Abstract
Conidiation patterning is evolutionarily complex and mechanism concerning conidiogenous cell differentiation remains largely unknown. Magnaporthe oryzae conidiates in a sympodial way and uses its conidia to infect host and disseminate blast disease. Arrestins are multifunctional proteins that modulate receptor down-regulation and scaffold components of intracellular trafficking routes. We here report an alpha-arrestin that regulates patterns of conidiation and contributes to pathogenicity in M. oryzae. We show that disruption of ARRDC1 generates mutants which produce conidia in an acropetal array and ARRDC1 significantly affects expression profile of CCA1, a virulence-related transcription factor required for conidiogenous cell differentiation. Although germ tubes normally develop appressoria, penetration peg formation is dramatically impaired and Δarrdc1 mutants are mostly nonpathogenic. Fluorescent analysis indicates that EGFP-ARRDC1 puncta are well colocalized with DsRed2-Atg8, and this distribution profile could not be altered in Δatg9 mutants, suggesting ARRDC1 enters into autophagic flux before autophagosome maturation. We propose that M. oryzae employs ARRDC1 to regulate specific receptors in response to conidiation-related signals for conidiogenous cell differentiation and utilize autophagosomes for desensitization of conidiogenous receptor, which transmits extracellular signal to the downstream elements of transcription factors. Our investigation extends novel significance of autophagy-associated alpha-arrestin signaling to fungal parasites.
Similar content being viewed by others
Introduction
Conidia are asexual spores used by quite many fungal pathogens for infection initiation and disease dissemination. At least twenty modes of conidiation have been described in conidial fungi1, including the causal agent of the rice blast disease Magnaporthe oryzae2,3, which generates conidia in a sympodial pattern4.
Foliar blast disease starts with aerial dispersal of sympodial conidia through wind and raindrop splash5. Conidia attach to host cuticles and produce germ tubes that differentiate into appressoria with huge turgor pressure6. Appressoria develop penetration peg to breach host surfaces allowing subsequent infection hyphae to ramify throughout surrounding epidermis cells7. Four to five days later, conidiophores emerge from infected sites and sympodial conidia are produced for secondary dissemination4,8.
Upon favorable environmental cues, the rice blast fungus differentiates conidiogenous cell at the tip of aerial conidiophore stalk. Mono-nucleus conidiogenous cell subsequently undergoes mitotic divisions to give a three-celled conidium. Then the active fertile site on the conidiophore stalk moves sideways to produce a second conidium. Several rounds of conidiation ultimately generate multiple sympodial conidia on a mature conidiophore4,9. In spite of such a clear mycological characterization of conidiogenous cell development in M. oryzae, very little is currently known regarding mechanism modulating spore patterning and its contribution to pathogenicity except for two transcription factors, ACR1 and CCA19,10. ACR1 encodes a transcription factor homologous to MEDA of Aspergillus nidulans and REN1 of Fusarium oxysporum, both of which been characterized as developmental regulators for correct conidial differentiation11,12,13. Δacr1 mutants produce conidia in an acropetal array, impair in appressorium formation and are nonpathogenic9. CCA1 was identified from a systematical analysis of 104 fungal-specific Zn2Cys6 transcription factors in M. oryzae. Δcca1 mutants sporulate in a way similar to that in Δacr1 mutants and could not bring about foliar blast disease because of defects in conidial germination and appressorium formation10. It was proposed that diverse modes of conidiation in conidial fungi arose through alteration in major genes controlling spore-specific gene expression9.
Arrestins are adaptor and scaffold proteins that assemble molecules involved in signaling events such as signal transduction and intracellular protein trafficking, gathering associated proteins within spatial proximity and thus facilitating their proper interaction14,15. This adaptor/scaffold function was initially characterized in the mammalian visual rhodopsin and beta-adrenergic system16,17, and the proteins were termed as visual/beta-arrestins for their capacity to bind to the activated G-protein-coupled receptors, preclude ligand-receptor interaction and arrest subsequent signaling18. It is now known that beta-arrestins also function in modulating other cell surface proteins such as growth factor receptors and ion-transport proteins19,20. In addition to the plasma membrane receptors, accumulating data show that beta-arrestins interact with a burgeoning list of cytosolic targets, for instances, component of mitogen-activated protein kinase cascades and regulatory proteins such as the tumor suppressor p5315,21, thereby involved in regulating a wide variety of cellular processes22.
Beyond the well-studied mammalian visual/beta-arrestins, a much wider and ancient family of arrestin proteins has been recently discovered in organisms from protists to mammals except plants, and referred to as alpha-arrestins23,24. Alpha-arrestins and visual/beta-arrestins lack sequence similarity between each other, but share the same tertiary structure of N- and/or C-terminal arrestin-domain25,26, and possess analogous function in trafficking steps of membrane-embedded proteins. However, as the more ancient branch that emerged relatively recently24, alpha-arrestins may simply function in a wider diversity of signaling routes than do visual/beta-arrestins22.
Members of mammalian alpha-arrestins include arrestin-domain containing protein 1–5 (ARRDC1-5) and TXNIP (Thioredoxin-interacting protein)25. Because of their recent discovery, the detail function of ARRDCs is less known and increasing data show that ARRDCs play various roles in regulation of membrane proteins and prominently function in the mediation of metabolism22,27. Alpha-arrestins in Saccharomyces cerevisiae or arrestin-related trafficking adaptors (ARTs) bind to cell surface receptors in response to stresses such as pH and heavy metals, and function by distinct mechanisms to internalize plasma membrane cargoes via either clathrin-mediated or clathrin-independent endocytosis28,29,30,31,32. Additionally, ARTs can negatively regulate yeast pheromone response via internalization and desensitization of the G-protein-coupled receptors Ste233.
PalF in A. nidulans is one of the filamentous fungal alpha-arrestins whose role is best understood34,35,36,37,38,39, which constitutes a critical component of the conserved ambient pH-signaling pathway involved in fungal pathogenicity40,41,42,43. PalF is activated in a signal and receptor-dependent pattern similar to that in the mammalian beta-arrestins, and activated PalF binds the pH receptor PalH and recruits the component of ESCRT compartments to the cortical pH signaling sites, which stimulates further process that mediates fungal adaptation to ambient pH. Lately, a systematic investigation of alpha-arrestins in A. nidulans has been documented in respect to growth, morphology, drug sensitivity and specifically turnover of the uric acid-xanthine transporter UapA. This study, in combination with a previously reported alpha-arrestin CreD, puts the basis for understanding critical roles of alpha-arrestins in down-regulation of cell surface receptors in the filamentous fungi44,45. Except for that in A. nidulans, characterization of alpha-arrestins other than PalF is barely documented in the remaining filamentous fungi.
Macroautophagy (herein referred as autophagy) is a highly conserved self-degradative pathway that regulates numerous physiological and pathological processes in eukaryotic cells46. The morphological hallmark of autophagy is incorporation of intracellular proteins and organelles into a double-membrane sequestering compartment termed the phagophore. The phagophore subsequently expands and matures into an autophagosome which eventually transports to and fuses with vacuoles/lysosomes for cargo degradation and recycling47. A subset of autophagy-related genes (ATGs) has been discovered that gathers at the pre-autophagosomal structures or phagophore assembly site (PAS), and cooperates with the forming phagophore to generate a mature autophagosome48,49. Upon vesicle completion, most of these ATG proteins are dismissed except for Atg8. Some Atg8 stays association with the completed autophagosome and enters into the vacuolar lumen, making Atg8 an ideal marker that tracks the autophagy flux50.
Here we describe characterization of an alpha-arrestin required for correct differentiation of conidiogenous cells in the filamentous fungus M. oryzae. We show that Δarrdc1 mutants conidiate in an acropetal way and dramatically reduce in pathogenicity. Fluorescent analysis strongly suggests that ARRDC1 is recruited into nascent autophagosomes during their biogenesis before maturation. We propose that ARRDC1 acts as an adaptor protein that internalizes its cargoes such as conidiogenous receptors on fertile sites of the conidiophore stalk, and assembles autophagosomes for endocytosis of conidiogenous receptor, which transmits conidiogenous signals to the downstream transcription factors. The identification of ARRDC1 unveils alpha-arrestins as a novel class of genes involved in a mycological morphogenesis insufficient of documentation and our discovery extends significance of autophagy-associated alpha-arrestin signaling to microbial pathogenicity.
Results
ARRDC1 putatively encodes is an arrestinC-domain containing protein
Using a BlastP analysis with arrestinN (pfam00339) and/or arrestinC (pfam02752) against the M. oryzae genome, we have identified seven arrestin-domain containing proteins referred to as ARRDC1-6 and MoPalF. The domain organization within proteins is schematically represented in Figure S1.
Because fungal alpha-arrestins have only been investigated in the yeast and Aspergillus species, we compared the rice blast fungal alpha-arrestins with that in S. cerevisiae and A. nidulans, respectively. In comparison with a relatively lower similarity between homologues in M. oryzae and S. cerevisiae (Table S1), alpha-arrestins are relatively well conserved in the two filamentous fungi (Table S2).
Here, we chose to focus on ARRDC1 because deletion of ARRDC1 generates mutants with striking defects not so often documented in the filamentous fungus (see below).
Construction of Δarrdc1 mutants
To investigate the cellular function of ARRDC1, we performed one-step gene replacement strategy (Figure S2A). The gene disruption vector was introduced into rice pathogenic strain KJ201, and hygromycin-resistant transformants were selected from two independent fungal transformation performances. DNA gel blot analysis identified six gene replacement mutants, of which three are shown (Figure S2B). All six replacement mutants display the same defective phenotypes, and one null mutant was used for phenotypic analysis and complementation assay.
ARRDC1 deletion causes defects in conidiogenous cell development
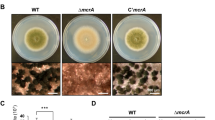
We investigated the role of ARRDC1 in colony growth and sporulation. We found that Δarrdc1 mutants grew slower than the wild-type KJ201 (Fig. 1A). Conidiation in Δarrdc1 mutants was dramatically reduced, and mutants typically produced 20-fold fewer spores compared with the wild-type (Fig. 1B). We then tested conidial germination by calculating germ tube emergence between the wild-type and mutant strains, and found that 97.50 ± 1.04% of the wild-type conidia germinated within 2 h of exposure to water, while only 73 ± 3.45% of mutant conidia retained the ability to germinate even after a prolonged inoculation of up to 48 h (Fig. 1C). And the wild-type typically produced uniform conidia with two septa that define three cells in a conidium, but conidia produced by Δarrdc1 mutants constantly enlarged and contained septa of variable numbers, as stained with Calcofluor White to visualize with cell walls and septa (Fig. 1D,E).
(A) Colony growth of M. oryzae strains on CM agar plates. The wild-type strain, Δarrdc1 and complementation strain arrdc-c were cultured at 25 °C for 10 days. The same capital letters on top of columns indicate non-significant differences as estimated by Duncan’s test (P ≤ 0.05). (B) Conidiation of M. oryzae strains. The strains were cultured at 25 °C for 10 days, and then conidiation recorded. The same capital letters on top of columns indicate non-significant differences as estimated by Duncan’s test (P ≤ 0.05). (C) The conidial germination rate of M. oryzae strains. Conidia were dropped on plastic coverslips and incubated at 25 °C for 48 h. The same capital letters on top of columns indicate non-significant differences as estimated by Duncan’s test (P ≤ 0.05). (D) The conidial morphology of the wild-type and Δarrdc1 mutants. Conidia from each strain were collected and stained with Calcofluor White. Bar = 20 μm. (E) Statistical analysis of conidia with numbers of septa. The Δarrdc1 mutants produce conidia with various numbers of septa. More than 300 conidia were counted for each strain. The same capital letters on top of columns indicate non-significant differences as estimated by Duncan’s test (P ≤ 0.05). (F) Conidiophore development of the wild-type and Δarrdc1 mutants. Δarrdc1 mutants gave less conidiophore and generated conidia in an acropetal way, and the wild-type produced conidiated in a sympodial pattern. Arrows indicate conidia produced in a head-to-tail way in the Δarrdc1 mutants. Bar = 20 μm. (G) Transcriptional expression patterns of conidiation-related transcription factors in the Δarrdc1 mutants. Relative expressions of CON7, COS1, COM1, ACR1 and CCA1 were measured in the Δarrdc1 mutants during conidiogenous cell development. Transcription levels were normalized to the endogenous gene beta-tubulin and relative values calculated by fold change (2−ΔΔCt) compared to each transcript in conidiogenous cell development of the wild-type. Total RNA was prepared from conidiogenous mycelia of the wild-type and Δarrdc1 mutants.
We then examined the process of sporulation in KJ201 and Δarrdc1 mutants. Aerial hyphae were erased with sterile distilled water, mycelia blocks cut and placed under constant illumination with high humidity for 24 h, and conidial development was observed. We found that the wild-type produced conidia in a sympodial pattern, in which multiple conidia emerge at several fertile sites on a single conidiophore stalk. A next round of conidial formation originates at the conidiophore stalk following termination of a previous round of conidiation. In contrast, conidiophore formation was reduced in the Δarrdc1 mutants and conidia were produced in an acropetal way. Although aerial conidiophore stalks undergo terminal swelling to produce the first conidium, a second conidium always shows up at the tip of the first one, and subsequently a third conidium emerges at the tip of the second conidium. Ultimately several conidia are linked in a chain and anchored on one fertile site of the conidiophore stalk (Fig. 1F). We conclude that the wild-type conidiates in a sympodial way and Δarrdc1 mutants produce conidia in an acropetal pattern.
Gene expression profile of CCA1 is affected in Δarrdc1 mutants
Many more pathogenicity-related transcription factors have been identified to be required for conidiation51, however, scarcity of investigation is involved in genes related to spore patterning except for ACR1 and CCA1. We thus investigated the expression profiles of ACR1 and CCA1 in Δarrdc1 mutants during conidiation. Other transcription factors we tested include CON7, COS1 and COM1. Absence of CON7 and COM1 results in abnormal conidial morphology52,53, and COS1 is required for conidiophore formation54. We found that CCA1 (0.16-fold) and COS1 (0.24-fold) were significantly down-regulated in the conidiogenous mycelia of Δarrdc1, as compared with the conidiogenous mycelia of the wild-type (Fig. 1G).
ARRDC1 is necessary for pathogenicity
To determine the role of ARRDC1 in pathogenesis, we inoculated two-week-old seedlings of a rice cultivar CO-39 with conidial suspensions of Δarrdc1 mutants and the wild-type. A representative result of four spraying assays is shown in Fig. 2A,B. While host of the wild-type displayed densely distributed leaf symptoms, dot lesions sparsely occurred on plants inoculated with Δarrdc1 mutants.
(A) Susceptible rice seedlings were sprayed with conidia suspensions (5 × 104/ml) of the wild-type, Δarrdc1 mutants and the complementation strains from 12-day-old CM plates. Photographs were taken at 5-day post-inoculation. (B) Data are presented as a bar chart showing percentage of lesion areas. The same capital letters on top of columns indicate non-significant differences as estimated by Duncan’s test (P ≤ 0.05). (C) Penetration peg formation and invasive growth were observed with intact and wounded barley leaves. 30 μl of conidial suspensions (5 × 104/ml) were inoculated on leaf explants. Photographs were recorded at 30 h after incubation in high humidity chamber. The triangle indicates invasive hyphae and the arrow indicates appressoria. Bar = 20 μm. (D) Statistical analysis of penetration peg formation. The same capital letters on top of columns indicate non-significant differences as estimated by Duncan’s test (P ≤ 0.05).
ARRDC1 is dispensable for appressorium formation and turgor generation
We performed a detailed examination of virulence-associated function to explore the reason why Δarrdc1 mutants dramatically reduce in pathogenicity. In vitro assays of appressorium formation showed that germ tubes in Δarrdc1 mutants formed appressoria similar to that of the wild-type (not shown). We then asked whether ARRDC1 is required for full turgor generation in the appressorium. Foliar blast infection in the rice blast fungus is a turgor-driven process associated with substantial glycerol accumulation in the appressoria55. We performed incipient cytorrhysis assays in which the rate of appressorium collapse is determined in the hyperosmotic concentration of glycerol56. We found no significant differences in appressorial turgor between Δarrdc1 mutants (76.00 ± 3.77% collapsed appressoria in the presence of 2 M glycerol) and the wild-type (77.72 ± 2.01% collapsed appressoria at 2 M glycerol).
ARRDC1 is required for penetration peg formation
Because barley is a natural host for M. oryzae, we used barley leaves to investigate penetration peg formation between Δarrdc1 mutants and the wild-type. Epidermal penetration assays were carried out to allow appressoria to form on both intact barley leaves and those in which the cuticles were removed firstly by abrasion. Formation of penetration peg was calculated 30 h after inoculation. We found that 14.60 ± 3.61% appressoria of null mutants produced penetration hyphae compared with 89.47 ± 2.01% in the wild-type appressoria. And a similar result was observed on abraded leaves (Fig. 2C,D). Thus, ARRDC1 is dispensable for appressorium formation and turgor generation, but required for penetration peg emergence.
Reintroduction of ARRDC1 restores colony growth, patterns of conidiation, penetration peg formation and pathogenicity
We performed a complementation assay to ask whether the phenotypes were attributable to ARRDC1 disruption. Complementation analysis was performed via introducing a 4.5-kb genomic fragment spanning the ARRDC1 gene into the null mutant. Transformants were selected, and the presence of a single copy of ARRDC1 was confirmed by DNA gel blot analysis (data not shown). Seedlings sprayed with conidia from transformants carrying the 4.5-kb fragment exhibited normal rice blast disease symptoms. And colony growth, sporulation, patterns of conidiation and penetration peg formation were all restored by complementation performance (Figs 1 and 2).
EGFP-ARRDC1 resides on different diameters of punctate sites
To elucidate the intracellular distribution of ARRDC1, we initially constructed four fusion proteins of EGFP tagged to both ends of ARRDC1 under control of its endogenous promoter and the H3 promoter to transform the Δarrdc1 mutants57. Both gene fusions were able to complement the defects of colony growth, spore patterning and pathogenicity in the Δarrdc1 mutants (data not shown). But sufficient signals could only be detected with gene fusions under the H3 promoter. Because EGFP tagged to both ends of ARRDC1 displays similar patterns of cellular distribution and EGFP-ARRDC1 gives more observable fluorescent signal, we thus used transformants expressing EGFP-ARRDC1 to perform confocal microscopy analysis.
In mycelia plugs collected from transformants on solid plates, we constantly captured multiple small dots of dim fluorescence distributed throughout the cytoplasm of the nascent conidia, and observed intensive punctate signals in the hyphae. We asked whether EGFP-ARRDC1 resides on different types of organelles or small dots undergo homotypic fusion to generate large-sized structures. We then inoculated conidia with CM media on microscope coverslips and carefully observed EGFP-ARRDC1 fluorescent signals during conidial development into vegetative hypha. In the dormant spore, EGFP-ARRDC1 was constantly captured to be distributed on several dim spots (Fig. 3A). After 8 h inoculation, fluorescent signal became strong and plenty of distinct punctate sites showed up in germinating conidia and germ tubes (Fig. 3B). Prolonging inoculation time up to 24 h, we found that EGFP-ARRDC1 fluorescence varied considerably in diameters and some dots became very large and were connected with smaller dots (Fig. 3C). We conclude that EGFP-ARRDC1 is distributed on multiple punctate vesicles of different diameters.
(A) In nascent conidia, multiple dim spots of EGFP-ARRDC1 among the entire cytoplasm were constantly captured. (B) EGFP-ARRDC1 signals become strong and plenty of distinct punctate sites were observed in the germinating conidium and its germ tube upon 8 h inoculation. (C) EGFP-ARRDC1 fluorescent dots vary considerably in diameters in the vegetative hypha developed from conidia prolonging inoculation time up to 24 h, and some dots become very large and are connected with smaller dots as indicated by the white triangles. Bar = 10 μm.
EGFP-ARRDC1 puncta are well colocalized with DsRed2-Atg8
The endocytic route is a major pathway responsible for recognition and transport of cargoes targeted to the vacuole and the Rab7-positive late endosome is a crucial intermediate during endocytosis in most cell types. In M. oryzae, Ramanujam, Naqvi and colleagues discovered that the late endosomal compartments, apart from membrane trafficking, regulate the biogenesis and spatio-temporal dynamics of cAMP signaling essential for pathogenesis through anchoring active G-protein signaling components58. Considering that alpha-arrestins possibly act as critical adaptors for turnover of cell surface proteins, we initially carried out colocalization assay between EGFP-ARRDC1 and MCHERRY-Rab7, but no overlapping was observed between the two fluorescent signaling (Figure S3).
Our previous study showed that multiple PAS sites in M. oryzae, indicated by colocalization of EGFP-Atg9 and DsRed2-Atg8, distributed on different diameters of puncta signals which constantly undergo homotypic fusion into larger structures59. Because the distribution pattern of EGFP-ARRDC1 bears a strong resemblance to the localization profile of PAS sites, we subsequently performed a colocalization analysis between EGFP-ARRDC1 and DsRed2-Atg8. We found that puncta of EGFP-ARRDC1 were well colocalized with DsRed2-Atg8, leaving other green signals distributing throughout the entire cytoplasm (Fig. 4) (79% of n = 106 ARRDC1 puncta colocalized with Atg8 dots).
Conidia coexpressed with EGFP-ARRDC1 and DsRed2-Atg8 were collected and inoculated on plastic coverslips with nutrient media allowing development of vegetative hypha. The white triangles indicate different diameters of colocalized sites between EGFP-ARRDC1 and DsRed2-Atg8 in the condia and vegetative hypha. Bar = 10 μm.
To further assess colocalization of EGFP-ARRDC1 with autophagic compartments. We generated Δatg9 mutants in the background of the wild-type stain KJ201 via targeted gene replacement using the deletion vector we previously constructed59. Δatg9 mutants in the background of KJ201 display similar defects as that in the background of another wild-type strain GUY11 as we previously reported (data not shown). We then co-expressed EGFP-ARRDC1 and DsRed2-Atg8 in Δatg9 mutants, and found that EGFP-ARRDC1 and DsRed2-Atg8 were still well colocalized with each other (Fig. 5) (75% of n = 100 ARRDC1 puncta colocalized with Atg8 dots). Because Δatg9 mutants are defective in autophagosome formation and introduction of ARRDC1 and/or Atg8 could not complement the autophagic defects in the Δatg9 mutants. We conclude that EGFP-ARRDC1 puncta is colocalized with the PAS structures and autophagosomes.
Discussion
In the current study, we have provided a novel documentation concerning alpha-arrestin of a fungal pathogen in virulence-associated cell differentiation, especially in conidiogenous cell development and penetration peg formation. As a first endeavor towards understanding alpha-arrestin pathway in the rice blast fungal pathogenicity, we have identified seven alpha-arrestins in the genome of M. oryzae. While deletion of arrestins such as ARRDC2 or ARRDC3 gives no obvious effect (not shown), possibly because of their functional redundancy, absence of ARRDC1 leads to pleiotropic defects underlining the indispensable significance of this alpha-arrestin. The most striking phenotype of Δarrdc1 mutants is its inability to establish a normal sympodial spore patterning and the mutants conidiate in an acropetal chain, i.e., a second conidium forms at the tip of a former one instead of a nearby fertile site on conidiophore stalks. Although germ tubes generate appressoria with normal turgor pressure, penetration peg development and invasive hypha growth are impaired and the mutants exhibit dramatically reduced disease symptoms.
Conidiogenous cell development is evolutionarily complex and characterization of genes concerning spore patterning is insufficiently documented and mainly concentrated on transcription factors1,9,12,13,60. In M. oryzae, only two transcription factors, CCA1 and ACR1, have been identified that are required for correct establishment of sympodial conidiation. And Δcca1 and Δacr1 mutants all produced conidia in an acropetal way similar to that observed in the Δarrdc1 mutants9,10. Because appressorium formation failed in Δcca1 and Δacr1 mutants, it was reasonably suggested that appressorium differentiation was impaired because of aberrant pattern of conidiogenous cell development9. In contrast, we found that although loss of ARRDC1 produced conidia with reduced germination rate, germ tubes normally gave appressoria comparable to the wild-type in respect of appressorium formation and turgor generation. We reason that CCA1 and ACR1 may function in multiple pathways including conidiation and appressorium formation, and our results imply that conidiogenous cell development and subsequent appressorium differentiation are not two intrinsically dependent happenings as suggested before.
Quite many phytopathogenic fungi, including M. oryzae, employ conidia as infectious propagules for disease initiation and dissemination. And conidia are produced in at least twenty patterns with mechanisms rarely known1. One way fertile hypha tips check status of vegetative growth and initiate conidiation is through remodeling plasma membrane proteins responsible for conidiogenous cell differenation. Because alpha-arrestins serve as critical regulators for turnover of cell surface proteins28,45,61, we suggest that upon favorable environmental cues, specific cell surface receptors transport to the fertile tip of conidiophore stalks to initiate conidiogenous cell development, and then recycle back to a second fertile site for next round of conidiation. Ultimately, M. oryzae produces several conidia directly linked to its center conidiophore stalk in a sympodial pattern, so that every nascent conidium is well nutritionally accommodated. Under natural conditions, ARRDC1 strictly modulates the conidiogenous receptor and prevents it from repeatedly catalyzing signaling activation of conidiation. In contrast, deletion of ARRDC1 disorganizes internalization and recycling of the conidiogenous receptor in the mutants, and the conidiogenous receptor is not promptly removed from fertile sites, prolonging signals energizing for conidiation. Thus, the next conidium generates from the tip of a former conidium, instead of another fertile tip in conidiophore stalks. We propose that M. oryzae employs alpha-arrestins like ARRDC1 to control specific conidiogenous receptors at fertile tips of conidiophore stalk and senses suitable stimulus to accurately initiate and terminate conidiation, possibly through controlling expression of downstream transcription factors such as CCA1, and the conidiogenous receptors have yet to be identified.
Autophagy is a strictly regulated process and autophagy activity is primarily modulated by number and size of autophagosomes. While the number of autophagosomes indicates the frequency of autophagosome formation, alteration in the diameters of autophagosomes allows sequestration of different quantities and dimensions of cargoes62. In the budding yeast cell, one PAS is regularly observed, in contrast with multiple PAS sites in the mammalian cell. Our previous study showed that in M. oryzae, multiple small Atg8-positive sites are easily detected in a single cell, and these structures constantly undergo homotypical fusion into large Atg8-positive structures59, resembling the homotypic fusion of two LC3-positive vesicles or Atg16L-positive autophagosome precursors in the mammalian cell63,64. Ultimately, these larger dimensions of Atg8-positive structures appear to merge into a center site, possible in concomitant with the rice blast fungal morphogenesis of small spherical vacuoles developing into a central vacuole. Here, we found that EGFP-ARRDC1 clearly colocalizes with DsRed2-Atg8 puncta and undergoes the same pattern of subcellular trafficking as that of Atg8-positive structures. And this pattern of colocalization is not altered in the Δatg9 mutants in which formation of autophagosomes is defective and DsRed2-Atg8 represents the PAS structures. We reason that ARRDC1 is not just transiently colocalized with autophagosomes, it enters into autophagic flux at the cargo recognition stage before maturation of phagophore into autophagosome.
Except for MVBs in the endocytic pathway, autophagosomes in the selective autophagy process emerge as alternative organelles responsible for recognition and transport of cargoes targeted to the vacuole65,66. These specific cargoes expand from entire organelles to signal molecules. For instances, the mammalian Notch signaling, a master regulator crucial for stem cell differentiation, is modulated by autophagy via uptake of the plasma membrane receptor Notch1 into ATG16L1-positive autophagosome precursor vesicles for degradation67. And by promoting degradation of Dishevelled, autophagy negatively regulates Wnt signaling which performs key functions in cell development, tissue self-renewal and tumorigenesis68. The Arabidopsis thalinana membrane-anchored TSPO is a multi-stress regulator that is transiently induced by abiotic stresses and its accumulation is strictly down-regulated through the autophagic pathway69,70. Here, we further propose that ARRDC1 acts as an adaptor/scaffolding protein that internalizes its cargoes like conidiogenous receptors on the fertile sites of the conidiophore stalk as we suggested above, and recruits autophagy-related vesicles for subsequent turnover of receptor proteins. Reciprocally, this autophagy-associated alpha-arrestin signaling route may provide a large pool of membrane materials and regulators on the plasma membrane for autophagosome formation without compromising other membrane trafficking events.
The identification of ARRDC1 unveils alpha-arrestins as an important catalog of genes involved in fungal parasites during their infection process. Further investigation of alpha-arrestins in the rice blast pathogen and other fungal parasites will auspiciously extend potential significance of alpha-arrestin mediated pathway to fungal pathogenicity and broaden insight into the molecular basis of microbial virulence.
Experimental Procedures
Fungal strains and growth conditions
M. oryzae strains were cultured on complete medium (CM) plates and preserved on filter paper disks as previously described71,72. KJ201 was used as the wild-type and all mutants mentioned in this study were generated from KJ201. CM plates were incubated at 25 °C under 12 h light/dark cycles.
Targeted gene replacement and fungal transformation
Construction of the replacement vector was performed using double-joint PCR as previously described73. Basically, two flanking sequences of the ARRDC1 locus were amplified from the genomic DNA of the M. oryzae wild-type KJ201 with primers 3241u1/u2 and 3241d1/d2, respectively. A 1.4 kb hygromycin B phosphotranferase gene (HPH) cassette was cloned from pCB1003 with primers Hphm1/m2. The three amplicons were linked together and served as the template for the final double-joint PCR amplified with nested primers 3241n1/n2. The double-joint PCR product was then inserted into the EcoRI/BamHI sites of pCAMBIA1300 to obtain the final gene replacement vector. The replacement vector was introduced into Agrobacterium tumefaciens strain AGL1 and Agrobacterium-tumefaciens mediated transformation (ATMT) was performed as previously described74. Resistant transformants were selected on CM plates supplemented with 250 μg/mL hygromycin B for gene replacement and 800 μg/mL G418 for complementation assay, respectively. The multiplex PCR with primers 3241ck1/ck2 and Hphck1/ck2 was used for the first round of mutant screening. The candidate mutants were purified from single conidium and southern blot analysis was performed to confirm the homology recombination and single integration event. For complementation assays, a 4.5-kb full-length copy of the ARRDC1 locus including its native promoter and terminator region was amplified from the genomic DNA using primers 3241c1/c2, and inserted into a modified pCAMBIA1300 vector containing a geneticin resistance gene. The resulting construct was randomly inserted into the genome of Δarrdc1 mutants via ATMT and southern blot analysis was carried out to verify successful single-copy integration. All primers used for gene replacement and complementation analysis are listed in Table S3.
Southern blot analysis
Genomic DNA was extracted by standard CTAB protocols. Agarose gel separation, restriction enzyme digestion and Southern hybridization analysis were performed following the standard procedure and the protocol provided within the digoxigenin (DIG) high prime DNA labelling and detection starter Kit I (Roche, Germany). For Southern blot, genomic DNA was digested with ScaI and labeled with the probe amplified using primers 3241sb1/sb2. The primers are listed in Table S3.
Phenotypic analysis
To visualize the conidiophore formation and pattern of conidiation, aerial hyphae from 5-day cultures growing on 90 mm CM plates were erased with sterile distilled water, and then mycelia blocks were cut and placed under constant illumination with 90% humidity for 24 h.
For sporulation test, strains were grown on three independent CM plates. After 10-day of growth, the spores were harvested and counted. To stain septa, appropriately diluted conidia (1 × 105/ml) collected from CM agar plates, were incubated on glass films in a moist chamber at 25 °C, samples were soaked with Calcofluor White in the dark for 2 min before epifluorescence microscopy examination.
Appressorial development assays were performed on hydrophobic microscope coverslips (Fisherbrand). Average values were determined from at least 200 spores and performed in triplicate. Appressorial turgor was estimated by incipient cytorrhysis assays55. Briefly, conidia of 20 μl droplets (5 × 105/ml) were incubated on microscope coverslips in a humid chamber for 24 h and induced to form appressoria. Water embracing appressoria was removed carefully and replaced with 20 μl of glycerol in concentration of 2 M. The number of appressoria that had collapsed after 5 min was recorded. Average values were determined from at least 200 spores and performed in triplicate.
Penetration peg formation was determined as previously described75. Simply, conidia were collected and resuspended to the concentration of 5 × 104/ml with 0.02% (wt/vol) Tween 20. A 20 μl droplet was deposited onto the upper side of leaves cut from 8-day-old barley (Hordeum vulgare cv. ZJ-8), and maintained on 4% (wt/vol) water agar plates at 25 °C for 30 h. The leaves that showed disease lesions were decolored by methanol, stained in lactophenol with 0.01% trypan blue-0.06% aniline blue, and observed under a microscope for host penetration. Images were taken using an Eclipse 80i microscope (Nikon).
Pathogenicity assays
Fourteen-day-old rice seedlings of the susceptible dwarf indica cultivar, CO-39, were infected with suspensions of M. oryzae conidia prepared in 0.02% tween 20, at a concentration of 5 × 104/ml, using Badger airbrush. Plants were incubated in plastic bags for 36 h to maintain high humidity and then transferred to controlled environmental chamber at 22 °C and 90% relative humidity for 72 h for full disease symptoms to develop. The infected leaves were collected at 5-day post-inoculation and images taken using an Epson Workforce scanner at a resolution of 2400 dpi.
Plasmid construction of pEGFP-ARRDC1, pMCHERRY-Rab7 and pDsRed2-Atg8
Fragment of ARRDC1 was amplified with primers arrdc1-xba1/2 from mycelia cDNA of the wild-type strain KJ201, and inserted into the XbaI site of pKD5GFP to generate the pEGFP-ARRDC1. The pMCherry-Rab7 was constructed via insertion of Rab7 cDNA amplified with rab7-xba1/2 into the Xba1 site of pKD3cmcherry, and the H3 promoter was replaced with the RP27 promoter amplified with primers rp27-1/2. Note that Rab7 expressed under the H3 promoter is mislocalized and signals diffusely distributed among the entire cytoplasm. All ligation reactions were performed with Infusion Kit from Clontech. The pKD5GFP and pKD3mcherry plasmids are kind gifts from Dr. Jianping Lu at College of Life Sciences, Zhejiang University57. pDsRed2-Atg8 was constructed according to our previous description59. Resistant transformants were selected on CM plates, supplemented with 800 μg/ml G418 for expression of pDsRed2-Atg8 and 500 μg/ml glufosinate ammonium for expression of pMCherry-Rab7, respectively. Transformants expressed pEGFP-ARRDC1 were selected on DCM (yeast nitrogen base without amino acid 1.7 g/l, NH4NO3 2 g/l, L-asparagine 1 g/l, glucose 10 g/l, and Na2HPO4 was used to adjust pH to 6.0) supplemented with 100 μg/mL chlorimuron-ethyl. The primers for fluorescent constructs are listed in Table S3.
Quantitative Real-Time PCR assay
RNA was isolated with the Trizol reagent (Invitrogen) following the manufacturer’s recommendation. A 2000 ng of total RNA was added to synthesize the first strand cDNA using SYBR ExScriptTM RT-PCR kit (Takara). Real-time PCR reaction was performed with SYBR Premix Ex Taq (Takara) on a C1000 thermal cycler (Bio-rad). Relative abundance of transcripts was calculated using the 2−ΔΔCt method with beta-tubulin (MGG00604) as the endogenous control. Thermocycler conditions were as follows: 3 min at 95 °C, followed by 45 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 15 s. A final dissociation cycle was incorporated to ensure the specificity of each primers. Data were collected from at least three independent experiments with three replicates, and one representative set of results was presented. Primers used for Real-Time PCR analysis are listed in Table S3.
Additional Information
How to cite this article: Dong, B. et al. Autophagy-associated alpha-arrestin signaling is required for conidiogenous cell development in Magnaporthe oryzae. Sci. Rep. 6, 30963; doi: 10.1038/srep30963 (2016).
Change history
30 September 2016
A correction has been published and is appended to both the HTML and PDF versions of this paper. The error has not been fixed in the paper.
References
Cole, G. T. Models of cell differentiation in conidial fungi. Microbiol Rev 50, 95–132 (1986).
Howard, R. J. & Valent, B. Breaking and entering: host penetration by the fungal rice blast pathogen Magnaporthe grisea . Annu Rev Microbiol 50, 491–512 (1996).
Couch, B. C. & Kohn, L. M. A multilocus gene genealogy concordant with host preference indicates segregation of a new species, Magnaporthe oryzae, from M. grisea . Mycologia 94, 683–693 (2002).
Ebbole, D. J. Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol 45, 437–456 (2007).
Dean, R. A. et al. The genome sequence of the rice blast fungus Magnaporthe grisea . Nature 434, 980–986 (2005).
Dagdas, Y. F. et al. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336, 1590–1595 (2012).
Kankanala, P., Czymmek, K. & Valent, B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724 (2007).
Wilson, R. A. & Talbot, N. J. Under pressure: investigating the biology of plant infection by Magnaporthe oryzae . Nat Rev Microbiol 7, 185–195 (2009).
Lau, G. W. & Hamer, J. E. Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genet Biol 24, 228–239 (1998).
Lu, J. P., Cao, H. J., Zhang, L. L., Huang, P. Y. & Lin, F. C. Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high-throughput gene knockout in the rice blast fungus. PLoS Pathog 10, e1004432 (2014).
Clutterbuck, A. J. A mutational analysis of conidial development in Aspergillus nidulans . Genetics 63, 317–327 (1969).
Busby, T. M., Miller, K. Y. & Miller, B. L. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics 143, 155–163 (1996).
Ohara, T., Inoue, I., Namiki, F., Kunoh, H. & Tsuge, T. REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum . Genetics 166, 113–124 (2004).
Lefkowitz, R. J., Rajagopal, K. & Whalen, E. J. New roles for beta-arrestins in cell signaling: not just for seven-transmembrane receptors. Mol Cell 24, 643–652 (2006).
Lefkowitz, R. J. Arrestins come of age: a personal historical perspective. Prog Mol Biol Transl Sci 118, 3–18 (2013).
Benovic, J. L. et al. Functional desensitization of the isolated beta-adrenergic receptor by the beta-adrenergic receptor kinase: potential role of an analog of the retinal protein arrestin (48-kDa protein). Proc Natl Acad Sci USA 84, 8879–8882 (1987).
Kuhn, H. & Wilden, U. Deactivation of photoactivated rhodopsin by rhodopsin-kinase and arrestin. J Recept Res 7, 283–298 (1987).
DeWire, S. M., Ahn, S., Lefkowitz, R. J. & Shenoy, S. K. β-Arrestins and cell signaling. Annual Review of Physiology 69, 483–510 (2007).
Lin, F. T., Daaka, Y. & Lefkowitz, R. J. Beta-arrestins regulate mitogenic signaling and clathrin-mediated endocytosis of the insulin-like growth factor I receptor. J Biol Chem 273, 31640–31643 (1998).
Szabo, E. Z., Numata, M., Lukashova, V., Iannuzzi, P. & Orlowski, J. Beta-Arrestins bind and decrease cell-surface abundance of the Na+/H+ exchanger NHE5 isoform. Proc Natl Acad Sci USA 102, 2790–2795 (2005).
Gurevich, E. V. & Gurevich, V. V. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol 7, 236 (2006).
Patwari, P. & Lee, R. T. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab 23, 216–222 (2012).
Kommaddi, R. P. & Shenoy, S. K. Arrestins and protein ubiquitination. Prog Mol Biol Transl Sci 118, 175–204 (2013).
Alvarez, C. E. On the origins of arrestin and rhodopsin. BMC Evol Biol 8, 222 (2008).
Aubry, L., Guetta, D. & Klein, G. The arrestin fold: variations on a theme. Curr Genomics 10, 133–142 (2009).
Aubry, L. & Klein, G. True arrestins and arrestin-fold proteins: a structure-based appraisal. Prog Mol Biol Transl Sci 118, 21–56 (2013).
Shea, F. F., Rowell, J. L., Li, Y., Chang, T. H. & Alvarez, C. E. Mammalian alpha arrestins link activated seven transmembrane receptors to Nedd4 family E3 ubiquitin ligases and interact with beta arrestins. PLoS One 7, e50557 (2012).
Lin, C. H., MacGurn, J. A., Chu, T., Stefan, C. J. & Emr, S. D. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 (2008).
Nikko, E., Sullivan, J. A. & Pelham, H. R. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep 9, 1216–1221 (2008).
Nikko, E. & Pelham, H. R. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 (2009).
O’Donnell, A. F., Apffel, A. & Gardner, R. G. Cyert MS. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol Biol Cell 21, 3552–3566 (2010).
Herrador, A., Herranz, S., Lara, D. & Vincent, O. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol Cell Biol 30, 897–907 (2010).
Alvaro, C. G. et al. Specific alpha-arrestins negatively regulate Saccharomyces cerevisiae pheromone response by down-modulating the G-protein-coupled receptor Ste2. Mol Cell Biol 34, 2660–2681 (2014).
Herranz, S. et al. Arrestin-related proteins mediate pH signaling in fungi. Proc Natl Acad Sci USA 102, 12141–12146 (2005).
Galindo, A., Calcagno-Pizarelli, A. M., Arst, H. N., Jr. & Penalva, M. A. An ordered pathway for the assembly of fungal ESCRT-containing ambient pH signalling complexes at the plasma membrane. J Cell Sci 125, 1784–1795 (2012).
Hervas-Aguilar, A., Galindo, A. & Penalva, M. A. Receptor-independent ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J Biol Chem 285, 18095–18102 (2010).
Lucena-Agell, D., Galindo, A. & Arst, H. N., Jr. Penalva MA. Aspergillus nidulans ambient pH signaling does not require endocytosis. Eukaryot Cell 14, 545–553 (2015).
Penalva, M. A., Lucena-Agell, D. & Arst, H. N., Jr. Liaison alcaline: Pals entice non-endosomal ESCRTs to the plasma membrane for pH signaling. Curr Opin Microbiol 22, 49–59 (2014).
Bussink, H. J. et al. Refining the pH response in Aspergillus nidulans: a modulatory triad involving PacX, a novel zinc binuclear cluster protein. Mol Microbiol 98, 1051–1072 (2015).
Landraud, P., Chuzeville, S., Billon-Grande, G., Poussereau, N. & Bruel, C. Adaptation to pH and role of PacC in the rice blast fungus Magnaporthe oryzae . PLoS One 8, e69236 (2013).
Ost, K. S., O’Meara, T. R., Huda, N., Esher, S. K. & Alspaugh, J. A. The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator. PLoS Genet 11, e1005159 (2015).
Cornet, M. & Gaillardin, C. pH signaling in human fungal pathogens: a new target for antifungal strategies. Eukaryot Cell 13, 342–352 (2014).
Penalva, M. A., Tilburn, J., Bignell, E. & Arst, H. N., Jr. Ambient pH gene regulation in fungi: making connections. Trends Microbiol 16, 291–300 (2008).
Boase, N. A. & Kelly, J. M. A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol Microbiol 53, 929–940 (2004).
Karachaliou, M. et al. The arrestin-like protein ArtA is essential for ubiquitination and endocytosis of the UapA transporter in response to both broad-range and specific signals. Mol Microbiol 88, 301–317 (2013).
Reggiori, F. & Klionsky, D. J. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361 (2013).
Reggiori, F. & Klionsky, D. J. Autophagosomes: biogenesis from scratch? Curr Opin Cell Biol 17, 415–422 (2005).
Suzuki, K. et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20, 5971–5981 (2001).
Kim, J., Huang, W. P., Stromhaug, P. E. & Klionsky, D. J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem 277, 763–773 (2002).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016).
Chung, H., Choi, J., Park, S. Y., Jeon, J. & Lee, Y. H. Two conidiation-related Zn(II)2Cys6 transcription factor genes in the rice blast fungus. Fungal Genet Biol 61, 133–141 (2013).
Odenbach, D. et al. The transcription factor Con7p is a central regulator of infection-related morphogenesis in the rice blast fungus Magnaporthe grisea . Mol Microbiol 64, 293–307 (2007).
Yang, J. et al. A novel protein Com1 is required for normal conidium morphology and full virulence in Magnaporthe oryzae . Mol Plant Microbe Interact 23, 112–123 (2010).
Zhou, Z., Li, G., Lin, C. & He, C. Conidiophore stalk-less1 encodes a putative zinc-finger protein involved in the early stage of conidiation and mycelial infection in Magnaporthe oryzae . Mol Plant Microbe Interact 22, 402–410 (2009).
Howard, R. J., Ferrari, M. A., Roach, D. H. & Money, N. P. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc Natl Acad Sci USA 88, 11281–11284 (1991).
Bhambra, G. K., Wang, Z. Y., Soanes, D. M., Wakley, G. E. & Talbot, N. J. Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea . Mol Microbiol 61, 46–60 (2006).
Li, H. J., Lu, J. P., Liu, X. H., Zhang, L. L. & Lin, F. C. Vectors building and usage for gene knockout, protein expression and fluorescent fusion protein in the rice blast fungus. J Agri Biotechnol 20, 94–104 (2012).
Ramanujam, R., Calvert, M. E., Selvaraj, P. & Naqvi, N. I. The late endosomal HOPS complex anchors active G-protein signaling essential for pathogenesis in magnaporthe oryzae . PLoS Pathog 9, e1003527 (2013).
Dong, B. et al. MgAtg9 trafficking in Magnaporthe oryzae . Autophagy 5, 946–953 (2009).
Chung, D. W. et al. acon-3, the Neurospora crassa ortholog of the developmental modifier, medA, complements the conidiation defect of the Aspergillus nidulans mutant. Fungal Genet Biol 48, 370–376 (2011).
Polo, S. & Di Fiore, P. P. Finding the right partner: science or ART? Cell 135, 590–592 (2008).
Jin, M. & Klionsky, D. J. Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett 588, 2457–2463 (2014).
Jahreiss, L., Menzies, F. M. & Rubinsztein, D. C. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 9, 574–587 (2008).
Moreau, K., Ravikumar, B., Renna, M., Puri, C. & Rubinsztein, D. C. Autophagosome precursor maturation requires homotypic fusion. Cell 146, 303–317 (2011).
Fader, C. M. & Colombo, M. I. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ 16, 70–78 (2009).
Zhuang, X., Cui, Y., Gao, C. & Jiang, L. Endocytic and autophagic pathways crosstalk in plants. Curr Opin Plant Biol 28, 39–47 (2015).
Wu, X. et al. Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat Commun 7, 10533 (2016).
Gao, C. et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol 12, 781–790 (2010).
Vanhee, C., Zapotoczny, G., Masquelier, D., Ghislain, M. & Batoko, H. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805 (2011).
Hachez, C. et al. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 26, 4974–4990 (2014).
Parker, D. et al. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat Protoc 3, 435–445 (2008).
Talbot, N. J., Ebbole, D. J. & Hamer, J. E. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell 5, 1575–1590 (1993).
Yu, J. H. et al. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41, 973–981 (2004).
Rho, H. S., Kang, S. & Lee, Y. H. Agrobacterium tumefaciens-mediated transformation of the plant pathogenic fungus, Magnaporthe grisea . Mol Cells 12, 407–411 (2001).
Liu, X. H. et al. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell 6, 997–1005 (2007).
Acknowledgements
We thank Dr. Jianping Lu at College of Life Sciences, Zhejiang University, for critical reading of this manuscript and providing the pKD5GFP and pKD3mcherry plasmids. We thank Dr. Yong-Hwan Lee at Seoul National University for providing the wild-type KJ201 strain. We would also like to thank Li Xe and Xijiao Song (Zhejiang Academy of Agricultural Sciences, Hangzhou, China) for technical assistance on the confocal microscopic analysis. This study was supported by the Major Projects Transgenic Program of the Ministry of Agriculture (Grant No: 2012ZX08009001), the Natural Science Foundation of China (Grant No: 31300129), 2013 Creative Projects of Zhejiang Academy of Agricultural Sciences, and State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (2010DS700124- KF1406/ZZ1607 and 2015-cxzt-20).
Author information
Authors and Affiliations
Contributions
B.D. wrote the manuscript text; B.D. prepared all figures and tables; B.D., X.X., G.C., D.Z., M.T. and F.X. conducted the experiments; B.D., X.X., G.C., D.Z., M.T., F.X., X.L., H.W. and B.Z. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Dong, B., Xu, X., Chen, G. et al. Autophagy-associated alpha-arrestin signaling is required for conidiogenous cell development in Magnaporthe oryzae. Sci Rep 6, 30963 (2016). https://doi.org/10.1038/srep30963
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep30963
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.