathway, Arabidopsis SDG8 (SET Domain Group 8) was shown to play a critical role against fungal pathogens Alternaria brassicicola and Botrytis cinerea16.
Furthermore, low or high temperature stress is one of serious environmental stresses affecting plant development. When Arabidopsis plants were exposed to cold temperature, H3K27me3 was significantly reduced in the area of chromatin containing COR15A (Cold-regulated15A) and ATGOLS3 (Galactinol Synthase 3) 17, which are cold stress response genes. In recent years, high temperature (HT) stress has gradually become a serious threat to crop production as global warming is getting worse. Cotton (Gossypium spp) is one of important crops in many parts of the world and is sensitive to HT stress18, which severely affects pollen formation, pollen germination, subsequent fertilization, and ovule longevity, leading to boll shedding and the significant reduction of cotton yield19. Therefore there is a great urge to screen and identify the potential genes conferring resistance to HT stress in molecular breeding of cotton. However, our understanding of mechanisms of resistance to HT in cotton is limited. The progenitor of Gossypium raimondii (G. raimondii) may be the putative contributor of the D-subgenome of Gossypium hirsutum (G. hirsutum) and Gossypium barbadense (G. barbadense) and, more importantly, provides lots of resistant genes20. In this study, we identified SET domain-containing proteins from whole genome of G. raimondii. Based on the analysis of phylogenetic tree, classification, gene structure and domain organization, gene expression profiling and response to HT stress, these results suggested the possible roles of different GrKMT and GrRBCMT genes in the development of G. raimondii and in response to HT. This study of SET domain-containing protein in G. raimondii have expanded understanding of the mechanism of epigenetic regulation in cotton and potentially provide some clues for discovering new resistant genes to HT stress in cotton molecular breeding.
Results
Identification of 52 SET domain-containing proteins in G. raimondii
To obtain all the member of SET domain-containing proteins in G. Raimondii, BLASTP analysis was performed using the sequence of SET domains of known Arabidopsis SET domain-containing protein against G. Raimondii genome Database. Fifty-two SET domain-containing members were identified in G. raimondii (Fig. 1, Supplementary Table S2, S3). Based on the KMT nomenclature and relationship to Arabidopsis homologs, each sequence was assigned to different KMT families (GrKMTs)9, and the candidate proteins similar to Rubisco methyltransferase family proteins were named as GrRBCMTs8.
Chromosomal distribution of GrKMT and GrRBCMT genes.
52 GrKTTs and GrRBCMTs have been mapped on chromosomes D01-D13 except GrRBCMT;9b (Gorai.N022300). The chromosome map was constructed using the Mapchart 2.2 program. The scale on the chromosome represents megabases (Mb) and the chromosome number is indicated at the top of each chromosome.
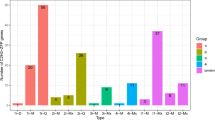
In total, 51 GrKMTs and GrRBCMTs have been mapped on chromosomes D01-D13 except for GrRBCMT;9b (Gorai.N022300) that is still on a scaffold (Fig. 1, Supplementary Table S2). In Chromosome D03, D05 and D08, there are at least six GrKMTs or GrRBCMTs; in chromosome D07, D12 and D13, there are less than six but more than one GrKMTs or GrRBCMTs, while chromosome D02 with 62.8Mb in length has only one member, GrS-ET;3.
According to the canonical criteria21,22, six pairs genes, GrKMT1B;2a/2b, GrKMT1B;3a/3d, GrKMT1B;3b/3c GrKMT2;3b/3c, GrKMT6A;1a/1b, GrRBCMT;9a/9b were diploid and GrKMT1A;4b/4c/4d were triploid. Most of duplicated genes are in class GrKMT1. Among them, GrKMT1B;3b/3c may be tandemly duplicated and others are more likely due to large scale or whole genome duplication except that GrRBCMT;9a/9b cannot be confirmed (Supplementary Table S4). In general, homologous genes are clustered together in the phylogenic tree and the duplicated genes share similar exon-intron structures, higher coverage percentage of full-length-CDS sequence and higher similarity of encoding amino acid (Figs 2 and 3; Supplementary Table S4).
Phylogenetic tree of KMT and RBCMT proteins.
This tree includes 52 SET domain-containing proteins from G. raimondii, 45 from A. thaliana and 44 from O. sativa. The 141 SET domain-containing proteins could be grouped into seven distinct classes, Class KMT1, KMT2, KMT3, KMT6, KMT7, S-ET and RBCMTs. KMT and RBCMT proteins sequences were aligned using Clustal W, and the phylogenetic tree analysis was performed using MEGA 6.0. The tree was constructed with the following settings: Tree Inference as Neighbor-Joining; Include Sites as Partial deletion option for total sequence analyses; Substitution Model: p-distance; and Bootstrap test of 1000 replicates for internal branch reliability. Gr, G. raimondii; At, A. thaliana; Os, O. sativa.
Gene structure of GrKMTs and GrRBCMTs.
The gene structure of GrKMTs and GrRBCMTs were constructed by Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/).
Phylogenetic analysis of SET domain-containing proteins
To analyze the characteristics of 52 SET domain-containing protein sequences in G. raimondii, 45 SET domain-containing protein sequences from A. thaliana and 44 SET domain-containing protein sequences from O. sativa (Supplementary Tables S2 and S3) were also extracted for the phylogenetic analysis. Based on canonical KMT proteins, the above 141 SET domain-containing proteins could be grouped into seven distinct classes (Fig. 2), class KMT1, KMT2, KMT3, KMT6, KMT7 and S-ET9, and class RBCMT once named SETD23. KMT1 exhibits H3K9 substrate specificities activity, KMT2/KMT7 for H3K4, KMT3 for H3K36 and KMT6 for H3K27. RBCMT possesses H3K4 and H3K36 methyltransferase activity in animals, but non-histone target specific proteins in plant8,10. The function of S-ET is still unclear. Furthermore, there are 18 members (10 in KMT1A and 8 in KMT1B) in Class KMT1 as the largest family of KMTs in the SET domain-containing proteins, following by 12 members in class RBCMT, while there is only one member in class KMT7 from each examined species.
Gene structure and domain organization of GrKMTs and GrRBCMTs
To understand the evolutionary origin and putative functional diversification, the gene structure of GrKMTs and GrRBCMTs was analyzed in their constitution of introns/exons. Our results showed that the number of introns/exons was various among different GrKMTs and GrRBCMTs. Most of GrKMT and GrRBCMT genes possess multiple exons, except GrKMT1A;2, GrKMT1A;4a/4b/4c/4d and GrS-ET;1/4a with only one (Fig. 3, Supplementary Table S2). Class GrKMT1A consists of relatively consistent exon number except GrKMT1A;1a/1b with fifteen, GrKMT1A;3a/3b with two and GrKMT1A;3c with four. Altogether, the number of exons in each class genes is greatly variable, and most of Class GrKMT2 genes contain the largest number of exons.
To explore the gene structure, the sequences of full-length GrKMTs and GrRBCMTs were deduced and their domain organization was examined. In GrKMTs, SET domain always locates at the carboxyl terminal of proteins, except Class S-ET and RBCMT. Among the same KMT class, the predicted GrKMTs and GrRBCMTs always share relatively conserved domain organization (Fig. 4, Supplementary Table S3).
Domain organization of GrKMT and GrRBCMT proteins.
Domain organization of SET domain-containing proteins in G. raimondii were detected by SMART and NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and the low-complexity filter was turned off, and the Expect Value was set at 10. The site information of domains was subjected to Dog2.0 to construct the proteins organization sketch map.
Based on the analysis of protein motifs in Class GrKMT1 proteins, they has mostly associated with SET motif and SRA (SET- and RING-associated) motif facilitating DNA accession and the binding of target genes at the catalytic center24. In Class GrKMT1 proteins, they also possess SET domain boundary domains, Pre-SET and Post-SET domains, which are usually present in other plant species25. Pre-SET is involved in maintaining structural stability and post-SET forms a part of the active site lysine channel26. Besides these typical domains, GrKMT1A;3c/4a also include additional AWS domain (associated with SET domain), which is highly flexible and involved in methylation of lysine residues in histones and other proteins27. Class KMT1B proteins also possess SET and Pre-SET domains except GrKMT1B;3a/3d, which are much shorter than the others and only has SET domain (Fig. 4, Supplementary Table S3). Other GrKMT proteins have some additional domain(s): Post-SET domain in GrKMT1B;2a; PB1 (a protein-protein interaction module) and Post-SET domain in GrKMT1B;2b; PWWP (Pro-Trp-Trp-Pro) that is a DNA binding domain and protein-protein interaction domain28, Zf-DBF that is predicted to bind to metal ions and Post-SET in GrKMT1B;3b/3c; F-box which is required for gene silence by means of interaction with core components29 and AWS domain in GrKMT1B;4. Class GrKMT2 proteins contain SET, post-SET and PHD (plant homeodomain) domain except GrKMT2;2a without PHD domain (Fig. 4, Supplementary Table S3). PHD domain has multiple functions by controlling gene expression as an epigenome reader through binding to nucleosomes30. GrKMT2;1 has additional PWWP and FYRN-FYRC (DAST, Domain associated with SET in Trithorax) domains as chromatin-associated proteins involved in histone modifications and a signature feature for the trithorax gene family respectively31. GrKMT2;2a has two GYF (glycine-tyrosine-phenylalanine) domains, which bind to lots of different proline-rich sequences (PRS)32. GrKMT2;3c has an additional SANT (SWI3-ADA2-N-CoR-TFIIIB) domain, which is mainly found in KMT6A. In Class GrKMT3, the SET-domain containing GrKMT3 proteins are more conserved in domain organization and all possess AWS, SET and post-SET domains except GrKMT3;3 with an additional PHD domain (Fig. 4, Supplementary Table S3). It is surprising that SET domain in GrKMT3;2 and GrKMT3;4 are located at the N-terminal or in the middle of the protein sequence, respectively. In Class GrKMT6, the SET-domain containing GrKMT6 proteins are also conserved in domain organization and proteins length (Fig. 4, Supplementary Table S3). GrKMT6A proteins possess SANT, AWS and SET domain except GrKMT6A;1b with an additional MyTH4 (Myosin Tail Homology) domain that can bind to microtubules in combination with FERM proteins (band 4.1, ezrin, radixin, moesin)33. SANT is a putative DNA-binding domain in many transcriptional regulatory proteins and is essential for histone acetyltransferase activity34. GrKMT6B proteins only include PHD and SET domain. In the class GrKMT7 proteins, there is only one member, GrKMT7;1, which is the longest GrKMT protein analyzed with F-box and SET domain.
S-ET proteins commonly have an interrupted SET domain and may be involved in H3K36me3 in human, but their functions are unknown in plant species8. GrS-ET family has 5 members with an interrupted SET domain with 194–264 aa in length. Compared to S-ET proteins in other plant species, they only contain a full interrupted SET domain except GrS-ET;1, which has two additional tandem TPR domains (tetratricopeptide repeat) acting as interaction scaffolds for the formation of multi-protein complexes35. GrRBCMT (plant SETD orthology groups) proteins include SET and Rubis-subs-bind domains except that GrRBCMT;1a/7c/9b only contains a SET domain and GrRBCMT;1b has TPR and SET domains (Fig. 4, Supplementary Table S3).
Tissue and organ expression of GrKMTs and GrRBCMTs
To explore the possible physiological functions of SET domain-containing proteins in G. raimondii, we designed gene-specific real-time quantitative RT-PCR primers (Supplementary Table S1) for detecting the expression patterns of 52 GrKMT and GrRBCMT genes in different tissues and organs, including root, stem, leaf, petal, anther, and ovary.
As indicated in Fig. 5, the SET domain-containing genes from G. raimondii showed diverse expression patterns in different tissues and organs. First, some genes from the same class differentially expressed in the six tissues and organs tested while other genes from different classes could also show similar expression patterns in different tissues and organs, indicating that dramatically functional divergence of GrKMT and GrRBCMT genes during plant development. Second, the expression patterns of GrKMTs and GrRBCMTs are obviously tissue and organ specific at a very low level of expression in reproductive organs and relatively high expression level in vegetative tissues. Furthermore, the majority of genes from different GrKMT and GrRBCMT classes were highly expressed in leaf and stem, indicating that they may play important roles in the development of leaf and stem (Fig. 5). In addition, GrKMT1A;4b and GrKMT1;3b, GrKMT1A;3b and GrS-ET;1 were highly expressed in anther and ovary, respectively, implying their specific functions in the corresponding tissues.
Tissue and organ expression of GrKMTs and GrRBCMTs.
A heatmap for gene expression patterns was generated with the software MultiExperiment Viewer (MeV). The expression patters of GrKMT and GrRBCMT genes are obviously tissue and organ special. Most of genes low express in petal, and high in leaf. Duplicated genes higher express in vegetative organs, and except that GrKMT1A;4b and GrKMT1B;3b strongly express in reproductive organs.
Seven pairs of duplicated genes from the GrKMTs and GrRBCMTs were also highly expressed in vegetative organs, leaf and stem, but with a low expression level in reproductive organs, except that GrKMT1A; 4b and GrKMT1B;3b highly expressed in anther (Fig. 5, Supplementary Figure S1). GrKMT6A;1a/1b and GrRBCMT;9a/9b showed similar expression patterns, while other duplicated genes differentially expressed in the six tissues and organs tested, suggesting that the expression patterns and functions of these genes are diverged during the evolution of gene duplication.
Expression profiles of GrKMTs and GrRBCMTs in response to high temperature stress
Molecular mechanism of epigenetic regulation is poorly understood in response to HT stress in cotton. In our current study, most of GrKMTs and GrRBCMTs were strongly expressed in leaf (Fig. 5). To better understand the roles of the SET domain-containing proteins in response to HT stress, after treatment at 38 °C, the expression profiles of GrKMT and GrRBCMT genes in leaves of seedlings were examined by real-time quantitative RT-PCR (Fig. 6), showing that the expression level of all the GrKMTs and GrRBCMTs genes were more or less affected by HT stress, but the change of their expression patterns were diverse. The expression of most of genes was shown to be decreased under HT conditions; only GrKMT1A;1b, GrKMT1B;3c, and GrKMT6B;2 were up-regulated and reached a peak at 12h after HT treatment. Of these examined genes, GrKMT1A;1a, GrKMT3;3 and GrKMT6B;1 were dramatically down-regulated after the HT treatment.
Expression of GrKMTs and GrRBCMTs in response to high temperature.
Many GrKMT and GrRBCMT genes are involved in high temperature response. Among them GrKMT1A;1a with H3K9 activity, GrKMT3;3 with H3K36 activity and GrKMT6B;1 with H3K27 activity maintain lower expression level at the process of the high temperature treatments. The error bars depict SD, and the asterisk shows the corresponding gene significantly up- or down-regulated by Student′s t test between the treatment and the control (P < 0.05).
All in all, upon exposure to HT, the transcript levels of seven members of GrRBCMT (GrRBCMT;1b/6a/7a/7c/8/9a/9b), five of GrKMT1 (GrKMT1A;1a/3a, GrKMT1B;2a/3c/4), two of GrKMT6 (GrKMT6A;1b/GrKMT6B;1), two of GrS-ET (GrS-ET;1/GrS-ET;2), GrKMT2;3b and GrKMT3;3, were significantly different from that of control at least at one time point (P < 0.05, Fig. 6).
Discussion
Classification and putative functions of GrKMTs and GrRBCMTs genes were predicted
Allotetraploid cotton G. hirsutum and G. barbadense are important economical crops and model plants for polyploids evolution studies. Genomes of G. hirsutum and G. barbadense may derived from allopolyploidization of D-subgenome (G. raimondii Ulbrich) and A-subgenome (G. herbaceum L)36. D-subgenome does not produce any spinnable fiber, but provides many fiber genes after merging with A genome37, contributing to stress tolerance during allotetraploid cotton domestication20. Nowadays, it is known that G. ramondii genome encodes 1004 resistant genes to Verticillium wilt38, 35 auxin response factors (ARFs)22 and 205 putative R2R3-MYB genes39 and so on.
In previous studies, it was shown that histone modifications played important roles in plant development11 and response to biotic and abiotic stress40. KMTs and KDMs tightly regulated the methylation status of lysine residues within histones41. Furthermore the status of histone lysine methylation links to the regulation of the expression of targeted genes. For example, H3K9 and H3K27 methylation is associated with gene silencing, whereas H3K4 and H3K36 methylation lead to gene activation42. It was known that histone lysine methyltransferases shared a highly conserved SET domain except Dot1 for H3K79 methylation43. SET domain-containing proteins could be divided into seven classes, based on their specificity for substrates9. In this study, we revealed that G. ramondii possessed 52 SET domain-containing proteins, which could be grouped to six KMT and one RBCMT classes (Fig. 2) including KMT1 (18), KMT2 (6), KMT3 (5), KMT6 (5), KMT7 (1), S-ET (5) and RBCMT (12). In SET domain-containing proteins of G. ramondii belonging to the first six classes, it was found that their domain organization was largely similar to the counterparts in Arabidopsis and Brassica rapa9. Besides SET domain and several associated domains, our results also showed that GrKMT1A, GrKMT2, GrKMT3, GrKMT6A, GrKMT6B and GrKMT7 proteins also contained SRA domain, PHD and PWWP domain, AWS domain, SANT domain, PHD domain, F-box domain respectively. Moreover, the domain organization of KMT1B is much more complex (see Fig. 4, Supplementary Table S3). Among these domain or motifs in the KMT proteins from G. ramondii, the intact SET domain in GrKMT proteins could insure the necessary methyltransferases activity, and other domains could provide more auxiliary roles.
G. ramondii was also found to have longer and interrupted SET domain in GrS-ET and Rubis-subs-bind domains in GrRBCMT. Previous report indicated that class S-ET proteins might lack methyltransferase activity. In animals, SETD3 containing SET and Rubis-subs-bind domains was found to have a H3K4/K36 methyltransferase activity44. Even though RBCMT proteins were identified in land plants and green algae, but their biological functions were still uncertain23. Ng et al. suggested that RBCMT class proteins had the weaker KMT activity from their similar and longer SET domain than that of canonical KMTs, but maintained the activity of non-histone substrate-specific methylation8. Ma et al. also found that LSMTs could trimethylate Rubisco in Fabaceae, Cucurbitaceae and Rosaceae, in addition to chloroplastic aldolases, which were only aldolases in most other plants10. However, possible biological functions of both GrS-ET and GrRBCMT proteins are still unclear in our current study.
Based on previous studies in SET domain-containing proteins in several plant species, we could predict the substrate specificities of different SET domain-containing proteins in G. ramondii: KMT1 for H3K9, KMT2 for H3K4, KMT3 for H3K36, KMT6 for H3K27 and KMT7 for H3K4 and also RBCMT for putative non-histone substrates.
GrKMTs and GrRBCMTs genes were involved in HT response
Genetic and epigenetic regulations of genes were demonstrated to play key roles in plant response to environmental high or low temperature. It was documented that histone methylation was the major epigenetic regulatory mechanism in response to biotic or abiotic stresses45. KMT proteins regulated the activity of target genes by methylating histone H3, such as, H3K4me and H3K36me associating with transcriptional activation, whereas H3K9me and H3K27me leading to gene silence13. It was also documented that drought stress14, pathogens46 and chilling17 response gene could be regulated by histone methylation. However, the roles of KMT proteins in HT stress were shown to be controversial at best: H3K4me1 of Chlamydomonas reinhardtii and H3K9me2 of OsFIE1 were sensitive to HT, while H3K9me2, H3K27me1/me2/me3 and H3K4me3 in Arabidopsis were not; a transcriptome analysis indicated that differential gene expressions between normal and high temperature conditions were directly related to epigenetic modifications, carbohydrate metabolism, and plant hormone signaling47. Our current results showed that many GrKMTs with histone methylation activity were involved in HT response (Fig. 6). Upon exposure to HT, up- or down- regulation of these genes might affect the status of methylation and further regulate the activity of target genes in response to HT. GrKMT1A;1a with H3K9 activity, GrKMT3;3 with H3K36 activity and GrKMT6B;1 with H3K27 activity maintain lower expression level during the HT response. AtKMT1A;1 (SDG33/SUVH4), homologous gene to GrKMT1A;1a is involved in host defense system by regulating target genes H3K9me48. KMT6B;1(SDG1/CLF) is one of core components of PRC2 and mainly contributes to the H3K27 activity49, whose increase at stress gene loci will repress heat shock response (HSR)50. However, the function of AtKMT3;3 (SDG4/ASHR/SET4) in resistance response is unknown. Therefore, we may infer that the lower level of H3K9 and H3K27 methylation will activate more target genes that are involved in HT responses, and the change of H3K27 activity is completely consistent with Kwon et al.17.
Plant reproductive tissues or organs contribute to seed set yield and are the most vulnerable parts to HT stress51. Our study predicted that GrKMT1A;4b, GrKMT1B;3b, GrKMT1A;3a and GrKMT1A;3b were presumed to be involved in H3K9me. These genes were found to be strongly expressed in anther or ovary, but at a low expression level in the vegetative organs. Among the genes in leaves dramatically regulated by HT stress, GrKMT1A;1a, GrKMT1A;2, GrKMT3;3, GrKMT6B;1, and GrKMT6B;2 highly expressed in anther and ovary (Figs 5 and 6), suggesting that if the roles of GrKMTs and GrRBCMTs were further investigated in reproductive tissues or organs, it would be able to mine novel resistant genes and provide new understanding for plant HT stress response.
Evolution of GrKMTs and GrRBCMTs impacts differentially on their functions
It has been our main interest how the evolution of duplicated genes affects their biological functions, since gene duplication has played a vital role in the evolution of new gene functions and is one of the primary driving forces in the evolution of genomes and genetic systems52. Gene families may evolve primarily through tandem duplication and polyploidy or large-scale segmental duplications52. Arabidopsis genome has undergone about two rounds of duplications before Arabidopsis/Brassica rapa split and after the monocot/dicot divergence53. The outcomes of duplicated genes include nonfunctionalization, neofunctionalization and subfunctionalization54. The nonfunctionalization of one copy is the most likely fate due to deleterious mutation, functionally redundant and dosage constraints54. G. ramondii undergone independent whole-genome duplication event approximately 13.3 to 20.0 million years ago, and shared one paleohexaploidization event with eudicots, but has a higher gene number and lower mean gene density compared with Arabidopsis36, meaning many genes were lost after duplication. We identified 46 KMTs and RBCMTs in Arabidopsis (2n = 10) and only 52 members in G. ramondii (2n = 26). Based on the canonical criteria21,22, seven pairs of GrKMT or GrRBCMT genes were created by the duplication of homologous genes. GrKMT1B;2a/2b, GrKMT1B;3a/3d, GrKMT2;3b/3c, GrKMT6A;1a/1b, GrRBCMT;9a/9b, GrKMT1A;4b/4c/4d might be due to ancient large-scale duplication event, while GrKMT1B;3b/3c may formed by tandem duplication (Supplementary Table S4). Even though GrKMT1B;3a was also shown to meet the parameters of duplicated genes for GrKMT1B;3b/3c/3d in NCBI, they were not considered as duplicated genes since GrKMT1B;3d is much shorter than GrKMT1B;3b/3c (Fig. 4; Supplementary Table S4). GrRBCMT;9a/9b as duplicated genes also could not be confirmed, because GrRBCMT;9b (Gorai. N022300) still not be mapped on any chromosome (Fig. 1).
Duplicated genes can generally be grouped into one clade of phylogenetic tree (Fig. 2); most of these genes exist in sister pairs or triplets and have similar gene structure with possible similar functions, whereas others are divergent in the distribution of introns/exons, suggesting the possibility of functional diversification22. We found that the gene structure was conserved in most of GrKMT genes, except GrKMT6A;1a/1b and GrRBCMT;9a/9b with one exon difference; domain organization of GrKMT1A;4b/4c/4d and GrKMT2;3b/3c were conserved, but GrKMT1B;2a/2b, GrKMT6A;1a/1b and GrRBCMT;9a/9b are divergent (Figs 3 and 4, Supplementary Table S3); only sisters genes of GrKMT6A;1a/1b and GrRBCMT;9a/9b showed similar expression patterns in different tissues and organs. For example, GrKMT1;3b/3c have same gene structure, domain organization, but GrKMT1;3b only highly expresses in anther, and is not involved in HT stress, and GrKMT1; 3c strongly expresses in root, stem and leaf and is sensitive to HT stress (Figs 3, 4, 5, 6; Supplementary Figures S1 and S2). Most duplicated genes also showed similar expression pattern in leaf except GrKMT1A;4b/4c/4d (Supplementary Figures S1 and S2), suggesting that some duplicated genes undergone functional differentiation but others not.
Methods
Identification of SET domain-containing proteins and construction of chromosome map
Sequences of SET domain-containing proteins from Arabidopsis thaliana were retrieved from the official website (https://www.arabidopsis.org/Blast/index.jsp). The sequences of SET domain of these sequences were used as queries to search G. raimondii homologs (http://www.phytozome.net, version 10.3) using the BLASTp. The sequence of SET domain-containing proteins of rice was extracted from Huang et al.9 and web http://www.phytozome.net (version 10.3). All the sequences were re-confirmed in SMART database (http://smart.embl-heidelberg.de/). The gene loci information of G. raimondii was used to generate the chromosome maps by the Mapchart 2.2 program55.
When candidate genes was found to be both >70% coverage of shorter full-length-CDS sequence and >70% identical in the sequence of their encoding amino acids, they were regarded as duplicated genes21. When the duplicated genes were located within 100 kb and were separated by ten or fewer non-homologues, they were defined as tandem duplicated genes22. The coverage of full-length-CDS sequence and the similarity of amino acid sequences were detected by Blastn/Blastp in NCBI.
Analysis of gene structure, domain organization and phylogenetic tree
The gene structure was reconstructed using Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). Domain organization was confirmed by SMART and NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and the low-complexity filter was turned off, and the Expect Value was set at 10. Then the site information of domains was subjected to Dog2.0 to construct the proteins organization sketch map56.
Multiple sequence alignments of SET domains were carried out by the Clustal W program57 and the resultant file was subjected to phylogenic analysis using the MEGA 6.0 program58. Based on the full-length protein sequences, the phylogenetic trees were constructed using Neighbor-Joining methods with Partial deletion and p-distance Method, Bootstrap test of 1000 replicates for internal branch reliability.
Plant material and high temperature treatment
G. raimondii seedlings were grown in greenhouse at 28 °C under a 10 h day/14 h night cycle. 5-week-old seedlings with 5–6 true leaves were placed in a growth chamber at high temperature condition (38 °C; 28 °C as a mock) for 12, 24, and 48 h. The leaves were harvested at the appropriate time points as indicated (triplicate samples were collected at each time point) for detecting genes expression in response to HT. The roots, stems and leaves were collected from plants at the stage of 5–6 true leaves and the petals, anther and ovary were sampled on the day of flowering for gene expression analysis of tissue/organ. The materials were quick frozen in liquid nitrogen and stored at −70 °C for further analysis.
RNA extraction and real-time quantitative RT-PCR
Total RNA was extracted from the materials mentioned above using TRIzol reagent kit (Invitrogen, Carlsbad, CA, US) according to the manufacturer’s specification. The yield of RNA was determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), and the integrity was evaluated using agarose gel electrophoresis stained with ethidium bromide. According to gene sequences of SET domain-containing proteins in G. raimondii (Supplementary Table S2), the primer pairs (Supplementary Table S1) used for real-time quantitative RT-PCR (RT-qPCR) were designed using Roche LCPDS2 software and synthesized by Generay Biotech (Generay, PRC). The amplified fragment lengths were between 75 bp and 200 bp, and the annealing temperature was between 58 °C and 60 °C. The cotton histone3 (AF024716) gene was used as the reference gene.
Quantification was performed with a two-step reaction process: reverse transcription (RT) and PCR. Each RT reaction consisted of 0.5 μg RNA, 2 μl of PrimerScript Buffer, 0.5 μl of oligo dT, 0.5 μl of random 6 mers and 0.5 μl of PrimerScript RT Enzyme Mix I (TaKaRa, Japan), in a total volume of 10 μl. Reactions were performed in a GeneAmp PCR System 9700 (Applied Biosystems, USA) for 15 min at 37 °C, followed by heat inactivation of RT for 5 s at 85 °C. The 10 μl RT reaction mix was then diluted × 10 in nuclease-free water and held at −20 °C. Real-time PCR was performed using LightCycler 480 Real-time PCR Instrument (Roche, Swiss) with 10 μl PCR reaction mixture that included 1 μl of cDNA, 5 μl of 2 × LightCycler 480 SYBR Green I Master (Roche, Swiss), 0.2 μl of forward primer, 0.2 μl of reverse primer and 3.6 μl of nuclease-free water. Reactions were incubated in a 384-well optical plate (Roche, Swiss) at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 30 s. Each sample was run in triplicate for analysis. At the end of the PCR cycles, melting curve analysis was performed to validate the specific generation of the expected PCR product. PCR efficiency (E) was determined from the slope produced by a RT-qPCR standard curve for each pair of primers using the following equation: E = 10(−1/slope) −1, and all the 53 gene primers yielded RT-qPCR data of good quality with a PCR efficiency >0.9 (Supplementary Table S1). The expression values of SET domain-containing proteins genes tested were normalized with the internal reference gene, and the relative expression levels in tissues and in response to HT stress were calculated with 2− ΔCT and 2− ΔΔCT methods59, respectively.
Additional Information
How to cite this article: Huang, Y. et al. Identification of SET Domain-Containing Proteins in Gossypium raimondii and Their Response to High Temperature Stress. Sci. Rep. 6, 32729; doi: 10.1038/srep32729 (2016).
References
Dupont, C., Armant, D. R. & Brenner, C. A. Epigenetics: definition, mechanisms and clinical perspective. Semin. Reprod. Med. 27, 351–357 (2009).
Grewal, S. I. & Rice, J. C. Regulation of heterochromatin by histone methylation and small RNAs. Curr. Opin. Cell Biol. 16, 230–238 (2004).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Herz, H. M., Garruss, A. & Shilatifard, A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 38, 621–639 (2013).
Tschiersch, B. et al. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13, 3822–3831 (1994).
Baumbusch, L. O. et al. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29, 4319–4133 (2001).
Springer, N. M. et al. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 132, 907–925 (2003).
Ng, D. W., Wang, T., Chandrasekharan, M. B., Aramayo, R., Kertbundit, S. & Hall, T. C. Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta 1769, 316–329 (2007).
Huang, Y., Liu, C., Shen, W. H. & Ruan, Y. Phylogenetic analysis and classification of the Brassica rapa SET-domain protein family. BMC Plant Biol. 11, 175 (2011).
Ma, S. et al. Molecular evolution of the substrate specificity of chloroplastic aldolases/Rubisco lysine methyltransferases in plants. Mol. Plant 9, 569–581 (2016).
Thorstensen, T., Grini, P. E. & Aalen, R. B. SET domain proteins in plant development. Biochim. Biophys. Acta 1809, 407–420 (2011).
Ay, N., Janack, B. & Humbeck, K. Epigenetic control of plant senescence and linked processes. J. Exp. Bot. 65, 3875–3887 (2014).
Berr, A., McCallum, E. J., Alioua, A., Heintz, D., Heitz, T. & Shen, W. H. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414 (2010).
Liu, N., Fromm, M. & Avramova, Z. H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant. 7, 502–513 (2014)
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R. & Avramova, Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113 (2007).
Wang, X. et al. Histone lysine methyltransferase SDG8 is involved in Brassinosteroid regulated gene expression in Arabidopsis thaliana. Mol. Plant 7, 1303–1315 (2014).
Kwon, C. S., Lee, D., Choi, G. & Chung, W. I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 60, 112–121 (2009).
Singh, R. P., Prasad, P. V. V., Sunita, K., Giri, S. N. & Reddy, K. R. Influence of high temperature and breeding for heat tolerance in cotton: a review. Adv. Agron. 93, 313–385 (2007).
Liu, Z., Yuan, Y. L., Liu, S. Q., Yu, X. N. & Rao, L. Q. Screening for high-temperature tolerant cotton cultivars by testing in vitro pollen germination, pollen tube growth and boll retention. J. Integrat. Plant Biol. 48, 706–714 (2006).
Zhang, T. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–577 (2015).
Yang, S., Zhang, X., Yue, J. X., Tian, D. & Chen, J. Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genomics 280, 187–198 (2008).
Sun, R. et al. Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genomics 15, 481–493 (2015).
Zhang, L. & Ma, H. Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 195, 248–263 (2012).
Liu, S. et al. Plant SET- and RING-associated domain proteins in heterochromatinization. Plant J. 52, 914–926 (2007).
Krajewski, W. A., Nakamura, T., Mazo, A. & Canaani, E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol. Cell. Biol. 25, 1891–1899 (2005).
Zhang, X. et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell 12, 177–185 (2003).
An, S., Yeo, K. J., Jeon, Y. H. & Song, J. J. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J. Biol. Chem. 286, 8369–8374 (2011).
Qin, S. & Min, J. Structure and function of the nucleosome-binding PWWP domain. Trends Biochem. Sci. 39, 536–547 (2014).
Pazhouhandeh, M. et al. F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc. Natl. Acad. Sci. USA 103, 1994–1999 (2006).
Sanchez, R. & Zhou, M. M. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 36, 364–372(2011).
Alvarez–Venegas, R. & Avramova, Z. Two Arabidopsis homologs of the animal trithorax genes: a new structural domain is a signature feature of the trithorax gene family. Gene 271, 215–221 (2001).
Kofler, M. M. & Freund, C. The GYF domain. FEBS J. 273, 245–256 (2006).
Hirano, Y., Hatano, T., Takahashi, A., Toriyama, M., Inagaki, N. & Hakoshima, T. Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 30, 2734–2747 (2011).
Boyer, L. A., Langer, M. R., Crowley, K. A., Tan, S., Denu, J. M. & Peterson, C. L. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10, 935–942 (2002).
Zhang, B. et al. OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 170, 1149–1161 (2016).
Wang, K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103 (2012).
Xu, Z. et al. Distribution and evolution of cotton fiber development genes in the fibreless Gossypium raimondii genome. Genomics 106, 61–69 (2015).
Chen, J. Y. et al. Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15, 148 (2015).
He, Q., Jones, D., Li, W., Xie, F., Ma, J., Sun, R., Wang, Q., Zhu, S. & Zhang, B. Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Sci. Rep. 6, 22980 (2016).
Luo, M., Liu, X., Singh, P., Cui, Y., Zimmerli, L. & Wu, K. Chromatin modifications and remodeling in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 129–136 (2012).
Black, J. C., Van Rechem, C. & Whetstine, J. R. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol. Cell 48, 491–507 (2012).
Jenuwein, T. The epigenetic magic of histone lysine methylation. FEBS J. 273, 3121–3135 (2006).
Farooq, Z., Banday, S., Pandita, T. K. & Altaf, M. The many faces of histone H3K79 methylation. Mutat. Res. 768, 46–52 (2016).
Kim, D. W., Kim, K. B., Kim, J. Y. & Seo, S. B. Characterization of a novel histone H3K36 methyltransferase setd3 in zebrafish. Biosci. Biotechnol. Biochem. 75, 289–294 (2011).
Kim, J. M., Sasaki, T., Ueda, M., Sako, K. & Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 6, 114 (2015).
Ding, B. & Wang, G. L. Chromatin versus pathogens: the function of epigenetics in plant immunity. Front. Plant Sci. 6, 675 (2015).
Min, L. et al. Sugar and auxin signaling pathways respond to high-temperature stress during anther development as revealed by transcript profiling analysis in cotton. Plant Physiol. 164, 1293–1308 (2014).
Castillo-González, C. et al. Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. Elife 4, e06671 (2015).
Huang, Y., Chen, D. H., Liu, B. Y., Shen, W. H. & Ruan, Y. Conservation and diversification of polycomb repressive complex 2 (PRC2) proteins in the green lineage. Brief. Funct. Genomics elw007 (2016).
Labbadia, J. & Morimoto, R. I. Repression of the heat shock response is a programmed event at the onset of reproduction. Mol. Cell 59, 639–650 (2015).
Sage, T. L., Bagha, S., Lundsgaard-Nielsen, V., Branch, H. A., Sultmanis, S. & Sage, R. F. The effect of high temperature stress on male and female reproduction in plants. Field Crop. Res. 182, 30–42 (2015).
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D. & May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10 (2004).
Blanc, G., Hokamp, K. & Wolfe, K. H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144 (2003).
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000).
Voorrips, R. E. MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78 (2002).
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y. & Yao, X. DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273 (2009).
Thompson, J. D., Higgins, D. G. & Gibson, T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids. Res. 22, 4673–4680 (1994).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using realtime quantitative PCR and the 2− ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
This work was supported by National Natural Science Foundation of China (U1403284), Natural Science Foundation of Hunan (13JJ8024), Open Fund of Innovation Platform (14K046) and Training Program for Youth Backbone Teacher (2015) in Universities of Hunan. We also thank Professor Hui Zhang from Key laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences for critical reading and editing the manuscript.
Author information
Authors and Affiliations
Contributions
L.Z. and Z.S. conceived, organized and planned the research, and drafted the manuscript. H.Y. conceived this study and participated in design, coordination and manuscript preparation. M.Y., C.P., M.F. and Y.X. contributed to plant materials preparation, high temperature treatments, RNA extraction, real-time PCR and data analysis. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Huang, Y., Mo, Y., Chen, P. et al. Identification of SET Domain-Containing Proteins in Gossypium raimondii and Their Response to High Temperature Stress. Sci Rep 6, 32729 (2016). https://doi.org/10.1038/srep32729
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32729
This article is cited by
-
Epigenetics: Toward improving crop disease resistance and agronomic characteristics
Plant Biotechnology Reports (2024)
-
Genome-wide analysis of SET domain genes and the function of GhSDG51 during salt stress in upland cotton (Gossypium hirsutum L.)
BMC Plant Biology (2023)
-
Identification of watermelon H3K4 and H3K27 genes and their expression profiles during watermelon fruit development
Molecular Biology Reports (2023)
-
Comprehensive molecular evaluation of the histone methyltransferase gene family and their important roles in two-line hybrid wheat
BMC Plant Biology (2022)
-
Systematic analysis of CCCH zinc finger family in Brassica napus showed that BnRR-TZFs are involved in stress resistance
BMC Plant Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.