Abstract
The tangerine pathotype of Alternaria alternata produces the A. citri toxin (ACT) and is the causal agent of citrus brown spot that results in significant yield losses worldwide. Both the production of ACT and the ability to detoxify reactive oxygen species (ROS) are required for A. alternata pathogenicity in citrus. In this study, we report the 34.41 Mb genome sequence of strain Z7 of the tangerine pathotype of A. alternata. The host selective ACT gene cluster in strain Z7 was identified, which included 25 genes with 19 of them not reported previously. Of these, 10 genes were present only in the tangerine pathotype, representing the most likely candidate genes for this pathotype specialization. A transcriptome analysis of the global effects of H2O2 on gene expression revealed 1108 up-regulated and 498 down-regulated genes. Expressions of those genes encoding catalase, peroxiredoxin, thioredoxin and glutathione were highly induced. Genes encoding several protein families including kinases, transcription factors, transporters, cytochrome P450, ubiquitin and heat shock proteins were found associated with adaptation to oxidative stress. Our data not only revealed the molecular basis of ACT biosynthesis but also provided new insights into the potential pathways that the phytopathogen A. alternata copes with oxidative stress.
Similar content being viewed by others
Introduction
Alternaria alternata is ubiquitously distributed in air, soil and various decaying plant materials1. Strains of this species can cause diseases on plants resulting in significant crop losses worldwide. Host-selective toxins (HSTs) are the essential pathogenic factors for virulent A. alternata. At least seven HSTs produced by A. alternata have been recognized, with each showing a specific toxicity to one host plant species, including the Japanese pear, strawberry, tangerine, apple, tomato, rough lemon and tobacco2. These HST-producing A. alternata pathogens produce conidia with similar morphologies and can be distinguished only based on their host preferences3. Thus, according to the characters and host range of HSTs, HST-producing A. alternata are usually assigned to seven pathotypes. Except for the tobacco pathotype, HSTs differing in chemical structures have been purified from other six A. alternata pathotypes. ACT produced by the tangerine pathotype, A. fries toxin (AFT) by the strawberry pathotype and A. kikuchiana toxin (AKT) by the Japanese pear pathotype share a 9,10-epoxy-8-hydroxy-9-methyl-decatrienoic acid (EDA) core moiety4,5,6. Genes required for EDA formation are organized in a similar manner among the three A. alternata pathotypes, while the compositions of other genes resided in the cluster are very different2.
The tangerine pathotype of A. alternata produces a host-selective ACT. Seven genes, ACTT2, ACTT3, ACTTR, ACTT5, ACTT6, ACTTS2 and ACTTS3 are required for the biosynthesis of ACT. RNA silencing or disruption of these genes led to the loss of ACT production and pathogenicity7,8,9,10,11. However, whether there are genes that are unique to the tangerine pathotype and if there are other additional candidate genes involved in the biosynthesis and regulation of ACT remain to be investigated.
In general, plants cells can rapidly generate large amount ROS in an oxidative burst as a defense response in the early events of plant-microbe interactions12. High ROS levels can cause a series of molecular damage such as DNA mutations, protein misfolding and lipid peroxidation, which can eventually lead to metabolic dysfunction and cell death13. To cope with the oxidative stress and colonize host plants, plant pathogens have evolved many strategies to neutralize ROS. Both enzymatic and non-enzymatic systems involving superoxide dismutase, peroxidases and glutathione, can scavenge intracellular toxic ROS14. The mitogen-activated protein kinase Hog1, a common stress response regulator with well characterized functions in response to hyperosmolality, has been found to be essential for oxidative stress resistance in Aspergillus fumigatus, Botryotinia fuckeliana and Cochliobolus heterostrophus (Du, Sarfati et al. 2006; Segmuller, Ellendorf et al. 2007; Igbaria, Lev et al. 2008). In B. cinerea, Ustilago maydis and Magnaporthe oryzae, the bZIP transcription factorYap1 was found to be the main regulator that mediates ROS detoxification (Molina and Kahmann 2007; Temme and Tudzynski 2009; Guo, Chen et al. 2011).
Recently, several outstanding studies have provided novel insights into the mechanisms for cellular protection against the toxicity of host ROS involved in A. alternata. It has been known that apart from HST, the ability to alleviate ROS by the tangerine pathotype of A. alternata is also crucial for pathogenesis to citrus15,16,17,18. Several genes which encode different kinds of proteins including the redox-responsive Yap1-like transcription factor, the Skn7 response regulator, the Hog1 MAP kinase, the Nox NADPH oxidases, the Nps6 non-ribosomal peptide synthetase and the Gpx3 glutathione peroxidase, have shown to be required for ROS detoxification and full virulence on citrus15,16,17,18,19. However, these findings were established only through the functional analysis and gene expression profiling in mutant strains15,16,17,18 and the interrelationships among these genes have not been established. In addition, the mechanisms responsible for other stresses and the genes involved in sporulation, which is an essential characteristic of the disease cycle of citrus brown spot, are completely unknown. For these reasons, we have fully sequenced the genome of a tangerine pathotype strain of A. alternata and performed a comparative genomics analysis. Furthermore, we carried out global transcriptome analysis of this fungus after H2O2 treatment to investigate the genes that are differentially expressed to help identifying the potential genes and metabolic pathways by which the fungus uses to cope with oxidative stress.
Results and Discussion
General features
The genome assembly of A. alternata strain Z7 was constructed using a combination of Illumina and Pacbio reads. The final assembly included 161 contigs (>1000 bp) with a total genome size of 34.41 Mb (Fig. 1A, Table 1). The genome size of A. alternata Z7 was approximately 11% larger than that of A. brassicicola, 25% smaller than that of A. destruens and comparable with other Alternaria species (Supplementary Table S1). The gene density of strain Z7 is similar to those of other sequenced Alternaria species, at ~351 genes per Mb. Among the sequenced Alternaria spp., A. solani has the highest density (377 genes per Mb) and A. destruens has the lowest (271 genes per Mb) (Supplementary Table S1). Large-scale genome synteny was found between A. alternata Z7 and the other Alternaria species with the exception of A. brassicicola (Fig. 1B). An orthoMCL analysis identified 11611 orthologous groups (containing 11660 proteins) in these seven pathotypes of Alternaria, many more than those between A. alternata Z7 and A. brassicicola (8180 proteins in 8003 orthologous groups). These results revealed a high degree of genome similarity across Alternaria strains living in different ecological niches and/or with different hosts specificities.
Genome sequence analysis.
(A) Genome organization and gene distribution in A. alternata Z7. The peripheral circle represents 103 contigs each with a size of over 5 Kb. The second and third circles of color bands show the genes that were up-regulated and down-regulated after H2O2 treatment, respectively. Higher intensity of the color represents a larger log2 fold change of gene expression. The fourth circle shows the GC content in 10 Kb windows with a step of 2 Kb. Gene duplications are shown in the center. (B) Genomic synteny of A. alternata Z7 with other 7 Alternaria species. The identity and length cutoff were set at 80% and 10 kb, respectively.
Phylogenetic analysis
A phylogenetic tree based on a combined analysis of 200 conserved single-copy orthologs randomly selected from 26 Alternaria species and Pyrenophora tritici-repentis was constructed and revealed highly close interspecific genetic relationships of the distinct pathotypes of A. alternata (Fig. 2). The systematics of Alternaria has been ambiguous. Simmons had assigned 77 Alternaria isolates from citrus into 10 species according to sporulation patterns20. However, researchers failed to delineate significant variation among those species based on DNA sequences of popular marker genes, such as those coding for calmodulin, translation elongation factor alpha, chitin synthase and 1, 3, 8-trihydroxynaphthalene reductase and actin21. Indeed, based on ITS, the small-spored, HST-producing A. alternata pathotypes of tangerine, rough lemon, strawberry, tomato, apple and pear could not be differentiated from each other or from several saprophytic isolates of A. alternata22. To help reduce taxonomic confusions, several sections within the genus Alternaria have been proposed23,24,25. In our phylogenetic tree, three clades Clade I, Clade II and Clade III corresponded well with the proposed section ‘Alternaria’, ‘Porri’ and ‘Brassicicola’, providing powerful support for the new systematics of Alternaria23. Still, many of the morphospecies within section Alternaria cannot be distinguished, even with sequence information from 200 marker genes. A. alternata Z7, A. citriarbusti, A. tangelonis can all produce ACT and cause citrus brown spot; however, they were grouped into separate branches in the phylogenetic tree. A similar situation was found with the apple pathotype species, for which A. mali BMP3063 and A. mali BMP3064 were grouped into different branches. Thus, phylogenetic relationships of Alternaria species are not completely correlated with their host ranges. Indeed, host specificity of the A. alternata isolates is determined by the HST which is encoded by a single cluster in the pathotypes and this cluster resides on the conditionally dispensable (CD) chromosomes2. By sequencing the CD chromosomes in three pathotype of A. alternata, one recent study identified large syntenic regions among the three CD chromosomes26. However, the HST clusters were unique to the respective pathotypes26. In contrast, A. alternata Z7 and A. turkisafria can both infect citrus and were clustered together in the phylogenetic tree, suggesting that they have speciated recently (Fig. 2).
Phylogenomic relationships of A. alternata Z7 with other fungi.
The maximum likelihood (ML) phylogenetic tree was built with the program MEGA6 using the Jones-Taylor-Thornton (JTT) amino acid substitution model. The corresponding host for each species was listed in the right column. Node supported as ML bootstraps (values ≥50%) are displayed above or below each branch.
Unique genes in the tangerine pathotype
As described above, although they have distinct host range, the small-spored, HTS-producing A. alternata could not be distinguished even with sequences from 200 genes. We wondered which genes represent the specific determinants for A. alternata to attack tangerines and the hybrids of tangerine and orange. To answer this question, we searched all the orthologous groups in the genomes of 26 Alternaria species. Those that were only present in all 4 tangerine pathotype strains and with their identities to each other over 80% were retrieved. Ten genes were identified to be uniquely present in the tangerine pathotype (Supplementary Table S2). These genes were predicted to encode several types of enzymes, such as enoylreductase, monooxygenase, thioesterase, reverse transcriptase, non-ribosomal peptide synthetase and polyketide synthase. Three of the ten genes encode hypothetical proteins. Intriguingly, these genes were found to be clustered in the Z7 genome. Subsequent analysis of the secondary metabolite biosynthetic gene clusters confirmed that they are important members of the ACT gene cluster (Fig. 3 and Supplementary Table S5).
ACT gene cluster
A total of 12 polyketide synthases (PKSs), 7 non-ribosomal peptide synthetases (NRPSs), 1 PKS-like and 5 NRPS-like genes were identified in the genome of A. alternata Z7. These genes have been found to be responsible for the biosynthesis of the backbone structure of many secondary metabolites. The number of backbone genes in strain Z7 is similar to that in Nectria haematococca but much smaller than those in other fungi, such as Magnaporthe grisea and Fusarium graminearum (Supplementary Table S3). The backbone genes in Z7 are organized into 18 secondary metabolite biosynthetic gene clusters or partial clusters. So far, several secondary metabolites such as alternariol, alternariol-9-methyl ether, dimethyl coprogen, alternapyrone and HSTs have been successfully identified2,27,28,29. However, the exact products of most of the clusters in Z7 remain largely unknown (Supplementary Table S4). The host selective ACT gene cluster in Z7 was found to be located in a 91.2 kb DNA fragment containing 25 genes and most of them have 2 or 3 copies, indicating the strong capacity to synthesize the ACT (Fig. 3). Proteins found in the ACT biosynthetic cluster include polyketide synthase PKS, NRPS, enoyl-CoA hydratase, HMG-CoA hydrolase, acyl-CoA dehydrogenases, cytochrome P450 monooxygenase and transcription factor (Fig. 3 and Supplementary Table S5). Previously reported genes, ACTT2, ACTT3, ACTTR,ACTT5, ACTTS2 and ACTTS3, were found in the cluster, while ACTT6 was not included in this cluster but was found in another contig (3503 bp in length), indicating the gene cluster responsible for ACT biosynthesis may be larger. These toxin-related genes except for AALTg11737, AALTg11744 and AALTg11742 also were found in A. citriarbusti, A. tangelonis and A. turkisafria. Both AALTg11737 and AALTg11744 are present only in Z7 and AALTg11742 is present in Z7 and A. citriarbusti only (Supplementary Table S5). All the above-mentioned 10 tangerine pathotype unique genes are included in this gene cluster, two of them have been previously reported as unique to the tangerine pathotype, i.e., enoylreductase ACTTS2 and polyketide synthase ACTTS310,11. Whether the remaining 8 specific genes play a critical role in ACT biosynthesis needs to be functionally elucidated.
Carbohydrate-active enzymes and secretomes
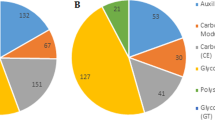
Carbohydrate-active enzymes (CAZymes) are responsible for the degradation of glycol-conjugated and oligo- and polysaccharides. They play important roles in the acquisition of nutrients from the environment for fungi. A total of 373 putative CAZyme genes were identified in Z7. These include 51 GlycosylTransferases (GTs), 161 Glycoside Hydrolases (GHs), 53 Carbohydrate Esterases (CEs), 11 Polysaccharide Lyases (PLs), 86 Auxiliary Activities (AAs) and 11 Carbohydrate-Binding Modules (CBMs). The types and numbers of CAZymes among different pathotypes of A. alternata are similar; however, A. gaisen has fewer CAZymes, i.e., 328 in A. gaisen versus 372~381 in other pathotypes (Fig. 4). A. alternata Z7 (53) has more CE than B. fuckeliana (35), Penicillium chrysogenum (21), F. graminearum (42) and M. grisea (51) but has fewer GH (161 versus 219~257) and GT (51 versus 97~106) than them (Fig. 4). Those different enzymes are mostly related to the degradation of plant cell wall components. Compared to B. fuckeliana, F. graminearum and M. grisea, Z7 has fewer (6 versus 17~24) xyloglucan transferases (GH16) responsible for degrading xylan and fewer (11 versus 16~21 and 7 versus 13~16) multifunctional catabolic enzymes (GH3, GH5) involved in the decomposition of plant pectin and hemicellulose30,31. Z7 lacks the α-glucuronidases (GH115) which is only active on xylan oligomers in ascomycetous fungi32 (Supplementary Table S6). However, Z7 has relatively high numbers of the AA3 family of cellobiose dehydrogenase (27), the AA7 family of glucooligosaccharide oxidase (17) and the AA9 (formerly GH61) family of polysaccharide monooxygenases (23) involved in degrading cellulose31,33 (Supplementary Table S6). The differences in the composition of CAZymes could help identify the CAZymes required for citrus infection in A. alternata Z7.
Identification of differentially expressed genes (DEGs)
Since ROS-detoxification is vital for A. alternata survival within plant hosts, cellular responses to oxidative stress are necessarily required for pathogenicity. To analyze the transcriptional response of A. alternata Z7 to H2O2, mycelia were treated with 15 mM H2O2 and samples were collected 30 minutes later. In total, we identified 1606 differentially expressed genes at 30 min post-treatment, 1108 of them displayed increased transcript levels and 498 of them displayed decreased transcript levels (Fig. 1A). To validate the DEG data, transcript levels of eighteen selected genes that showed different expression patterns in response to the H2O2 treatments were confirmed using quantitative reverse transcription PCR (qRT-PCR). While the magnitude of fold changes differed between the two methods for some of the genes, all 18 tested genes showed similar trends in transcript accumulation between the transcriptome and qRT-PCR results (Supplementary Fig. S2). These results showed that our DEG data was reliable for further analysis.
Functional analysis of DEGs
Of the differentially expressed genes, 659 up-regulated and 271 down-regulated genes could be assigned into functional categories of the gene ontology (GO): biological process, molecular function and cellular component. GO terms predominantly enriched in both the up-regulated and down-regulated genes were involved in the following biological processes: ‘primary metabolic process’ (GO:0044238), ‘cellular metabolic process’ (GO:0044237),‘oxidoreductase activity’ (GO:0016491), ‘nitrogen compound metabolic process’ (GO:0006807),‘macromolecule metabolic process’ (GO:0043170), ‘establishment of localization’ (GO:0051234) and ‘transport’ (GO:0006810) (Fig. 5). Moreover, the category ‘biosynthetic process’ (GO:0009058) was also over-represented for the up-regulated genes (Fig. 5). There were 149 genes exhibiting differential expression in the category ‘oxidoreductase activity’, with105 being induced and 44 being repressed under oxidative stress. These genes belonged to a diversity of dehydrogenases and oxidases and they mainly participate in primary metabolic processes like amino acid synthesis and degradation, glucose and lipid metabolism (Supplementary Table S7). The three most up-regulated genes in this category encoded 2,4-dichlorophenol 6-monooxygenase (AALTg3456, log2FC 5.1), alcohol dehydrogenase (AALTg3691, log2FC 4.7) and glutathione reductase (AALTg2809, log2FC 3.3). The three enzymes with the most down-regulation were uricase (AALTg5212, log2FC -2.56), nitrite reductase (AALTg4186, log2FC -3.31) and C-5 sterol desaturase (AALTg6701, log2FC -3.53).
Differential expression of antioxidation genes
ROS homeostasis in most organisms is maintained through the balance between ROS production and ROS scavenging. Most organisms have evolved oxidative stress response mechanisms to scavenge elevated intracellular ROS levels by antioxidant enzymes. Those enzymes include catalase, glutathione peroxidase, ascorbate peroxidase and superoxide dismutase, which are known as universal antioxidant enzymes involved in ROS detoxification in all living organisms. We examined the expression of genes encoding those scavengers. Transcript levels of 12 peroxidases were significantly up-regulated after H2O2 treatment, including three catalases, one catalase-peroxidase, four cysteine peroxiredoxin, one ascorbate peroxidase, one glutathione peroxidase (AaGpx3), one carboxymuconolactone decarboxylase and one hybrid ascorbate-cytochrome C peroxidase (Table 2). Our result is consistent with the recent finding that glutathione peroxidase AaGpx3 in A. alternata was essential for the detoxification of cellular stresses induced by ROS18. Notably, three Fe-Mn type superoxide dismutases and four Fe-Cu type superoxide dismutases were identified in the A. alternata Z7 genome, however, none of them showed significantly different expression during H2O2 stress.
Non-enzymatic defense systems against ROS include compounds that are oxidized by ROS and thereby reduce oxidant levels in cells34. Thioredoxin is a class of small proteins that act as electron donors to ribonucleotide reductases and peroxidases35. We examined the expression of all the thioredoxin encoding genes and found that seven of them were significantly up-regulated (Table 2). Thioredoxins in Cryptococcus neoformans and Ascochyta rabiei are also known to be associated with oxidative stress tolerance, suggesting the conserved role thioredoxin played in fungi36,37. Glutathione is a thiol-containing tripeptide which maintains the intracellular redox homeostasis by reducing cellular disulfide bonds38. Besides the AaGpx3, 10 other genes involved in glutathione metabolism were also highly induced (Table 2). It is worth mentioning that no gene in this pathway was down-regulated in transcript abundance. The enrichment of up-regulated genes in this pathway suggests that the glutathione system plays an important role in the elimination of ROS in A. alternata.
Kinases
Protein kinases are responsible for the phosphorylation of proteins and participate in various cell processes. A total of 137 kinases were identified in the genome of A. alternata Z7. Among these, the transcript levels of 10 kinases were significantly elevated and six were down-regulated during H2O2 stress (Supplementary Table S8). In some fungi including A. alternata, the transcript levels of the oxidative stress response genes are controlled by the mitogen-activated protein kinase (MAPK) Hog139,40,41. In our data, the expression of AaHog1 (AALTg10096, log2FC 1.2) genes in A. alternata Z7 were significantly up-regulated during H2O2 treatment, confirming its important role in ROS scavenging. Moreover, several genes involved in the Hog1 MAPK signaling pathway were also up-regulated in their expression. These include the histidine phosphotransfer protein Ypd1 involved in the osmolarity two-component sensing and response system (AALTg9320, log2FC 1.1), protein phosphatase Ptc1 (AALTg9981, log2FC 1.1) and tyrosine-protein phosphatase Ptp2 (AALTg5082, log2FC 2.2). These genes are known to be associated with development, stress response, signal transduction and virulence at varying degrees in different fungal species42,43,44. However, there has been little evidence that these genes are involved in ROS elimination. Another differentially expressed kinaseis the sucrose non-fermenting protein Snf1, which is a serine/threonine protein kinase and plays a key role in controlling carbon source utilization45,46. The Snf1 protein kinase is also involved in regulating a broad range of cellular and morphogenetic processes, such as spore formation, filamentation and invasive growth, autophagy, virulence, as well as response to environmental stresses including oxidative stress, heat shock and alkaline pH46,47,48,49,50. Induced expression of this important kinase (AALTg3740, log2FC 1.3) was observed in our transcriptome data, suggesting a potential role of Snf1 in ROS-detoxification in A. alternata.
Transcription factors
The expression of the oxidative stress response genes can also be controlled by distinct transcription factors. For example, homologs of the yeast YAP1-like transcription factor are the main regulators of ROS resistance in many filamentous fungi19,51,52. The A. alternata Z7 genome contains 283 transcription factors belonging to 19 subfamilies. The largest subfamily is theZn2/Cys6, which includes 147 members while the second largest is the C2H2 zinc-finger subfamily with 47 genes. These two subfamilies account for about 70 percent of the total transcription factors in A. alternata Z7 (Supplementary Table S9). After exposure of the A. alternata strain Z7 to H2O2 for 30 min, 33 differentially expressed transcription factors were discovered with the expressions of 19 being up-regulated and 14 being down-regulated, respectively (Supplementary Table S10). The transcript level of the AaYap1 gene (AALTg912, log2FC 1.5) was significantly induced as was expected. However, the heat shock factor type DNA-binding transcription factor AaSkn7 (AALTg8622), which was recently revealed to be involved in cellular resistance to oxidative stress and pathogenicity to citrus in A. alternata, did not show significantly different expression between the two treatments in our investigation53. The results suggest that AaSkn7 is essential but not specifically involved in response to H2O2 stress. Interestingly, we discovered that the transcript level of the nitrate-specific transcription factor NirA homolog (AALTg8635, log2FC, 1.3) was apparently increased after H2O2 treatment. NirA activates the expression of the nitrate assimilation genes when nitrate or nitrite is present54. Most of the remaining transcription factors belong to the subfamilies of Zn_clus, zf-C2H2, Myb_DNA-binding and HLH and the functions of their encoding genes are largely unknown in oxidative stress response (Supplementary Table S10).
MFS and ABC transporters
The major facilitator superfamily (MFS) and the ATP-binding cassette (ABC) transporters are the top two biggest classes of transporters in fungi. Members of the former mainly play roles in nutrient uptake and drug efflux while the latter transport a broad range of compounds like ions, drugs and sugars55. Involvement of these two families of transporters in multidrug resistance (MDR) has been widely investigated. The MDR transporter ABC3 in M. grisea was also revealed to play an important role in pathogenesis and in response to intracellular oxidative stress56. After exposure of A. alternata Z7 to H2O2, 28 and 14 genes encoding MFS were significantly induced and repressed, respectively, while the expressions of seven and one ABC transporter genes were up-regulated and down-regulated, respectively (Supplementary Table S11). Of the 28 up-regulated MFS genes in A. alternata Z7 after H2O2 treatment, six were predicted to be putatively involved in MDR. These six MFS genes were AALTg5693 (quinidine resistance, log2FC 2.8), AALTg9681 (benomyl/methotrexate resistance, log2FC 3.4), AALTg6207 (multidrug resistance, log2FC 2.5), AALTg10470 (multidrug resistance, log2FC 2.0), AALTg9513 (benomyl/methotrexate resistance, log2FC 3.1) and AALTg8610 (gliotoxin efflux transporter, log2FC 1.8). One ABC transporter gene AALTg8644 (log2FC 1.2) was also up-regulated and it showed a high level similarity to the ABC multidrug transporter Mdr2 in A. fumigatus57. Interestingly, most of the down-regulated MFS genes are associated with sugar transport, indicating that the absorption of sugar nutrients may be retarded under oxidative stress.
P450 and ergosterol biosynthesis
Cytochrome P450 (CYP) proteins are a type of monooxygenases which play essential roles in the biosynthesis of secondary metabolites and in the detoxification of toxic compounds58. We identified 13 CYPs whose expression was induced or repressed after H2O2 treatment (Supplementary Table S12). Specially, the 14-alpha sterol demethylase ERG11B gene (AALTg7699), which plays an essential role in ergosterol biosynthesis and drug resistance in many fungi59,60, showed a decreased expression by about 2.5 fold. Other genes required in ergosterol biosynthesis were then investigated. Interestingly, AALTg8874 (Sterol desaturase family, log2FC 1.5), AALTg10933 (sterol O-acyltransferase, log2FC 1.0), AALTg6701(Delta(7)-sterol 5(6)-desaturase, ERG3, log2FC -2.6) and AALTg1316 (squalene epoxidase, log2FC -2.0) showed changes in transcriptional level during H2O2 stress. Furthermore, the sterol regulatory element binding Sre1(AALTg8325, log2FC -1.1), which was reported to activate genes required for sterol biosynthesis under low oxygen, was significantly down-regulated61. In another experiment, a distinct expression pattern of sterol synthesis genes in response of Cryptococcus neoformans to H2O2 treatment was found, in which ERG11was induced while ERG3 did not alter the transcript level at 30 min after H2O2 treatment36. These results may indicate a cross-talk between ergosterol biosynthesis and ROS resistance. However, the underlying mechanisms for the putative relationships between the two pathways may be different among fungi.
HSPs and ubiquitin
Some general stress response proteins such as ubiquitin and heat shock proteins (HSPs) are known to play critical roles in fungal survival under various stresses. HSPs are constitutively expressed but can be induced to high levels under certain stresses in all living organisms. These proteins primarily function as molecular chaperones and maintain protein homeostasis in routine biological processes and under various stressful conditions62. For example, HSPs positively respond to ROS accumulation caused by thermal stress in yeast63. In this investigation, we found that 5 HSP40 (AALTg4905, log2FC 1.1, AALTg10836, log2FC 3.0, AALTg2843, log2FC 1.7, AALTg5344, log2FC 1.5 and AALTg5883, log2FC 1.1) and 2 HSP12 (AALTg4088, log2FC 4.2 and AALTg5164, log2FC 4.0) increased their level of mRNA expression in the H2O2 stress condition (Supplementary Table S13). Considering that the expression of HSPs may be specific to different conditions, these HSPs may be involved in H2O2 tolerance in A. alternata. Degradation of proteins by the ubiquitin dependent proteosome pathway is vital for maintaining cellular homeostasis. In mammalian cells, permanently oxidized proteins are recycled through the ubiquitin dependent proteasomal pathway64. Similar mechanism was also found in C. neoformans as the amount of ubiquitin conjugated proteins in the cell lysate is positively correlated the H2O2 concentration36. Besides, previous studies have found that both the ubc8 deletion strain in C. neoformans and the ubi4 deletion strain in Candida albicans displayed increased sensitivity to H2O265,66. Through comparative transcriptome analysis, two ubiquitin-conjugating enzymes (AALTg6960, log2FC 1.8 and AALTg4835, log2FC 1.5) and one E3 ubiquitin-protein ligase (AALTg10966, log2FC 1.8) showed increased expression in response to H2O2 stress, suggesting a potential role of ubiquitin related processes in oxidative stress tolerance in A. alternata.
Conclusions
In this study, we sequenced the genome of a tangerine pathotype of the plant fungal pathogen A. alternata Z7 and identified 19 novel genes associated with the biosynethesis of ACT. Ten genes were only found in the tangerine pathotype and they likely represented host-specificity determinants. Our analyses not only revealed the putative molecular basis of ACT biosynthesis but also provided potential molecular signatures for developing new methods of rapidly and efficiently detecting the tangerine pathotype of A. alternata. We also performed comparative global transcriptional studies of A. alternata Z7 to H2O2 stress and provided a broad-based analysis of gene expression linked to ROS resistance. The transcriptome data presented here will pave the way for future research on detoxification mechanisms of A. alternata towards ROS and further help reveal the underlying pathogenic mechanisms of this economically important fungal pathogen.
Methods
Genome sequencing and assembly
A. alternata strain Z7 was selected for genome sequencing using the long reads PacBio technology and the HiSeq 2000 platform67. A total of 1.6 Gb PacBio data, 1.1 Gb pair-end data and 4.0 Gb mate-pair data were generated in the sequencing process, which correspond to ~200 fold of sequence depth. The genome assembly was accomplished following a previously used method with the HGAP.2 assembler and the CLC Genomics Workbench program68,69. The assembled A. alternata Z7 genome has been deposited in GenBank under the accession number LPVP00000000 and genome information of other Alternaria species was downloaded from the Alternaria genomes database70.
Gene prediction and annotation
Ab initio gene predictions of the genomic sequences of Alternaria species were performed with a combination of Augustus and GeneMark-ES71,72. The resulting prediction was refined using TopHat2 and Cufflinks on the RNA-seq libraries73,74. These predicted genes were primarily annotated based on BLASTp search against the NCBI (http://www.ncbi.nlm.nih.gov/) nr database from 13/06/15(E < 1 × 10−5 identity >25%, query coverage >50%). The tRNAs were identified by the tRNAscan-SE program75 and genome repetitive elements were defined using RepeatMasker76.
Synteny, orthology and phylogenomic analysis
Syntenic analysis was performed by Circos with an identity and length cutoff set at 80% and 10 kb, respectively77. Orthologous gene relationship among species were determined using OrthMCL and reciprocal BLASTp with identity >50% and query coverage >50%78. The amino acid sequences from 200 random selected orthologous groups with only one gene in each species were concatenated and aligned with ClustalW279. The maximum likelihood phylogenetic tree was subsequently built with the program MEGA6 using the Jones-Taylor-Thornton (JTT) model80. Statistical support for the phylogenetic tree was performed by non-parametric bootstrap analysis with 1000 replicates.
Protein family classifications
The whole genome protein families were classified by InterproScan and Pfam analysis81,82. Fungal secondary metabolite pathways were predicted using the web-based analytical tool SMURF83.
Transcriptome analysis
The A. alternata Z7strain was grown in liquid PDB at 25 °C in a shaker incubator for 2 days. H2O2 was added to the cultures to the final concentration of 15 mM with shaking for 30 min. The pure culture of A. alternata Z7 was used as a negative control. Mycelia were then collected for total RNA extraction using an AxyPrepTM multisource total RNA miniprep kit. RNA-Seq was conducted for two biological replicates of each sample. The libraries were performed using an IlluminaTruSeq RNA Sample Preparation Kit and were sequenced on an Illumina Hiseq 2000 platform, generating 50 bp single-end reads. Index of the A. alternata Z7 genome was built using Bowtie2 and clean reads were mapped to the reference genome using TopHat274,84. The reads numbers mapped to each gene was counted by HTSeq and the resulting transcript count tables were subjected to DESeq R package for differential expression analysis85,86. Transcripts with an adjusted P value less than 0.01 and a log2 (Fold change) greater than 1 were determined as differentially expressed. The differentially expressed genes were annotated by blast search against the NCBI nr databases. Gene Ontology (GO) enrichment analysis of DEGs was conducted using Blast2GO87. The transcriptome data reported in this study have been deposited in NCBI’s Sequence Read Archive (SRA) with accession number SRP071688.
qRT-PCR analysis
To validate the transcriptome data obtained by RNA sequencing, qRT-PCR was carried out on eighteen A. alternata DEGs. 10 μg of each RNA sample was used for reverse transcription with the Prime Script RT reagent kit (TakaRa Biotechnology, Co., Dianlian, China). Relative expression of the selected genes was quantified in triplicate on a 7300 Real Time PCR system (ABI, USA). Primers used in this study were listed in Supplementary Table S14. The actin-encoding gene (KP341672) was used as an internal control and the resulting data was normalized using the comparative 2−ΔΔCT as described previously59.
Additional Information
How to cite this article: Wang, M. et al. Genomic and transcriptomic analyses of the tangerine pathotype of Alternaria alternata in response to oxidative stress. Sci. Rep. 6, 32437; doi: 10.1038/srep32437 (2016).
References
Thomma, B. P. Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol 4, 225–236 (2003).
Tsuge, T. et al. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 37, 44–66 (2013).
NISHIMURA, S. Host-specific toxins from alternaria alternata problems and prospects. Proceedings of the Japan Academy, Series B 56, 362–366 (1980).
Nakashima, T. et al. Isolation and structures of AK-toxin I and II, host-specific phytotoxic metabolites produced by Alternaria alternata Japanese pear pathotype. Agric. Biol. Chem. 49, 807–815 (1985).
Nakatsuka, S.-i. et al. Structure of AF-toxin II, one of the host-specific toxins produced by alternariaalternata strawberry pathotype. Tetrahedron Lett. 27, 2753–2756 (1986).
Kohmoto, K. et al. Isolation and biological activities of two host-specific toxins from the tangerine pathotype of Alternaria alternata. Phytopathology 83, 495–502 (1993).
Masunaka, A. et al. Distribution and Characterization of AKT Homologs in the Tangerine Pathotype of Alternaria alternata. Phytopathology 90, 762–768 (2000).
Miyamoto, Y. et al. Functional analysis of a multicopy host-selective ACT-toxin biosynthesis gene in the tangerine pathotype of Alternaria alternata using RNA silencing. Mol Plant Microbe Interact 21, 1591–1599 (2008).
Miyamoto, Y. et al. Function of genes encoding acyl-CoA synthetase and enoyl-CoA hydratase for host-selective act-toxin biosynthesis in the tangerine pathotype of Alternaria alternata. Phytopathology 99, 369–377 (2009).
Ajiro, N. et al. Role of the host-selective ACT-toxin synthesis gene ACTTS2 encoding an enoyl-reductase in pathogenicity of the tangerine pathotype of Alternaria alternata. Phytopathology 100, 120–126 (2010).
Miyamoto, Y. et al. ACTTS3 encoding a polyketide synthase is essential for the biosynthesis of ACT-toxin and pathogenicity in the tangerine pathotype of Alternaria alternata. Mol Plant Microbe Interact 23, 406–414 (2010).
Nanda, A. K., Andrio, E., Marino, D., Pauly, N. & Dunand, C. Reactive oxygen species during plant-microorganism early interactions. J Integr Plant Biol 52, 195–204 (2010).
Beckman, K. B. & Ames, B. N. The free radical theory of aging matures. Physiol Rev 78, 547–581 (1998).
Heller, J. & Tudzynski, P. Reactive oxygen species in phytopathogenic fungi: signaling, development and disease. Annu. Rev. Phytopathol. 49, 369–390 (2011).
Chung, K. R. Stress Response and Pathogenicity of the Necrotrophic Fungal Pathogen Alternaria alternata. Scientifica 635431, 10 (2012).
Chen, L. H., Lin, C. H. & Chung, K. R. A nonribosomal peptide synthetase mediates siderophore production and virulence in the citrus fungal pathogen Alternaria alternata. Mol Plant Pathol 14, 497–505 (2013).
Chen, L. H., Yang, S. L. & Chung, K. R. Resistance to oxidative stress via regulating siderophore-mediated iron acquisition by the citrus fungal pathogen Alternaria alternata. Microbiology 160, 970–979 (2014).
Yang, S. L., Yu, P. L. & Chung, K. R. The glutathione peroxidase-mediated reactive oxygen species resistance, fungicide sensitivity and cell wall construction in the citrus fungal pathogen Alternaria alternata. Environ Microbiol 16, 1462–2920 (2015).
Lin, C. H., Yang, S. L. & Chung, K. R. The YAP1 homolog-mediated oxidative stress tolerance is crucial for pathogenicity of the necrotrophic fungus Alternaria alternata in citrus. Mol Plant Microbe Interact 22, 942–952 (2009).
Simmons, E. G. Alternaria themes and variations (226–235) classification of citrus pathogens. Mycotaxon 70, 263–323 (1999).
Peever, T. L., Su, G., Carpenter-Boggs, L. & Timmer, L. W. Molecular systematics of citrus-associated Alternaria species. Mycologia 96, 119–134 (2004).
Kusaba, M. & Tsuge, T. Phylogeny of Alternaria fungi known to produce host-specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Curr Genet 28, 491–498 (1995).
Lawrence, D. P., Gannibal, P. B., Peever, T. L. & Pryor, B. M. The sections of Alternaria: formalizing species-group concepts. Mycologia 105, 530–546 (2013).
Woudenberg, J., Groenewald, J., Binder, M. & Crous, P. Alternaria redefined. Studies in Mycology 75, 171–212 (2013).
Woudenberg, J. H. et al. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud Mycol 82, 1–21 (2015).
Tsuge, T. et al. Evolution of pathogenicity controlled by small, dispensable chromosomes in Alternaria alternata pathogens. Physiol. Mol. Plant Pathol. 95, 27–31 (2016).
Fujii, I., Yoshida, N., Shimomaki, S., Oikawa, H. & Ebizuka, Y. An iterative type I polyketide synthase PKSN catalyzes synthesis of the decaketide alternapyrone with regio-specific octa-methylation. Chem. Biol. 12, 1301–1309 (2005).
Saha, D. et al. Identification of a polyketide synthase required for alternariol (AOH) and alternariol-9-methyl ether (AME) formation in Alternaria alternata. PLoS One 7, e40564 (2012).
Bills, G. et al. New insights into the echinocandins and other fungal non-ribosomal peptides and peptaibiotics. Natural product reports 31, 1348–1375 (2014).
Noda, J., Brito, N. & Gonzalez, C. The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol 10, 1471–2229 (2010).
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42, 21 (2014).
Chong, S. L. et al. The alpha-glucuronidase Agu1 from Schizophyllum commune is a member of a novel glycoside hydrolase family (GH115). Appl Microbiol Biotechnol 90, 1323–1332 (2011).
Correa, T. L., Dos Santos, L. V. & Pereira, G. A. AA9 and AA10: from enigmatic to essential enzymes. Appl Microbiol Biotechnol 17, 17 (2015).
Jamieson, D. J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14, 1511–1527 (1998).
Arner, E. S. & Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267, 6102–6109 (2000).
Upadhya, R., Campbell, L. T., Donlin, M. J., Aurora, R. & Lodge, J. K. Global transcriptome profile of Cryptococcus neoformans during exposure to hydrogen peroxide induced oxidative stress. PLoS One 8, 28 (2013).
Singh, K., Nizam, S., Sinha, M. & Verma, P. K. Comparative transcriptome analysis of the necrotrophic fungus Ascochyta rabiei during oxidative stress: insight for fungal survival in the host plant. PLoS One 7, 12 (2012).
Pompella, A., Visvikis, A., Paolicchi, A., De Tata, V. & Casini, A. F. The changing faces of glutathione, a cellular protagonist. Biochem Pharmacol 66, 1499–1503 (2003).
Du, C., Sarfati, J., Latge, J. P. & Calderone, R. The role of the sakA (Hog1) and tcsB (sln1) genes in the oxidant adaptation of Aspergillus fumigatus. Med Mycol 44, 211–218 (2006).
Lin, C. H. & Chung, K. R. Specialized and shared functions of the histidine kinase- and HOG1 MAP kinase-mediated signaling pathways in Alternaria alternata, a filamentous fungal pathogen of citrus. Fungal Genet Biol 47, 818–827 (2010).
Herrero, E., Ros, J., Belli, G. & Cabiscol, E. Redox control and oxidative stress in yeast cells. Biochim Biophys Acta 11, 15 (2008).
Arino, J., Casamayor, A. & Gonzalez, A. Type 2C protein phosphatases in fungi. Eukaryot Cell 10, 21–33 (2011).
Lee, K. T. et al. Distinct and redundant roles of protein tyrosine phosphatases Ptp1 and Ptp2 in governing the differentiation and pathogenicity of Cryptococcus neoformans. Eukaryot Cell 13, 796–812 (2014).
Fassler, J. S. & West, A. H. Histidine phosphotransfer proteins in fungal two-component signal transduction pathways. Eukaryot Cell 12, 1052–1060 (2013).
Hardie, D. G. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8, 774–785 (2007).
Zhang, T. et al. PdSNF1, a sucrose non-fermenting protein kinase gene, is required for Penicillium digitatum conidiation and virulence. Appl Microbiol Biotechnol 97, 5433–5445 (2013).
Hong, S. P. & Carlson, M. Regulation of snf1 protein kinase in response to environmental stress. J Biol Chem 282, 16838–16845 (2007).
Cullen, P. J. & Sprague, G. F. Jr. Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci USA 97, 13619–13624 (2000).
Honigberg, S. M. & Lee, R. H. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol Cell Biol 18, 4548–4555 (1998).
Wang, Z., Wilson, W. A., Fujino, M. A. & Roach, P. J. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase and the cyclin-dependent kinase Pho85p. Mol Cell Biol 21, 5742–5752 (2001).
Lessing, F. et al. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell 6, 2290–2302 (2007).
Guo, M. et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog 7, e1001302 (2011).
Chen, L. H., Lin, C. H. & Chung, K. R. Roles for SKN7 response regulator in stress resistance, conidiation and virulence in the citrus pathogen Alternaria alternata. Fungal Genet Biol 49, 802–813 (2012).
Strauss, J., Muro-Pastor, M. I. & Scazzocchio, C. The regulator of nitrate assimilation in ascomycetes is a dimer which binds a nonrepeated, asymmetrical sequence. Mol Cell Biol 18, 1339–1348 (1998).
Perlin, M. H., Andrews, J. & Toh, S. S. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv Genet 85, 201–253 (2014).
Sun, C. B., Suresh, A., Deng, Y. Z. & Naqvi, N. I. A multidrug resistance transporter in Magnaporthe is required for host penetration and for survival during oxidative stress. Plant Cell 18, 3686–3705 (2006).
Tobin, M. B., Peery, R. B. & Skatrud, P. L. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene 200, 11–23 (1997).
Cresnar, B. & Petric, S. Cytochrome P450 enzymes in the fungal kingdom. Biochim Biophys Acta 1, 29–35 (2011).
Sun, X., Wang, J., Feng, D., Ma, Z. & Li, H. PdCYP51B, a new putative sterol 14alpha-demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl Microbiol Biotechnol 91, 1107–1119 (2011).
Buied, A., Moore, C. B., Denning, D. W. & Bowyer, P. High-level expression of cyp51B in azole-resistant clinical Aspergillus fumigatus isolates. J Antimicrob Chemother 68, 512–514 (2013).
Chang, Y. C., Bien, C. M., Lee, H., Espenshade, P. J. & Kwon-Chung, K. J. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol Microbiol 64, 614–629 (2007).
Tiwari, S., Thakur, R. & Shankar, J. Role of Heat-Shock Proteins in Cellular Function and in the Biology of Fungi. Biotechnol Res Int 132635, 31 (2015).
Moraitis, C. & Curran, B. P. Reactive oxygen species may influence the heat shock response and stress tolerance in the yeast Saccharomyces cerevisiae. Yeast 21, 313–323 (2004).
Jung, T. & Grune, T. The proteasome and its role in the degradation of oxidized proteins. IUBMB Life 60, 743–752 (2008).
Leach, M. D., Stead, D. A., Argo, E., MacCallum, D. M. & Brown, A. J. Molecular and proteomic analyses highlight the importance of ubiquitination for the stress resistance, metabolic adaptation, morphogenetic regulation and virulence of Candida albicans. Mol Microbiol 79, 1574–1593 (2011).
Ko, Y. J. et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 8, 1197–1217 (2009).
Huang, F. et al. Identification of a novel phylogenetic lineage of Alternaria alternata causing citrus brown spot in China. Fungal Biol 119, 320–330 (2015).
Chin, C. S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569 (2013).
Taniguti, L. M. et al. Complete Genome Sequence of Sporisorium scitamineum and Biotrophic Interaction Transcriptome with Sugarcane. PLoS One 10 (2015).
Dang, H. X., Pryor, B., Peever, T. & Lawrence, C. B. The Alternaria genomes database: a comprehensive resource for a fungal genus comprised of saprophytes, plant pathogens and allergenic species. BMC Genomics 16, 015–1430 (2015).
Borodovsky, M. & Lomsadze, A. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinformatics 4, 1–10 (2011).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, 2013–2014 (2013).
Schattner, P., Brooks, A. N. & Lowe, T. M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33, W686–W689 (2005).
Zhi, D., Raphael, B. J., Price, A. L., Tang, H. & Pevzner, P. A. Identifying repeat domains in large genomes. Genome Biol 7, 31 (2006).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009).
Chen, F., Mackey, A. J., Stoeckert, C. J., Jr. & Roos, D. S. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res 34, D363–D368 (2006).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30, 2725–2729 (2013).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Finn, R. D. et al. Pfam: the protein families database. Nucleic Acids Res 42, 27 (2014).
Khaldi, N. et al. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol 47, 736–741 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics btu638 (2014).
Anders, S. & Huber, W. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL) (2012).
Gotz, S. et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36, 3420–3435 (2008).
Acknowledgements
This work was supported by the National Foundation of Natural Sciences of China (31371961), China Agriculture Research System (CARS-27) and the Special Fund for Agro-Scientific Research in the Public Interest (201203034).
Author information
Authors and Affiliations
Contributions
M.W., D.Y. and H.L. designed the research. M.W., D.Y. and X.S. analyzed the data. M.W. and H.L. wrote the paper. K.C. and J.X. gave important suggestions and critically read this manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, M., Sun, X., Yu, D. et al. Genomic and transcriptomic analyses of the tangerine pathotype of Alternaria alternata in response to oxidative stress. Sci Rep 6, 32437 (2016). https://doi.org/10.1038/srep32437
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32437
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.