Abstract
Small proteins characterized by a double-glycine (GG) secretion motif, typical of secreted bacterial antibiotics, are encoded by the genomes of diverse cyanobacteria, but their functions have not been investigated to date. Using a biofilm-forming mutant of Synechococcus elongatus PCC 7942 and a mutational approach, we demonstrate the involvement of four small secreted proteins and their GG-secretion motifs in biofilm development. These proteins are denoted EbfG1-4 (enable biofilm formation with a GG-motif). Furthermore, the conserved cysteine of the peptidase domain of the Synpcc7942_1133 gene product (dubbed PteB for peptidase transporter essential for biofilm) is crucial for biofilm development and is required for efficient secretion of the GG-motif containing proteins. Transcriptional profiling of ebfG1-4 indicated elevated transcript levels in the biofilm-forming mutant compared to wild type (WT). However, these transcripts decreased, acutely but transiently, when the mutant was cultured in extracellular fluids from a WT culture and biofilm formation was inhibited. We propose that WT cells secrete inhibitor(s) that suppress transcription of ebfG1-4, whereas secretion of the inhibitor(s) is impaired in the biofilm-forming mutant, leading to synthesis and secretion of EbfG1-4 and supporting the formation of biofilms.
Similar content being viewed by others
Introduction
Cyanobacteria in nature often reside in biofilms, bacterial assemblages encased in a self-produced extracellular matrix1,2. Such microbial assemblages can lead to economic loss, for example due to material decay or blockage of flow through membranes in desalination plants3,4. However, phototrophic biofilms may be beneficial in other processes, for example when employed in wastewater purification systems and bioremediation processes5,6,7,8. Additionally, biofilm-based biofuel production systems that are efficient to harvest have been shown to generate high product yields with minimal water and nutrient inputs9,10. Furthermore, since grazing by small protistan predators imposes a major difficulty in growing cyanobacteria in open ponds, the biofilm, which serves as a physical barrier against these predators, has the potential for crop protection11,12,13.
In comparison with vast knowledge on the molecular mechanisms that underlie biofilm development in heterotrophic bacteria (see reviews14,15,16,17,18,19,20,21,22,23,24,25 and references therein), relatively little is known about these processes in cyanobacteria. Recent studies, however, have provided new insight into cyanobacterial aggregation processes and biofilm development26,27,28,29,30. For example, inactivation of Synechocystis PCC 6803 homologs of ATP binding cassette (ABC) transporters causes flocculation of the cultures and adherence to glass culture tubes30. An exoprotein of Anabaena sp. PCC 7120 is required for filament adhesion and aggregation31. Involvement of the signaling molecule cyclic dimeric GMP (c-di-GMP) in biofilm formation has been shown for Synechocystis26 and Thermosynechococcus elongatus32. In the latter case, blue light, acting through the cyanobacteriochrome SesA, elevates c-di-GMP and triggers a sessile phenotype at 31 °C, a relatively low temperature for T. elongatus32. SesA, together with two additional cyanobacteriochromes, provides a color-sensitive system for c-di-GMP-dependent cell aggregation27. Constitutive expression of the c-di-GMP producing enzyme, diguanylate cylase, in Synechocystis results in biofilm formation, whereas expression of phosphodiesterase to reduce c-di-GMP causes enhanced buoyancy26.
We recently demonstrated that the constitutive planktonic growth of the cyanobacterium Synechococcus elongatus PCC 7942 under standard laboratory conditions is due to a self-suppression mechanism that relies on secreted inhibitor(s)33. Here, we identify genes that enable biofilm development and provide evidence for the extracellular presence of four gene products denoted EbfG1-4 (enable biofilm formation possessing a double-glycine motif). Using a mutational approach, we demonstrate the requirement of the peptidase domain of the Synpcc7942_1133 product for efficient secretion of EbfG1-4. These data, together with transcriptional analysis of ebfG1-4, indicate that mediation of biofilm development in S. elongatus relies on extracellular inhibitors that repress expression of extracellular components required for biofilm formation.
Results
Small proteins characterized by a double glycine motif enable biofilm development
Previously, we demonstrated that inactivation of genes encoding homologs of type II protein secretion systems or the type IV pilus assembly apparatus (components of these complexes share a high degree of similarity34,35,36,37) impairs a biofilm inhibitory mechanism of S. elongatus and enables biofilm development33. Inactivation of synpcc7942_2071, which encodes a homolog of subunit E of type II secretion systems (T2SE), results in a mutant (T2SEΩ) that adheres to the growth tube in contrast to the planktonic phenotype of the WT strain (Fig. 1a). Analyses by fluorescence microscopy (Fig. 1b) and cryo-scanning electron microscopy (cryo-SEM, Fig. 1c) substantiate biofilm development by T2SEΩ (see Schatz et al.33 for additional cryo-SEM as well as environmental-SEM analyses).
We previously demonstrated that the gene Synpcc7942_1134 is required for biofilm formation by S. elongatus33. The product of this gene exhibits an N-terminal secretion motif (denoted GG-motif) typical of bacterial antibiotics, e.g. microcins. Such proteins undergo maturation during secretion by cleavage of the secretion motif after a conserved glycine-glycine (or glycine-alanine, as in the case of Synpcc7942_1134) motif 38,39. Wang et al.40 identified three previously unannotated open reading frames located immediately upstream of Synpcc7942_1134, the putative products of which are also characterized by a GG-motif40 (also see Fig. 2a,b). These gene products are referred to here as EbfG1-4. The putative precursors encoded by these genes are approximately 9–10 kDa, with 6–7 kDa remaining after their maturation by removal of the secretion motif40. Proteins possessing GG-motifs are prevalent in cyanobacteria40; however, mature EbfG1-4 of S. elongatus do not show homology to proteins from other cyanobacteria or exhibit domains of known function.
Small proteins characterized by double glycine motifs are required for biofilm development.
(a) Genomic region of ebfG1-4, genes whose products are characterized by a double-glycine secretion motif. Deletion of these four genes is indicated by ∆4. Arrows indicate PCR primers used for detection of the four-gene transcript (see Fig. 3b). Bar graph presents percentage of total chlorophyll in the suspended cells (average of three independent biological repeats ± standard deviation). Strains analyzed include: wild type (WT), the biofilm-forming mutant T2SEΩ and deletion of ebfG1-4 in T2SEΩ (T2SEΩ/∆4). /comp designates addition into neutral site (NS) 1 of the indicated DNA fragment, covering ebfG1 through Synpcc7942_1131, for complementation experiments (see T2SEΩ/∆4/comp). Similar fragments into which mutations changing the conserved GG-secretion motif to double alanine were introduced into the T2SEΩ/∆4 strain (for example, T2SEΩ/∆4/EbfG1m). Each motif conversion is indicated by a dot positioned at the cognate ORF in the genomic map. The ‘Quad’ fragment contains motif conversions in all four proteins. For statistical analysis we employed analysis of variance (ANOVA) and post analysis multiple comparison contrasts. In the bar graph, different letters assign statistical significance (see Table S1 for p-values). (b) Sequence of the double-glycine motif of EbfG1-4 and selected secreted peptides. Cerein7B of Bacillus cereus, EnterocinA and EnterocinB of Enterococcus faceum, MicrocinE492 of Klebsiella pneumonia and Microcin24, MicrocinH47 and ColicinV of Escherichia coli. Black shading indicates the double-glycine or glycine-alanine present just prior to the peptide cleavage site (arrow). Positions typically occupied by hydrophobic or hydrophilic amino acids are indicated by a circle or a triangle, respectively.
To test whether the GG-motifs of EbfG1-4 are required for biofilm development we combined mutations in these loci with mutation of Synpcc7942_2071 (T2SEΩ strain). The T2SEΩ mutation alone results in a culture in which about 5% of the total chlorophyll is present in suspended cells (Fig. 2a), while the remaining chlorophyll resides in biofilm-forming cells attached to the bottom and sides of the culture vessel. Deletion of ebfG1-4 in the context of the T2SEΩ mutation completely abolished biofilm formation, with 100% of the chlorophyll present in suspended cells, similar to the WT (Fig. 2a, T2SEΩ/Δ4). Recombination of a DNA fragment bearing the genes ebfG1-4 into the genome at neutral site 1 (NS1) complements the phenotype of T2SEΩ/Δ4, restoring biofilm formation to the level observed in T2SEΩ (Fig. 2a, T2SEΩ/Δ4/comp).
To examine whether the GG-motifs in EbfG1-4 are required for biofilm formation, we altered the trans-complementing DNA fragment to separately modify each GG-motif to AA. For example, introduction of DNA encoding a modified EbfG1 GG-motif (with EbfG2-4 sequences unchanged) only partially restored biofilm development to the completely planktonic T2SEΩ/Δ4 strain (Fig. 2a, T2SEΩ/Δ4/EbfG1m). On average, 60% of the total chlorophyll was found in suspended cells, significantly more than the 2–5% found inT2SEΩ or T2SEΩ/∆4/comp (see Table S1 for p-values). Similarly, modification of the GG-motif of EbfG2 or EbfG4 to AA (Fig. 2a, T2SEΩ/Δ4/EbfG2m and T2SEΩ/Δ4/EbfG4m, respectively) in the complementing DNA fragment partially restored biofilm development to the T2SEΩ/Δ4 strain (50–70% chlorophyll in suspended cells). In contrast, mutation of the GG-motif in EbfG3 only slightly interfered with biofilm development; in this case just 15% of the chlorophyll was in the planktonic cells (Fig. 2a, compare T2SEΩ/Δ4/EbfG3m and T2SEΩ/∆4/comp). When the GG-motifs in all four proteins were mutated simultaneously, biofilm development was completely abolished (Fig. 2a, T2SEΩ/Δ4/Quad). In summary, these data support the suggestion that each of the GG-motifs contributes to biofilm formation. Furthermore, because the combination of the four mutations completely abrogated biofilm development, whereas each individual mutation only partially interfered with biofilm formation, we suggest functional redundancy between EbfG1-4.
Transcripts of ebfG1-4 are highly abundant in T2SEΩ
Differences in transcript levels from the ebfG1-4 genes between the WT and T2SEΩ were followed using RT-qPCR. Initially, T2SEΩ grows planktonically; however, biofilms start forming, usually on the third day of growth. Transcript levels of ebfG1-4 in T2SEΩ 1 day after inoculation into fresh medium were 14–37 fold higher compared to WT (Fig. 3a, day 1 Fresh Medium). Once biofilms formed, the sessile as well as the planktonic cells of the mutant were characterized by significantly higher levels of these transcripts compared to 1 day old mutant cultures (Fig. 3a, T2SEΩ Fresh Medium). The fact that transcripts of ebfG1-4 are similarly elevated in planktonic and biofilm forming cells of T2SEΩ at 3 and 6 days (Fig. 3a, 3P, 3BF, 6P, 6BF) suggests that high transcript levels of these genes are not by themselves sufficient to drive biofilm development.
Transcript level of ebfG1-4 is elevated in T2SEΩ compared to WT and is reduced under conditioned medium from a WT culture.
(a) RT-qPCR analyses of ebfG1-4 in cells grown for 1, 3 and 6 days. Cultures were initiated in fresh medium or conditioned medium from a WT culture. When biofilms were present (T2SEΩ grown in fresh medium for 3 and 6 days), the transcript level is shown for planktonic cells (3P, 6P) and cells in the biofilm (3BF, 6BF). Transcript levels of psbC, encoding CP43, a chlorophyll binding protein of photosystem II, served to normalize total RNA levels. Transcript abundance is indicated as fold change relative to WT on the first day of growth in fresh medium. Data are presented on a log2 scale. Bars indicate average of three independent biological repeats ± standard deviation). For statistical analysis we employed three factors ANOVA and post analysis multiple comparison contrasts. In each bar graph, different letters assign statistical significance (see Table S2 for p-values). Similar trends in transcript level were previously reported for ebfG4 using semiquantitative RT-qPCR33. (b) Gel image of PCR products representing a four-gene polycistronic transcript of ebfG1-4. Primers used for the PCR are indicated in Fig. 2a and Table S4.
Extracellular fluids from a WT culture (hereafter, conditioned medium) inhibit biofilm development of T2SEΩ33. Inoculation of the T2SEΩ mutant into conditioned medium significantly reduced ebfG1-4 transcript levels during the first day of growth, as compared to T2SEΩ at the same growth stage in fresh medium. Further growth of T2SEΩ in conditioned medium resulted in increased transcript levels of ebfG1-4 compared to day 1, but did not reach the levels detected in biofilm forming cells of this mutant (Fig. 3a). In two cases, ebfG1 and ebfG4, the transcript levels after 6 days were not significantly different between planktonic T2SEΩ cells grown in fresh medium and those grown in conditioned medium, indicating that conditioned medium results in expression levels of these genes consistent with planktonic cells but not with biofilm forming cells.
The similar trends of transcriptional changes of ebfG1-4 suggested that these genes are co-regulated. Using the primers indicated in Fig. 2a for RT-PCR, we detected a polycistronic transcript containing ebfG1-4 (Fig. 3b). Likely due to low expression in WT, this transcript was not previously identified in RNA-seq of S. elongatus by Vijayan et al.41. The mechanism underlying regulation of these genes is yet unknown.
A cysteine peptidase is required for efficient secretion of EbfG1-4
Using insertional inactivation, we previously demonstrated that the gene Synpcc7942_1133 is essential for biofilm development33. The protein encoded by this gene is characterized by a peptidase domain of the ‘C39 family’ (Fig. 4a), named after the conserved cysteine residue essential for transport and maturation of precursor substrates that possess a GG-motif42,43. Akin to the analysis performed with the GG-motifs, a mutational complementation approach was employed to examine the requirement for a functional peptidase domain to enable biofilm development. The double mutant T2SEΩ/1133Ω exhibits a planktonic phenotype33 (Fig. 4b). Biofilm development was restored when a DNA fragment bearing Synpcc7942_1133 was combined with the T2SEΩ/1133Ω double mutation (Fig. 4b, T2SEΩ/1133Ω/comp). Replacement of the conserved cysteine residue with alanine in this trans-complementing DNA fragment, however, completely abolished biofilm formation (Fig. 4b, T2SEΩ/1133Ω/PteBm), in agreement with the hypothesis that the protein encoded by Synpcc7942_1133 (denoted PteB for peptidase transporter essential for biofilm) is involved in the secretion of EbfG1-4.
The conserved cysteine in the peptidase domain of PteB is essential for biofilm development.
(a) Amino acid alignment of the N-terminal domain of C39 peptidases and the product of Synpcc7942_1133 from S. elongatus. CvaB of Escherichia coli, LagD and LcnC of Lactococcus lactis, ComA of Streptococcus pneumonia, PlnG of Lactobacillus plantarum and PedD of Pediococcus acidilactici. Identity or similarity between at least 50% of the aligned sequences is indicated by black or grey shading, respectively. The asterisk denotes the conserved cysteine residue typically involved in processing of a double-glycine containing substrate. (b) A kanamycin resistance cassette (KmR) was inserted into Synpcc7942_1133 and combined with inactivation of t2sE (T2SEΩ/1133Ω). /comp designates addition into neutral site (NS) 1of the indicated DNA fragment, covering ebfG1 through Synpcc7942_1131, for complementation experiments (see T2SEΩ/1133Ω/comp). A similar fragment into which a mutation changing the conserved cysteine to alanine (indicated by a diamond) was introduced into the T2SEΩ/1133Ω strain (T2SEΩ/1133Ω/PteBm). Bar graphs indicate percentage of chlorophyll in suspended cells (average of three independent biological repeats ± standard deviation). For statistical analysis we employed ANOVA and post analysis multiple comparison contrasts. In the bar graph, different letters assign statistical significance (see Table S3 for p-values).
To test whether PteB is required for secretion of the small GG-motif proteins of S. elongatus, we analyzed the extracellular fluids of the strains T2SEΩ/1133Ω/comp and T2SEΩ/1133Ω/PteBm for the presence of these secreted proteins. The control strain T2SEΩ/1133Ω/comp forms biofilms similarly to T2SEΩ and likely has high transcript levels for ebfG1-4. Comparative analyses by mass spectrometry indicated significantly higher levels of peptides derived from EbfG1-4 in the conditioned media of the biofilm forming strain T2SEΩ/1133Ω/comp as compared to that of the planktonic strain T2SEΩ/1133Ω/PteBm, which possesses the cysteine to alanine mutation (Fig. 5). The presence of some of the peptides in extracellular fluids from T2SEΩ/1133Ω/PteBm cultures may be due to cell lysis. It is also possible that the cysteine to alanine mutation does not completely abrogate PteB function and some secretion and maturation does occur in the mutant. This potential activity, however, is insufficient to support biofilm development because the T2SEΩ/1133Ω/PteBm grows planktonically (Fig. 4b).
The conserved cysteine in the peptidase domain of PteB is required for efficient secretion of EbfG1-4, small proteins exhibiting a double glycine motif.
Mass spectrometry analyses of conditioned media from 2 d old cultures of the control strain (T2SEΩ/1133Ω/comp) and the mutant, in which the conserved cysteine in the peptidase domain was mutated to alanine (T2SEΩ/1133Ω/PteBm). Samples were digested prior to analyses using trypsin alone or in combination with chymotrypsin. Table provides normalized areas of detected peptides for three independent biological replicates. Normalized area is proportional to the peptide level. Zero values indicate the inability to detect a peptide. Graph on the left presents the log2(t + 1) transformation of the data for each peptide (indicated by protein and number) as box plots, with the box representing the second and third quartiles, the horizontal line indicating the median and the whisker bars representing the maximum and minimum values. Each individual data point is plotted as a circle (Control = blue, Mutant = red). Note that on the box plot of the mutant, the bottom dot is a zero value, which actually represents multiple observation as is indicated in the table. The differences between the control and the mutant strain for peptide EbfG1#2 and EbfG4#3 are significant at a p-value of 0.039 and for all other peptides at a p-value of 0.05, as calculated using a one-tailed Mann-Whitney test for two independent samples.
Of note, the peptides TTYNPPSYPYPSYPK (EbfG1), SGYSIPTYPK (EbfG2) and LLQTANSVAAAIAK (EbfG4) were detected by mass spectrometry in conditioned media. Unlike all other peptides detected in these analyses, which represent cleavage by the trypsin or chymotrypsin peptidases used for sample digestion prior to analysis, these three peptides likely represent cleavage of the secretion motif, because in the putative precursor protein they are preceded by GG (EbfG1 and EbfG2) or GA in the case of EbfG4 (see Fig. S1) and not by an amino acid typically preceding trypsin or chymotrypsin cleavage sites. These data provide support for maturation and secretion of these small proteins by removal of the secretion motif. In summary, the mutational approach and the mass spectrometry analyses support the suggestion that efficient secretion of EbfG1-4 is required for biofilm development and that the peptidase domain of PteB is involved in the secretion process.
Discussion
The data suggest that S. elongatus employs a GG-secretion motif, typically associated with microcins and a transport system involving a cysteine peptidase for secretion and maturation of the small proteins encoded by ebfG1-4. Wang et al. identified similar gene clusters throughout the vast diversity of sequenced cyanobacterial genomes and suggested that these gene products represent a potential source for natural products40. To our knowledge, the function of these small cyanobacterial proteins has not been experimentally examined prior to this work. This study, which implicates the small proteins of S. elongatus in biofilm development, assigns a novel function to proteins possessing a microcin-like secretion motif.
Cyanobacterial proteins with a microcin-like secretion motif may represent a large variety of functions. In this vein, it should be noted that HetC of Anabaena sp. PCC 7120, which is required for normal differentiation of heterocysts, exhibits a domain organization similar to the cysteine peptidase PteB. It was suggested that HetC may be involved in processing of PatS, the small protein that is required for normal heterocyst pattern formation44, although PatS is not characterized by a GG-secretion motif.
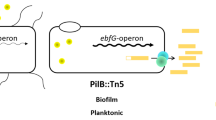
Our experiments revealed that transcript abundance of ebfG1–4, which are cotranscribed, is highly elevated in the biofilm forming strain T2SEΩ as compared to WT (Fig. 3). Inoculation of the T2SEΩ mutant in conditioned medium from a WT culture prevents biofilm formation33 and substantially reduces the transcript levels of ebfG1-4 at the initial stage of growth (Fig. 3 day 1 and day 3). These data, together with the results supporting secretion of EbfG1-4 and involvement of PteB in secretion, allow us to refine our previous model for biofilm development by S. elongatus (Fig. 6). Using a T2S-like system, the WT deposits a biofilm inhibitor to the extracellular milieu and consequently represses transcription of ebfG1-4 (Fig. 3) and pteB (see data for Synpcc7942_113333). Suppression of the expression of these genes, even transiently, was associated with biofilm inhibition. In T2SEΩ, secretion of the biofilm inhibitor is impaired, allowing high levels of transcription of ebfG1-4 (Fig. 3) and pteB33 and the small proteins encoded by ebfG1-4 are synthesized at high levels. Adequate secretion and maturation of EbfG1-4 by a transport system in which the cysteine peptidase (PteB) takes part, is required for biofilm formation.
Transcription of ebfG1-4 and secretion of the small proteins encoded by these genes is governed by extracellular biofilm inhibitor(s).
Using a T2S-like system, WT cells secrete an inhibitor (red) which suppresses transcription of the genes ebfG1-4 (this study) and pteB33. The biofilm-forming mutant, T2SEΩ, is most likely impaired in secretion of the inhibitory factor and therefore expresses ‘biofilm-genes’ at a higher level. Consequently, the small proteins that possess GG-secretion motif are produced at increased levels compared to the WT and are secreted and processed by a transport system in which the cysteine peptidase, PteB takes part. Dashed and thick arrows represent low and high transcript levels, respectively. The hatched regions of the secreted proteins represent the GG-motif. Biofilm development or its absence is indicated by plus (+) or minus (−) symbols, respectively. The T2S system may not be directly involved in secretion of the inhibitor.
Methods
Strains, culture conditions, biofilm quantification and microscopy
Synechococcus elongatus PCC 7942 and all derived strains were grown essentially as described previously33. Specifically, 25 ml cultures were grown in round-bottom Pyrex tubes (20 cm in length, 3 cm in diameter). Cotton-plugged Pasteur pipette (23 cm long) inserted through a sponge-plug served for bubbling of 3% CO2 in air into the cultures. The tip of the Pasteur pipette was placed 1–2 cm above the bottom of the tube. Prior to bubbling into the culture, the gas was humidified by passing through a bottle with double distilled water and filtered using 0.22 μm filter. BG11 medium45 served for culturing; however, to reproducibly observe biofilm formation, it was important to add the ferric ammonium citrate and citric acid components (final concentrations 0.0226 and 0.0312 mM, respectively) from freshly made stocks. Autoclaved BG11 was used within 4 days to inoculate cultures. Cultures were grown at 30 °C and under incandescent light (20–30 μmol photons m−2 sec−1).
For assessment of biofilm formation, cells were cultured under continuous bubbling. Experiments were initiated by diluting cultures at the exponential phase of growth to an optical density at 750 nm of 0.5. Biofilm development under this setting typically initiated following 2–3 days of growth and quantification was performed after 7 days to allow assessment of fully developed biofilms (longer growth time did not increase biofilm development). Percentage of chlorophyll in suspended cells served to quantity biofilm formation as follows. The suspended fraction was sampled for chlorophyll determination by extraction in 80% acetone (final concentration). In cases were the suspended fraction appeared especially dense (planktonic strains or poor biofilm formers), a 0.2 mL sample was used for extraction. When robust biofilms were formed, 15 mL of planktonic cells were removed and concentrated 3 to 6-fold by centrifugation (5000 g, 10 min) prior the chlorophyll extraction in 80% acetone. Determination of chlorophyll in the biofilm was performed following removal of the planktonic cells using a pipette and addition of 80% acetone to the sessile cells. Extraction was carried out over-night in the refrigerator. Dilution in 80% acetone was performed to reach the linear range of the calibration curve and chlorophyll was quantified based on absorbance at 663 nm. For statistical analysis of comparisons of chlorophyll in suspension we employed analysis of variance (ANOVA) and post analysis multiple comparison contrasts (see statistical grouping in Figs 2 and 4 and Tables S1 and S3 for p-values).
For collection of conditioned medium, cultures were centrifuged (5000 g, 10 min) at room temperature and the supernatant was removed and passed through a 0.22 μm filter. Supplementation with nutrients by addition of medium stock solutions as in the preparation of fresh growth medium was performed when the conditioned medium served for cyanobacterial growth.
For microscopic analysis of biofilms, microscope slides were inserted into growth tubes containing T2SEΩ cultures (for 6 days) so that biofilms formed on the slides. A Leica TCS SPE DM2500 was employed for imaging by fluorescence microscopy using objective HCX APO l 40x/0.80 (excitation 532 nm, emission 620–650 nm). 3D image reconstruction was performed using Imaris. Analysis by SEM was described previously33.
Genetic manipulations of cyanobacterial cells
Insertional inactivation of genes Synpcc7942_2071 (t2sE), Synpcc7942_1134 (ebfG4) and Synpcc7942_1133 was described previously33. In the case of deletion of ebfG1 through ebfG4 (∆4), disruption was obtained by deletion of the fragment between the NheI and ClaI sites. Resulting constructs were introduced into S. elongatus using standard transformation methods that take advantage of its natural competence46. Mutant cyanobacterial clones resistant to the appropriate antibiotic were confirmed for double homologous recombination (allele replacement) and complete chromosomal segregation using PCR on genomic DNA. Primers and additional cloning information are provided in Table S4. Inactivation of t2SE impaired the natural DNA competence of S. elongatus; thus, to obtain T2SEΩ strains with additional genetic changes, we initially introduced the other genetic modifications and subsequently inactivated t2sE.
Vectors used to introduce the WT and mutated complementation DNA fragments into NS1 were generated using the GeneArt® Seamless Cloning and Assembly Kit (Life Technologies) on PCR-generated cloning fragments and standardized devices (a chloramphenicol resistance device and a NS1 device with tetracycline resistance) derived via ZraI or EcoRV-HF (New England Biolabs) restriction digestion of CYANO-VECTOR donor plasmids as described by Taton et al.47. To improve the efficiency of cloning, the 6-kb WT DNA fragment was constructed from two 3-kb fragments amplified with the Q5® High-Fidelity DNA polymerase (New England Biolabs). To introduce glycine- or cysteine to alanine mutations, the appropriate cloning fragment was assembled from subfragments; these segments were generated using seamless assembly cloning primers designed to introduce the appropriate base mutations at the center of the overlap between subfragments. Combining multiple such subfragments or generating subfragments from previously mutated vectors allowed the generation of the quadruple GG-mutation vector. The sequence of each vector was confirmed. These vectors were used to transform S. elongatus strains as described above. For additional information see Table S4 and Fig. S2.
Mass spectrometry analysis
Conditioned medium was harvested as described above. Mass spectrometry was performed by the de Botton Institute for Protein Profiling at The Nancy and Stephen Grand Israel National Center for Personalized Medicine (Weizmann Institute of Science).
Sample preparation
Culture supernatants were concentrated ~20 fold on 3 kDa molecular weight cutoff-filters. Proteins were reduced by incubation with dithiothreitol (5 mM; Sigma) for 45 min at 60 °C and alkylated with 10 mM iodoacetamide (Sigma) in the dark for 45 min at 21 °C. Proteins were then subjected to digestion with trypsin (Promega; Madison, WI, USA, ratio of 50:1 protein amount:enzyme amount) for 4 h at 37 °C followed by digestion with chymotrypsin (Sigma, ratio of 50:1 protein amount:enzyme amount) for 16 h at 37 °C. The digestions were stopped by trifluroacetic acid (1%). Following digestion, peptides were desalted using solid-phase extraction columns (Oasis HLB, Waters, Milford, MA, USA). The samples were stored in −80 °C until further analysis.
Liquid chromatography
ULC/MS grade solvents were used for all chromatographic steps. Each sample was loaded using split-less nano-Ultra Performance Liquid Chromatography (10 kpsi nanoAcquity; Waters, Milford, MA, USA). The mobile phase was: A) H2O + 0.1% formic acid and B) acetonitrile +0.1% formic acid. Desalting of the samples was performed online using a reversed-phase C18 trapping column (180 μm internal diameter, 20 mm length, 5 μm particle size; Waters). The peptides were then separated using a T3 HSS nano-column (75 μm internal diameter, 250 mm length, 1.8 μm particle size; Waters) at 0.35 μL/min. Peptides were eluted from the column into the mass spectrometer using the following gradient: 4% to 30% B in A (vol/vol) in 105 min, 35% to 90% B in A in 5 min, maintained at 95% for 5 min and then back to initial conditions.
Mass Spectrometry
The nano Ultra Performance Liquid Chromatography was coupled online through a nanoESI emitter (10 μm tip; New Objective; Woburn, MA, USA) to a quadrupole orbitrap mass spectrometer (Q Exactive Plus, Thermo Scientific) using a FlexIon nanospray apparatus (Proxeon). For initial identification, data were acquired in Data-Dependent Acquisition (DDA) mode, using a Top20 method. MS1 resolution was set to 70,000 (at 400 m/z) and maximum injection time was set to 20 msec. MS2 resolution was set to 17,500 and maximum injection time of 60 msec. For targeted analysis, data was acquired in Parallel Reaction Monitoring (PRM) mode, monitoring previously identified peptides from EbfG1-4. MS2 resolution was set to 35,000 (at 400 m/z) and maximum injection time of 100 msec.
Data processing and analysis
For identification purposes, raw data was first processed using Proteome Discoverer v1.41. MS/MS spectra were searched using Mascot v2.4 (Matrix Sciences) and Sequest HT. Data were searched against the S. elongatus protein database as downloaded from UniprotKB (http://www.uniprot.org/), appended with EbfG1-4, along with 125 common laboratory contaminant proteins. Fixed modification was set to carbamidomethylation of cysteines and variable modification was set to oxidation of methionines. Search results were then imported back to Expressions to annotate identified peaks. Proteins were then grouped based on shared peptides and Identifications were filtered such that the global false discovery rate was maximum of 1%. For PRM analysis, raw data and DDA search results were imported into the Skyline software (https://skyline.gs.washington.edu/). The software was used for retention time alignment, peak detection of peptide fragments and their quantification.
Statistical analysis used the Mann-Whitney U test, a non-parametric test, because the majority of the data (or any viable transformation of it) are not normally distributed.
RNA preparation and RT-PCR
RNA was prepared as previously described48 and treated with DNase (TURBO DNase, Ambion). Random hexamers (Promega) were used to prime cDNA using 1.5 μg RNA and reverse transcriptase (RevertAid, Fermentas). PCR amplification was performed using Fast SYBR Green Master Mix (Thermo Fisher Scientific) with addition of betaine pH 9.0 (final concentration of 0.525M) and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Melting curve analysis was performed (65 to 95 °C, 0.5 °C/5s) to confirm amplification of a single cDNA sequence for each gene. Primer dimers or unexpected amplicons were not observed. No signals were detected in the negative controls (samples in which reverse transcriptase was not added). Specific primers for RT-qPCR and RT-PCR of the ebfG1-4 transcript are indicated in Table S4. Transcript levels of psbC, encoding CP43, a chlorophyll binding protein of photosystem II, served to normalize total RNA levels.
For statistical analysis we employed three factor ANOVA to calculate the covariance (normality and equality of variances were tested using Kruskal-Wallis & Levene’s tests respectively, both found to be >0.05). Next, we used multiple comparison contrasts to evaluate the differences of each pair of comparison. In each bar graph in Fig. 3a, different letters assign statistical significance (see Table S2 for p-values).
Additional Information
How to cite this article: Parnasa, R. et al. Small secreted proteins enable biofilm development in the cyanobacterium Synechococcus elongatus. Sci. Rep. 6, 32209; doi: 10.1038/srep32209 (2016).
References
Gorbushina, A. A. Life on the rocks. Environ. Microbiol. 9, 1613–1631 (2007).
Rossi, F. & De Philippis, R. Role of cyanobacterial exopolysaccharides in phototrophic biofilms and in complex microbial mats. Life 5, 1218–1238, doi: 10.3390/life5021218 (2015).
Baker, J. S. & Dudley, L. Y. Biofouling in membrane systems-A review. Desalination 118, 81–89, doi: 10.1016/S0011-9164(98)00091-5 (1998).
Flemming, H. C. Biofouling in water systems-cases, causes and countermeasures. Appl. Microbiol. Biotechnol. 59, 629–640, doi: 10.1007/s00253-002-1066-9 (2002).
Ivnitsky, H. et al. Bacterial community composition and structure of biofilms developing on nanofiltration membranes applied to wastewater treatment. Water Res. 41, 3924–3935 (2007).
Egan, S., Thomas, T. & Kjelleberg, S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 11, 219–225, doi: S1369-5274(08)00052-0 [pii] 10.1016/j.mib.2008.04.001 (2008).
Roeselers, G., Loosdrecht, M. C. & Muyzer, G. Phototrophic biofilms and their potential applications. J. Appl. Phycol. 20, 227–235, doi: 10.1007/s10811-007-9223-2 (2008).
Wagner, M. et al. Microbial community composition and function in wastewater treatment plants. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology 81, 665–680 (2002).
Gross, M., Jarboe, D. & Wen, Z. Biofilm-based algal cultivation systems. Appl. Microbiol. Biotechnol. 99, 5781–5789, doi: 10.1007/s00253-015-6736-5 (2015).
Heimann, K. Novel approaches to microalgal and cyanobacterial cultivation for bioenergy and biofuel production. Curr. Opin. Biotechnol. 38, 183–189, doi: 10.1016/j.copbio.2016.02.024 (2016).
Matz, C. et al. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102, 16819–16824, doi: 10.1073/pnas.0505350102 (2005).
DePas, W. H. et al. Biofilm formation protects Escherichia coli against killing by Caenorhabditis elegans and Myxococcus xanthus. Appl. Environ. Microbiol. 80, 7079–7087, doi: 10.1128/AEM.02464-14 (2014).
Simkovsky, R., Effner, E. E., Iglesias-Sanchez, M. J. & Golden, S. S. Mutations in novel lipopolysaccharide biogenesis genes confer resistance to amoebal grazing in Synechococcus elongatus. Appl. Environ. Microbiol. doi: 10.1128/AEM.00135-16 (2016).
Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nature Rev. Microbiol. 2, 95–108 (2004).
Stanley, N. R. & Lazazzera, B. A. Environmental signals and regulatory pathways that influence biofilm formation. Molec. Microbiol. 52, 917–924 (2004).
Branda, S. S., Vik, S., Friedman, L. & Kolter, R. Biofilms: the matrix revisited. Trends Microbiol. 13, 20–26, doi: 10.1016/j.tim.2004.11.006 (2005).
Parsek, M. R. & Greenberg, E. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13, 27–33 (2005).
Kolter, R. & Greenberg, E. P. Microbial sciences - The superficial life of microbes. Nature 441, 300–302 (2006).
Karatan, E. & Watnick, P. Signals, regulatory networks and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347, doi: 73/2/310 [pii] 10.1128/MMBR.00041-08 (2009).
Landini, P., Antoniani, D., Burgess, J. G. & Nijland, R. Molecular mechanisms of compounds affecting bacterial biofilm formation and dispersal. Appl. Microbiol. Biotechnol. 86, 813–823 (2010).
McDougald, D., Rice, S. A., Barraud, N., Steinberg, P. D. & Kjelleberg, S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nature Rev. Microbiol. 10, 39–50 (2011).
Elias, S. & Banin, E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol. Rev. 36, 990–1004, doi: 10.1111/j.1574-6976.2012.00325.x (2012).
Rendueles, O. & Ghigo, J. M. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol. Rev. 36, 972–989, doi: 10.1111/j.1574-6976.2012.00328.x (2012).
Nagar, E. & Schwarz, R. To be or not to be planktonic? Self-inhibition of biofilm development. Environ. Microbiol. 17, 1477–1486, doi: 10.1111/1462-2920.12583 (2015).
Petrova, O. E. & Sauer, K. Escaping the biofilm in more than one way: desorption, detachment or dispersion. Curr. Opin. Microbiol. 30, 67–78, doi: 10.1016/j.mib.2016.01.004 (2016).
Agostoni, M., Waters, C. M. & Montgomery, B. L. Regulation of biofilm formation and cellular buoyancy through modulating intracellular cyclic di-GMP levels in engineered cyanobacteria. Biotechnol. Bioeng, doi: 10.1002/bit.25712 (2015).
Enomoto, G., Ni-Ni-Win, Narikawa, R. & Ikeuchi, M. Three cyanobacteriochromes work together to form a light color-sensitive input system for c-di-GMP signaling of cell aggregation. Proc. Natl. Acad. Sci. USA 112, 8082–8087, doi: 10.1073/pnas.1504228112 (2015).
Jittawuttipoka, T. et al. Multidisciplinary evidences that Synechocystis PCC6803 exopolysaccharides operate in cell sedimentation and protection against salt and metal stresses. PloS one 8, e55564, doi: 10.1371/journal.pone.0055564 (2013).
Schwarzkopf, M., Yoo, Y. C., Hueckelhoven, R., Park, Y. M. & Proels, R. K. Cyanobacterial Phytochrome2 Regulates the Heterotrophic Metabolism and Has a Function in the Heat and High-Light Stress Response. Plant Physiol. 164, 2157–2166, doi: 10.1104/pp.113.233270 (2014).
Fisher, M. L., Allen, R., Luo, Y. & Curtiss, R. 3rd . Export of extracellular polysaccharides modulates adherence of the Cyanobacterium Synechocystis. PloS one 8, e74514, doi: 10.1371/journal.pone.0074514 (2013).
Oliveira, P., Pinto, F., Pacheco, C. C., Mota, R. & Tamagnini, P. HesF, an exoprotein required for filament adhesion and aggregation in Anabaena sp PCC 7120. Environ. Microbiol. 17, 1631–1648, doi: 10.1111/1462-2920.12600 (2015).
Enomoto, G. et al. Cyanobacteriochrome SesA Is a Diguanylate Cyclase That Induces Cell Aggregation in Thermosynechococcus. J. Biol. Chem. 289, 24801–24809, doi: 10.1074/jbc.M114.583674 (2014).
Schatz, D. et al. Self-suppression of biofilm formation in the cyanobacterium Synechococcus elongatus. Environ. Microbiol. 15, 1786–1794, doi: 10.1111/1462-2920.12070 (2013).
Peabody, C. R. et al. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149, 3051–3072 (2003).
Filloux, A. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta 1694, 163–179 (2004).
Cianciotto, N. P. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13, 581–588 (2005).
Johnson, T. L., Abendroth, J., Hol, W. G. & Sandkvist, M. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255, 175–186 (2006).
Riley, M. A. & Wertz, J. E. Bacteriocins: evolution, ecology and application. Ann. Rev. Microbiol. 56, 117–137, doi: 10.1146/annurev.micro.56.012302.161024 (2002).
Dirix, G. et al. Peptide signal molecules and bacteriocins in Gram-negative bacteria: a genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides 25, 1425–1440 (2004).
Wang, H., Fewer, D. P. & Sivonen, K. Genome mining demonstrates the widespread occurrence of gene clusters encoding bacteriocins in cyanobacteria. PloS one 6, e22384, doi: 10.1371/journal.pone.0022384 PONE-D-11-07449 [pii] (2011).
Vijayan, V., Jain, I. H. & O’Shea, E. K. A high resolution map of a cyanobacterial transcriptome. Genome Biol. 12, R47, doi: 10.1186/gb-2011-12-5-r47 (2011).
Michiels, J., Dirix, G., Vanderleyden, J. & Xi, C. Processing and export of peptide pheromones and bacteriocins in Gram-negative bacteria. Trends Microbiol. 9, 164–168 (2001).
Lin, D. Y. W., Huang, S. & Chen, J. Crystal structures of a polypeptide processing and secretion transporter. Nature 523, 425–U492, doi: 10.1038/nature14623 (2015).
Corrales-Guerrero, L., Flores, E. & Herrero, A. Relationships between the ABC-Exporter HetC and Peptides that Regulate the Spatiotemporal Pattern of Heterocyst Distribution in Anabaena. PloS one 9, doi: ARTN e104571 10.1371/journal.pone.0104571 (2014).
Stanier, R. Y., Kunisawa, R., Mandel, M. & Cohen-Bazire, G. Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev. 35, 171–205 (1971).
Clerico, E. M., Ditty, J. L. & Golden, S. S. Specialized techniques for site-directed mutagenesis in cyanobacteria. In Methods in Molecular Biology, ed. E. Rosato, Humana Press, Inc., Totowa, NJ, 155–171 (2007).
Taton, A. et al. Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 42, e136, doi: 10.1093/nar/gku673 (2014).
Tu, C. J., Shrager, J., Burnap, R. L., Postier, B. L. & Grossman, A. R. Consequences of a deletion in dspA on transcript accumulation in Synechocystis sp. strain PCC6803 J. Bacteriol. 186, 3889–3902 (2004).
Simkovsky, R. et al. Impairment of O-antigen production confers resistance to grazing in a model amoeba-cyanobacterium predator-prey system. Proc. Natl. Acad. Sci. USA 109, 16678–16683, doi: 10.1073/pnas.1214904109 (2012).
Acknowledgements
Rakefet Schwarz and Susan Golden are supported by the program of the National Science Foundation and the US-Israel Binational Science Foundation (NSF-BSF 2012823). This study was also supported by a grant from the Israel Science Foundation (ISF 1406/14) to Rakefet Schwarz. We thank Yishai Levin, Meital Kupervaser and Alon Savidor at the de Botton Institute for Protein Profiling, The Nancy and Stephen Grand Israel National Center for Personalised Medicine (Weizmann Institute of Science) for MS analyses. We thank Shlomi Dagan for statistical analysis.
Author information
Authors and Affiliations
Contributions
R.P., E.N., E.S., Z.R., R. Simkovsky, S.G. and R. Schwarz designed research; R.P., E.N., E.S. and R. Simkovsky, performed research; R.P., E.N., E.S., R. Simkovsky, S.G. and R. Schwarz analyzed the data; R.P., E.N., E.S., Z.R., R. Simkovsky, S.G. and R. Schwarz wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Parnasa, R., Nagar, E., Sendersky, E. et al. Small secreted proteins enable biofilm development in the cyanobacterium Synechococcus elongatus. Sci Rep 6, 32209 (2016). https://doi.org/10.1038/srep32209
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep32209
This article is cited by
-
Cell specialization in cyanobacterial biofilm development revealed by expression of a cell-surface and extracellular matrix protein
npj Biofilms and Microbiomes (2023)
-
Linking microbial slime community structure with abiotic factors and antifouling strategy in hydroelectric cooling systems
Brazilian Journal of Microbiology (2023)
-
Protist impacts on marine cyanovirocell metabolism
ISME Communications (2022)
-
Hydrodynamic conditions affect the proteomic profile of marine biofilms formed by filamentous cyanobacterium
npj Biofilms and Microbiomes (2022)
-
Exploring the Role of Bacterial Extracellular Polymeric Substances for Sustainable Development in Agriculture
Current Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.