Abstract
Although plastid genomes of flowering plants are typically highly conserved regarding their size, gene content and order, there are some exceptions. Ericaceae, a large and diverse family of flowering plants, warrants special attention within the context of plastid genome evolution because it includes both non-photosynthetic and photosynthetic species with rearranged plastomes and putative losses of “essential” genes. We characterized plastid genomes of three species of Ericaceae, non-photosynthetic Monotropa uniflora and Hypopitys monotropa and photosynthetic Pyrola rotundifolia, using high-throughput sequencing. As expected for non-photosynthetic plants, M. uniflora and H. monotropa have small plastid genomes (46 kb and 35 kb, respectively) lacking genes related to photosynthesis, whereas P. rotundifolia has a larger genome (169 kb) with a gene set similar to other photosynthetic plants. The examined genomes contain an unusually high number of repeats and translocations. Comparative analysis of the expanded set of Ericaceae plastomes suggests that the genes clpP and accD that are present in the plastid genomes of almost all plants have not been lost in this family (as was previously thought) but rather persist in these genomes in unusual forms. Also we found a new gene in P. rotundifolia that emerged as a result of duplication of rps4 gene.
Similar content being viewed by others
Introduction
Ericaceae is a large and diverse family of eudicots comprising approximately 120 genera and is divided into seven subfamilies1. Many species are economically important (crop and ornamental) plants. Most members of Ericaceae form associations with mycorrhizal fungi, on which they are dependent for such processes as nitrogen and phosphorus acquisition2. This dependence on fungi is most notable in species from the subfamily Monotropoidae, the members of which are partially or completely mycoheterotrophic, obtaining carbon through fungal interactions. Recent characterization of the plastid genomes of the photosynthetic Ericaceae Vaccinium macrocarpon and Arbutus unedo3,4 revealed many unique shared traits, such as a high repeat content, a drastic reduction in small single-copy regions, the putative pseudogenization of several essential genes (accD, clpP, rps16, ycf1, ycf2) and multiple rearrangements.
Here, we characterize the plastid genomes of two non-photosynthetic Ericaceae, Monotropa uniflora and Hypopitys monotropa, as well as that of their photosynthetic relative Pyrola rotundifolia. These are the first plastid genomes from mycoheterotrophic dicots reported to date and the findings allow comparison with both parasitic dicots and mycoheterotrophic monocots. Among monocots, the most notable examples of mycoheterotrophy are orchids; indeed, virtually all members of this large and diverse family are dependent on fungi during germination and some continue to gain carbon from fungi during their entire life cycle. It has been noted that mycoheterotrophic Ericaceae and Orchidaceae are very similar concerning their ecology5. The plastid genomes of a number of non-photosynthetic orchid species have been characterized and the results have shown a wide spectrum of structures, from the early stages of degradation in Corallorhiza6 to the highly reduced genome in Epipogium7. Characterization of the M. uniflora and H. monotropa plastid genomes will reveal whether such parallelism exists at the molecular level with regard to the structure and mode of reduction of the plastome. Recently, DNA hybridization was applied to survey the gene content of plastid genomes in mycoheterotrophic Ericaceae (including M. uniflora and H. monotropa)8, with the results suggesting that photosynthesis-related genes have been lost from both species. However, DNA hybridization is not an exhaustive technique and can give both false positives (due to signals from nuclear or mitochondrial pseudogenes) and false negatives (due to the high divergence of plastid gene sequences).
A usually implicit assumption regarding the plastid genomes of non-photosynthetic plants is that their structure reflects the absence of selection acting on photosynthetic genes (thus leading to their loss or pseudogenization) and the retention of housekeeping genes. In this case, all non-photosynthetic plant plastomes should have a similar structure: highly reduced, with few genes responsible for housekeeping functions being retained. Indeed, a study of the highly reduced plastome of the orchid Rhizanthella gardneri revealed a striking similarity in gene content with the plastid genomes of unrelated parasitic plants9. However, with the increased availability of information on plastid genome sequences, it has become evident that the situation is more complex. Barrett and Davis10 proposed a model of gradual gene loss, in which photosynthesis-related genes are lost first and ribosomal RNA, ribosomal proteins, transfer RNAs and several housekeeping genes probably are among the last to be lost. This model is based primarily on orchid studies and recent information on two highly reduced orchid genomes of Epipogium species7 appears to support it. Regardless, independent testing using data from unrelated taxa of mycoheterotrophic plants is required. The endpoint of plastid genome reduction in non-photosynthetic plants could be the complete loss of the plastome11. However, such cases seem to be extremely rare, only one is known in higher plants12. Retention of plastid genomes in photosynthetic plants is explained by a hypothesis which postulates that colocation of genes and their products in mitochondria and chloroplasts is beneficial for the redox regulation of gene expression (CoRR hypothesis, reviewed in ref. 13). However, this does not readily explain genome retention in non-photosynthetic plants; other hypotheses were put forward (reviewed in ref. 14). In order to assess their support by the data, more extensive sampling of non-photosynthetic plants is required.
To sum up, our study has two aims: first, to provide a more complete survey of gene content and plastome structure in Ericaceae, a group previously shown to have very unusual plastomes3,4 and second, focused on mycoheterotrophic Ericaceae, to complement the current knowledge on non-photosynthetic plant plastomes.
Results and Discussion
Structure and gene content of the Pyrola rotundifolia plastome
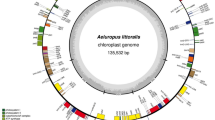
The plastid genome of P. rotundifolia is a circular molecule 168,995 bp in length. It exhibits a typical quadripartite structure: large and small single-copy (LSC and SSC) regions of 109,174 and 11,945 bp, respectively and two parts of an inverted repeat (IR) of 23,937 bp each. The size of the small single-copy region is in contrast to what is observed in other Ericaceae, in which the small single-copy region is highly reduced, at ~3 kb (Fig. 1, Table 1). Additionally, the P. rotundifolia plastome is longer than that of most flowering plants due to an increased proportion of non-coding DNA represented by repeats and pseudogenes. The same is characteristic for V. macrocarpon3 but not for A. unedo, the plastome of which is ~150 kb4. The P. rotundifolia plastome contains 113 genes (considering duplicated genes as one gene), 20 of which are located in the IR. All of the photosynthesis-related genes usually found in the plastomes of photosynthetic angiosperms (encoding components of photosystems I and II, cytochrome b6/f complex, rbcL, ATP synthase complex) are present. There are, however, some deviations from a typical gene set: ycf1 and ycf2 are pseudogenes (both are present in the plastome as a small fragment, ~20% of intact gene length) and a novel gene combining the rps4 N-terminal domain and transmembrane domains has evolved (see below). Three genes encoding components of the NADH dehydrogenase complex (ndhD, ndhA, ndhF) are represented by obvious pseudogenes (i.e., they are truncated and harbour frameshift mutations) and others (ndhB, ndhC, ndhE, ndhH, ndhI, ndhJ) contain an in-frame stop codon. This may indicate the early stages of pseudogenization or RNA editing; given that other genes of this group are undoubtedly pseudogenes, the second possibility is less probable. In other Ericaceae studied to date, these genes are either intact – as in A. unedo4, or only few (ndhG, ndhI and ndhK) are pseudogenes - as in V. macrocarpon3. The NDH complex is involved in chlororespiration and cyclic electron transport in photosystem I (reviewed in ref. 15) and is hypothesized to balance reactive oxygen species (ROS) levels to alleviate ROS-induced stress16. NDH genes are absent from non-streptophyte lineages of Archaeplastida, while present in most Streptophyta; together with their function this suggests that their retention in the plastome was the key adaptation to terrestrial way of life17. However, secondary loss of ndh genes from the plastome is known from several plant lineages (e. g., Geraniaceae, some gymnosperms, many orchids and some other monocot lineages). A phylogeny-wide transcriptome survey shows that this is a complete loss of the ndh gene complex, rather than the replacement of plastid genes by nuclear and/or mitochondrial genes18. Tobacco plants with knocked-out ndh genes are viable under optimal growth conditions but susceptible to water stress19,20 and mutations in ndhF were found to affect photosynthesis efficiency under changing light intensities21. These results suggest that the NDH complex plays a role in plant-environment interaction and that its dispensability depends on ecological factors (discussed, e.g. in ref. 22). Pseudogenization of ycf2 and ycf1, mentioned above, is a very intriguing case. Both genes were found to be essential in tobacco23, but are absent in grasses (e.g. in ref. 24). Function of ycf2 remains unknown; ycf1 was recently identified as a component of the translocon on the inner envelope of chloroplasts25. This function was postulated based on experiments on Arabidopsis thaliana. The question of how universal the mechanism of protein translocation involving ycf1 is disputable26,27. de Vries et al.26 note that the loss of ycf1 usually co-occurs with the loss of accD. Ericaceae seemed to support this observation. However, we argue that accD is intact at least in several Ericaceae (see below). As for ycf1, ycf1-like sequence in the P. rotundifolia plastome contains a truncated ORF (~200 amino acids) and is thus unlikely to be functional. The same is true for two other photosynthetic Ericaceae – V. macrocarpon and A. unedo.
Plastid genome maps of Pyrola rotundifolia, Monotropa uniflora and Hypopitys monotropa.
Genes shown inside the circle are transcribed clockwise; those outside the circle are transcribed counterclockwise. Blue and orange lines indicate direct and inverted repeats, respectively. The filled orange area indicates the inverted repeat region.
Structure and gene content of the Monotropa uniflora and Hypopitys monotropa plastomes
In both species, the plastid genome is highly reduced, at ~46 kb in M. uniflora and 35 kb in H. monotropa; the difference in length is because the latter lacks the IR, whereas the former retains an IR of ~9.5 kb (Fig. 1, Table 1). As in A. unedo and V. macrocarpon, M. uniflora has a very short small single-copy region of ~800 bp which encodes a single gene, trnF-GAA. The GC-content is 34.4% in H. monotropa and 29.4% in M. uniflora. The latter is much less than observed in other plants and is due to the presence of numerous AT-rich repeats (see below) and to an AT-bias in protein-coding sequences. As expected, the gene set is highly reduced in both species, with all genes encoding components of the photosynthetic apparatus having been lost or pseudogenized (Table 2). Genes encoding a bacterial type RNA polymerase (PEP – plastid encoded polymerase) subunits were also lost; the same is observed in most non-photosynthetic plants except for the liverwort Aneura mirabilis28. Although in model plants PEP is active across the whole organism, including non-green parts, its main function is the transcription of photosynthesis-related genes, while housekeeping genes are transcribed by another, nuclear encoded polymerase (NEP)29. Thus PEP function is dispensable in non-photosynthetic plants (see, e.g. ref. 30). Notably, ATP synthase genes are also lost in both species (in H. monotropa, atpB is present as pseudogene) while in many other non-photosynthetic plants they are retained6,31,32,33. Recently Kamikawa et al.34 proposed that the ATP synthase complex is necessary for the function of TAT (twin-arginine translocator) system which translocates proteins into thylakoids. The characterization of H. monotropa and M. uniflora transcriptome will show whether this system is active in these plants. In view of Kamikawa et al.34 hypothesis, we expect that it is not. The only class of genes not influenced by the reduction is ribosomal RNA genes; most ribosomal protein genes are also intact. Despite the longer length of the M. uniflora plastome, it contains fewer genes than H. monotropa. Of nine genes encoding ribosomal proteins for the large subunit, which are generally present in plastid genomes (reviewed in ref. 35), two (rpl32 and rpl23) have been lost from M. uniflora; in contrast, H. monotropa encodes a complete set. In both species, the status of rpl20 is unclear; compared to A. unedo it has a premature stop codon which results in ~70 bp shorter ORF. In V. macrocarpon, rpl20 ORF is present (though not annotated) and is 42 nt shorter than in A. unedo. Regarding small subunit ribosomal RNA proteins, two (rps16 and rps15) have been lost from both species. rps16 is known to be lost from the plastome and functionally replaced by nucleus-encoded copy in several photosynthetic plants36 and rps15 is non-essential, according to knock-out studies35. In addition, the number of transfer RNA genes is reduced compared to photosynthetic plants. Notably, all intron-containing tRNA genes are absent in both species, though intron-containing protein-coding genes (rpl2, rpl16) are intact. Despite a larger size, M. uniflora plastome has a more reduced set of tRNA genes: in addition to the above-mentioned genes trnG-GCC, trnL-CAA, trnL-UAG, trnR-ACG, trnR-UCU, trnS-GGA and trnT-UGU are also absent and trnV-GAC and trnM-CAU are pseudogenes (they maintain conserved tRNA secondary structure but exhibit several nucleotide substitutions, including in the anticodon). H. monotropa possesses intact trnL-UAG, trnM-CAU, trnR-ACG and trnR-UCU genes. Remarkably, in both species trnE-UUC – the gene that is hypothesized to be the main reason for plastome conservation because of its necessity for heme biosynthesis14 - is intact. matK is present but trnK intron and exons are lost; infA is intact. ycf1 and ycf2 are absent in both species. accD and clpP genes persist but are highly diverged (see details below).
Both species display a gene order that deviates from the typical and from those of A. unedo and V. macrocarpon. The most surprising finding is that the ribosomal RNA operon, which is highly conserved in its structure and position and is located in the inverted repeat region in most plants37, is divided into two parts. The organization of rRNA genes into a single operon facilitates their coordinated expression, which is important for plastid ribosome biogenesis. However, in M. uniflora, rrn16 is located in the IR, together with a few other genes (rps4, rps14, trnS-UGA, and accD) and other rRNA genes are located in the LSC region. rrn16 is also located distantly from other rRNA genes in H. monotropa. Experimental data show that the rRNA operon can be transcribed by both nucleus- and plastid-encoded RNA polymerases (reviewed in ref. 37), which are presumably differentially utilized during plant development (e.g. in ref. 38). Although the operon is structurally stable, its promoter region is labile; for example, parasitic plants from the genus Cuscuta lacking the PEP enzyme also have lost PEP promoter motifs30. As both rrn16 and rrn23 are expressed in H. monotropa (unpublished data), we suggest that the rrn23-rrn4.5-rrn5 gene cluster has acquired a new NEP promoter. The largest block that is colinear with the plastomes of other plants is that containing most ribosomal protein genes: the S10 operon.
As mentioned above, the gene contents of M. uniflora and H. monotropa were previously assessed using DNA hybridization8. Although the results are largely consistent, there are a few exceptions, e.g., hybridization indicated the presence of several ndh and RNA polymerase genes, which we demonstrated to be absent from the plastome. Presumably, this discrepancy can be explained by the presence of plastid-derived sequences in the mitochondrial or nuclear genome, which may generate positive signals in hybridization experiments.
When this article was in review, a note describing the plastid genome of Hypopitys monotropa was published39. It is mostly congruent with our findings on H. monotropa plastome in terms of length and gene content (with the exception of two genes – accD and clpP - that will be discussed in detail below) but the gene order is different. Also, the sequences are highly divergent (10% for protein coding genes at amino acid level) indicating intraspecific polymorphisms or the existence of cryptic species.
Repetitive sequences in Ericaceae plastomes
We explored the repeat contents of five Ericaceae plastomes and found a large increase of repetitive DNA fraction in all species, regardless of their photosynthetic capacity. The results for V. macrocarpon and A. unedo are consistent with previous reports3,4. The highest fraction of repeats was found in P. rotundifolia (Supplementary Table 1) and V. macrocarpon. The V. macrocarpon and P. rotundifolia repeats differ in sequence, are located at different sites and are not homologous, emphasizing the tendency of Ericaceae to accumulate repeats. In P. rotundifolia, the longest repeats originate from a series of tandem duplications in the accD gene (see discussion below) and the plastome region containing the accD gene in M. uniflora also harbours multiple repeats (Supplementary Fig. 1). The entire region is extremely AT-rich and of low DNA complexity. Similarly, abundant AT-rich repeats were recently reported in a phylogenetically unrelated plant, the orchid Cypripedium japonicum40. These sites have proven to be informative markers for studies of population genetics in this species and M. uniflora repeats are potentially useful for this purpose too.
GC content and codon usage in Ericaceae plastomes: contrasts between two non-photosynthetic species
Previous studies of the plastomes of non-photosynthetic plants suggest that they exhibit the following tendencies with regard to the molecular evolution of their protein-coding genes: high AT-content, exhibiting in the most extreme cases codon and amino acid usage bias7,9,20; increased rates of nucleotide substitutions without relaxation of selection in housekeeping genes7,21; and neutral evolution of photosynthesis-related genes.6,7,9 Furthermore, recent studies of highly reduced plastomes7,41,42,43 suggest that the degree of manifestation of these tendencies correlates with the degree of plastome reduction. Thus, we expected to observe these features in H. monotropa and M. uniflora, which have highly reduced plastomes. However, our comparative analysis revealed strong evidence for such patterns only in M. uniflora; in H. monotropa, the patterns are more similar to those observed in photosynthetic Ericaceae. The total GC content of photosynthetic Ericaceae members ranges between 36 and 38%, which is typical for most plants. In H. monotropa it is only slightly lower and notably lower in M. uniflora (Table 3). In general, the most GC-rich region of the plastome is the IR. The M. uniflora IR shows the lowest GC content due to the presence of a highly AT-rich repetitive region in accD and the unusual localization of rrn23 outside the IR. With regard to codon usage, we found no apparent alterations in H. monotropa compared to photosynthetic Ericaceae, though M. uniflora seems to be biased toward AT-rich codons (Supplementary Table 2). Rates of substitution accumulation in shared protein-coding genes differ significantly between all pairs of the studied Ericaceae (p-value < 10−3 by Tajima’s relative rate test with Bonferroni correction). The rates of substitutions in non-synonymous and synonymous positions change proportionally, so the dN/dS ratio is similar in all branches, except for two very short branches, where it is hard to estimate (Fig. 2). The rate of substitution accumulation is highest in a lineage of M. uniflora. In a lineage of H. monotropa it is closer to the values in photosynthetic lineages (Fig. 2, Supplementary Table 3). As mentioned above, the M. uniflora plastome exhibits an unusually high number of low-complexity AT-rich repeats and as in C. japonicum40, these abundant AT-rich repeats might serve as markers for population genetic analyses in Monotropa.
Phylogenetic tree of Ericaceae reconstructed based on plastid rDNA, plastid protein-coding and nuclear rDNA sequences.
Branch lengths correspond to dN, dS and dN/dS values. The high dN/dS (3.83) value for Pyrola/Vaccinium branch is presumably an artefact caused by an insufficient number of synonymous substitutions.
Pseudogenes in Ericaceae plastomes
Pseudogenes are quite rare in plant plastomes; a few notable exceptions are most non-photosynthetic plants and several green plants (for review see ref. 44). Among the latter, the most prominent and well-studied are Geraniaceae; these plants contain highly rearranged plastomes, in which many regions are duplicated and additional copies of genes arise from these duplications45,46. Similar events have occurred in Trachelium (Campanulaceae)47. In both cases, the genome duplications are correlated with a high repeat content and an increased substitution rate.
Similar to the abovementioned plants, species of Ericaceae, photosynthetic or not, possess an unusually large number of pseudogenes (Table 2). These pseudogenes can be divided into three categories: (1) pseudogenes of photosynthesis-related genes in non-photosynthetic Ericaceae; (2) presumably non-functional copies of intact genes; and (3) pseudogenes of genes that are dispensable or became dispensable due to the presence of a functional copy in the nuclear genome. In contrast to non-photosynthetic plants in the early stages of plastome reduction, which retain pseudogenes for most of photosynthesis-related genes (e.g. refs 28,32) pseudogenes of the first category are present only in H. monotropa (atpB, psaB, ccsA) but have been completely lost from M. uniflora. Pseudogenes resulting from the degradation of duplicated genes are numerous in P. rotundifolia and V. macrocarpon (psaA, rps18, rps3 in V. macrocarpon3 and rrn16, rpl2, petD and accD in P. rotundifolia) while in A. unedo the only example is ndhA4. Notably, P. rotundifolia and V. macrocarpon have the highest repeat content among Ericaceae (Supplementary Table 1). Repeats are known to favour structural changes, including duplications, by the mechanism of illegitimate recombination48. We hypothesize that this mechanism gave rise to the above-mentioned pseudogenes. The third category includes ycf1, ycf2 and several ndh genes. However, the reason for the pseudogenization of genes in this category (either dispensability or functional replacement) is less clear because it should ideally be inferred from genome-wide data on the presence of genes in the nuclear genome and on functional genetic studies. One of the few examples of such a study thus far is that of Rousseau-Gueutin and coworkers49, who explored the replacement of plastid accD in Campanulaceae by a nuclear gene arising via a transfer from the plastome.
Unusual genes in Ericaceae plastid genomes
Although many genes in Ericaceae have been pseudogenized, a close examination of two (accD and clpP) that were previously reported to be pseudogenes shows that they may indeed represent intact genes. clpP encodes a subunit of the clp-protease complex, which is necessary for removing undesirable proteins within plastids (reviewed in ref. 50). accD encodes a subunit of acetyl-CoA carboxylase, which participates in fatty acid synthesis. clpP and accD are even present in highly reduced plastid genomes7,33,41,42, from which genes related to photosynthesis as well as many housekeeping genes have been lost. Therefore, it is sometimes hypothesized that the main reason for the retention of plastid genomes in heterotrophic plants is for the coding potential of these genes and that all genes of the translation machinery (i.e., encoding ribosomal components and tRNAs) are retained for translation of their mRNA (e.g. refs 7,9). Surprisingly, it was reported that the V. macrocarpon3 and A. unedo4 plastomes lack functional clpP and accD genes. Additionally, Actinidia chinensis, a plant from Actinidiaceae (order Ericales), a relative of Ericaceae, is reported to have lost clpP51. Herein, we argue in support of an idea that both clpP and accD are functional in most of the Ericaceae sequenced to date (and Actinidia may also have functional clpP). While investigating a short fragment of accD located in the plastid genomes of H. monotropa, M. uniflora, P. rotundifolia and A. unedo, we found that it is a part of a long open reading frame (ORF) in all these genomes: 1905 bp in H. monotropa, 4383 bp in M. uniflora, 3141 bp in A. unedo and 2466 bp in P. rotundifolia. For comparison, the accD gene in most plants is approximately 1500 bp long. The closest matches of this ORF to the NCBI nr protein database using BLASTP are to the plastid accD gene from Ericales and other angiosperms. According to BLASTP alignments, two regions in each of these ORFs align to accD of photosynthetic Ericales: the first is approximately 600 bp (200 aa) long and the other is approximately 210 bp (70 aa). No matches were retrieved for the remaining part of these ORFs, which consists of multiple AT-rich repeat blocks (Supplementary Fig. 2). To assess whether this ORF may be intact and encodes a protein possibly performing the functions of accD, we tested for selection by a site model using PAML, which allows to evaluate the selective pressure acting on each codon of a gene. The results are presented in Fig. 3a. Among 326 columns in a multiple alignment (with each column corresponding to a codon) with no gaps, 67%, 32% and 1% of the codons are under negative selection, neutral evolution and positive selection, respectively. According to the likelihood ratio test, the p-value of the hypothesis that this ORF has sites evolving under positive selection is 0.13; thus, we suppose that the sequence evolves primarily through a combination of negative selection and neutral evolution. This result provides circumstantial evidence that accD is not a pseudogene in Ericaceae. We did not find a long accD-containing ORF in V. macrocarpon suggesting that accD has indeed been lost from the plastome of this species and that it is presumably functionally replaced by a nuclear-encoded gene. However, an unannotated region of ~2800 bp in length that contains accD-like sequence is still present. An alternative explanation is that apparent pseudogenization of accD in V. macrocarpon plastome is an artefact of the sequencing/assembly that created non-triplet indels within the accD ORF. It was sequenced using 454 pyrosequencing technology, which has an increased error rate in homopolymeric regions. Such regions are abundant in the accD ORF of other Ericaceae. A more evident example of such artefact is the annotation of accD as pseudogene in Gruzdev et al.39. In their plastome assembly, the accD ORF has a frameshift due to 1-bp indel in homopolymeric (6A) region. Notably, the sequence of another isolate of H. monotropa from the same authors (not published but available in the GenBank, accession number KU640957) has an intact accD ORF, strongly suggesting that the frameshift in the published sequence is an error.
Mode of selection for accD (a) and clpP (b) in Ericaceae. The dots indicate the probability of negative selection, neutral evolution and positive selection for codons in genes accD (a) and clpP (b). The probabilities are estimated by the Empirical Bayes method in PAML. Green lines above the accD diagram designate regions homologous to parts of accD from plants with typical accD structure.
In most plants, clpP is split into three exons, though loss of the first intron can be observed in several species7,52,53. We found that this gene is present in M. uniflora, H. monotropa, V. macrocarpon and P. rotundifolia and A. chinensis (from which it was also thought to be lost), but in an intronless form of a single long exon arising from the joining of the three typical exons (intronless clpP is also found in several parasitic Orobanchaceae33). Its divergent sequence is most likely the reason why this gene was previously thought to be a pseudogene. Site model analysis using PAML suggested that 37%, 57% and 6% of codons are under negative selection, neutral evolution and positive selection, respectively. According to a likelihood ratio test, the p-value of the hypothesis that this gene has codons under positive selection is 0.011. Sites under negative and positive selection and those evolving neutrally are evenly distributed along the length of the gene (Fig. 3b).
However, the results of the site-wise selection analysis for clpP and accD should be treated with caution. First, these genes are highly divergent in Ericaceae (Supplementary Table 4) and multiple alignment of divergent sequences may contain alignment mistakes that distort selection analysis54. Although a study54 indicates that alignments by PRANK result in the most precise estimates of selective pressure, we also tested other multiple alignment tools. They produced similar estimates of proportion of sites under negative selection, positive selection and neutral evolution, although the significance of the presence of positive selection varies (Supplementary Table 5) Second, the number of sequences (four for accD and five for clpP) is the minimum that can be used in the site model. Thus sampling of additional sequences from Ericaceae is necessary to ascertain the mode of selection of accD and clpP this family.
Search within the H. monotropa transcriptome (unpublished data) demonstrates that clpP and accD are expressed (supplementary note 1). Interestingly, in H. monotropa these genes are situated close to each other and, probably, are expressed as a single polycistronic RNA. Although it is split in the transcriptome assembly into two contigs with lengths 3577 bp and 1230 bp, with the break situated in the middle of accD, a continuity of RNA-seq read coverage in the corresponding plastome region and a similarity of coverage between these two transcripts (supplementary note 1) suggests that this is a misassembly, and, in fact, the transcript is single. Regarding M. uniflora, transcriptome data are available for BLAST searching within “1000 Plant Genomes Project”55 ( https://www.bioinfodata.org/Blast4OneKP/), showing that clpP and accD are also expressed. These results also support the results of our computational analysis that clpP and accD are intact.
Another unusual hypothetical gene is an rps4-like gene found in the P. rotundifolia plastome. The rps4 gene encodes the ribosomal protein S4, one of the components of the plastid ribosome, which is present in all sequenced plastid genomes, including the most reduced42. The gene length is approximately 600 bp. In addition to a normal, full-length copy of rps4 in the P. rotundifolia plastid genome, a short fragment (114 bp) corresponding to the rps4 5′ region is also present. This fragment is the 5′ end of a long ORF of 1074 bp (herein, it will be referred to as ORF357, as it potentially encodes a 357 amino acid polypeptide). A search of similar sequences in the draft assembly of the plastid genome of Orthilia secunda (unpublished data, GenBank accession number KU588419), a close relative of P. rotundifolia, revealed the presence of this ORF (sequence similarity of 93%). BLAST alignment of the 3′ ends of this ORF from both species to GenBank protein and nucleotide databases yielded no significant matches at e-value <1. Thus, we suggest that the 3′ end is not homologous to any gene and has arisen from an intergenic spacer. Domain analysis by InterPro with TMHMM embedded for predicting transmembrane domains suggests that this 3′ region contains 6 transmembrane domains (Fig. 4a). To evaluate whether the presence of transmembrane domains could be an artefact of domain prediction, we performed computational simulations, generating random ORFs with different GC-contents and estimating by TMHMM software the fraction of amino acids in these ORFs that form transmembrane domains. The result for ORF357 was higher than predicted for a random sequence with the same GC-content (Fig. 4b), lying above the 99th percentile of the randomly generated ORFs both for P. rotundifolia and O. secunda. Furthermore, according to the BLASTN self-alignment of ORF357, these transmembrane domains are not similar to each other. Thus, we suggest that the domains did not arise via tandem duplications of a single ancestral sequence or at least this duplication was so ancient, that traces of similarity no longer exist. RT-PCR analysis indicated that ORF357 is expressed in P. rotundifolia (Supplementary Fig. 3), which supports the hypothesis that this ORF is functional. To explore the mode of selection acting on ORF357, we estimated the ratio of non-synonymous to synonymous substitutions. Pairwise analysis between ORF357 from P. rotundifolia and O. secunda revealed a dN/dS of 0.83 ± 0.23 for the entire sequence (hereafter, a value after ± denotes the standard error), 0.53 ± 0.48 for the rps4-like 5′ end and 0.89 ± 0.25 for the putative transmembrane 3′ end. To test whether the values of dN/dS are significantly different from 1, we performed likelihood ratio tests, which showed that in all three cases the difference is insignificant under significance criterion of p-value less than 0.05. Although dN/dS values close to 1 are considered to be a sign of neutral evolution, they are often a combination of negative selection acting on some sites and positive selection acting on others. As ORF357 sequences are only currently available for two species, it is not yet possible to estimate selection acting on specific codons. We expect that this ORF is present in other species of tribe Pyrolae (the group of Ericaceae to which P. rotundifolia and O. secunda belong); if so, sequencing will allow for a detailed conclusion regarding the selection acting on this new gene.
(a) Domain structure of the hypothetical product of ORF357; (b) transmembrane domain prediction for ORF357 and random ORFs with different GC contents. Blue dots denote mean values of parts of those random ORFs that are predicted to form transmembrane helices by TMHMM, whiskers represent the standard deviation.
Conclusions
We characterized the plastid genomes of three species of Ericaceae: non-photosynthetic M. uniflora and H. monotropa and their photosynthetic relative P. rotundifolia. Comparative analysis of an expanded set of Ericaceae plastomes allowed to re-examine the gene content and to assume that clpP and accD that were supposed to be pseudogenes in this family, are functional. Plastomes of non-photosynthetic Ericaceae exhibit extensive gene loss, rearrangements, nucleotide and amino acid biases, a high substitution rate and genes in unusual forms. These traits are a combination of features that are linked with the heterotrophy (loss of photosynthesis-related genes) and features typical for Ericaceae (extensive rearrangements, high repeat content, nucleotide composition bias, transformation of several genes).
With such unusual structure of plastid genomes, Ericaceae are a valuable model for the study of plastome evolution and furthermore, may provide insight into an intriguing question about the nuclear determinants of plastome structure and coevolution of nuclear and plastid genomes. Now the data on this subject begins to accumulate; e.g. recent study by Zhang et al.56 that suggests the role of DNA recombination, replication and repair systems in creating plastome complexity. This study was focused on Geraniaceae, the group which is not closely related to Ericaceae, but has many convergences in plastid genome structure (rearrangements, high repeat content, pseudogenes). We expect that characterization and analysis of more plastid genomes and nuclear genes from Ericaceae will allow to test, expand and complement these findings.
Materials and Methods
DNA extraction, library preparation, sequencing
DNA was extracted using the CTAB method57. For each species, two types of libraries, standard shotgun and long-insert (mate pair), were prepared. The shotgun libraries were prepared using the TruSeq DNA sample prep kit and the mate pair library using the Nextera Mate pair sample prep kit (Illumina) following the manufacturer’s instructions for agarose gel based size selection (Supplementary Table 6). The libraries were sequenced using HiSeq2000 and Miseq instruments (see Supplementary Table 6 for sequencing parameters and output).
RNA manipulations
RNA was extracted using the RNEasy mini kit (Qiagen) with the addition of the Plant RNA Isolation Aid solution (Ambion) to the lysis buffer. RNA was treated twice with RNase-free DNAse (Qiagen). cDNA synthesis was performed using the MMLV reverse transcription kit (Evrogen). Following reverse transcription, PCR was performed using primers Pyrrot-rps4-F (5′-GGGAGCTTTACCAGGACTAAC-3′) and Pyrrot-rps4-div-R (5′-AATGTTAGTGGACGGTGGTATC-3′) for rps4-like ORF357. As a positive control, we also amplified two photosynthesis-related genes – petB (Pyrrot-petB-F 5′-ATGAGTAAAGTCTACGATTGGTTC-3′ and Pyrrot-petB-R 5′-AAACGAGTCAAAGTGGATTGTC-3′) and psaB (Pyrrot-psaB-F-5′-ACTCGTCGTATTTGGTTTGGGATT-3′ and Pyrrot-psaB-R 5′-TTATTCCATCGGACAAACTCTCC-3′). A negative control with no reverse transcriptase added was also included.
Preprocessing of reads
Prior to de novo assembly, junction adapters were removed from the mate-pairs reads and the reads were classified into true mate-pairs and paired-end reads using NextClip 0.858, the latter were discarded. Low-quality regions and sequencing adapters were removed from all reads with Trimmomatic 0.3259. The long reads produced by MiSeq were trimmed from the 3′ and 5′ ends using a minimum quality of 20 and trimmed from the 3′ end by a sliding window of size 5 with a minimum average quality of 20. Reads with an average quality lower than 20 were discarded. The shotgun libraries reads produced by HiSeq were trimmed to remove bases with a quality lower than 3 from the 3′ end and reads with an average quality lower than 20 were discarded. Reads shorter than 30 bases were discarded. We also used Kmernator 1.2 ( https://github.com/JGIBioinformatics/Kmernator) to remove all k-mers (of length 21) with coverage less than 5.
Plastid genome assembly
Assembly was performed using SPAdes 3.560, with default parameters, which performs preliminary error correction and assembly with k-mers with lengths 21, 33, 55, 77, 99, 127 (the latter only when long MiSeq reads are used in assembly), with the addition of the ‘careful’ parameter. Identification of contigs of plastid origin was performed by BLASTN 2.2.29+ and TBLASTX 2.2.29+61 searches of protein-coding genes and proteins from the plastid genome of Arbutus unedo (GenBank accession JQ067650), Vaccinium macrocarpon (GenBank accession NC_019616) and Camellia sinensis (GenBank accession NC_020019) with a maximum e-value of 10−5. Contigs corresponding to plastid genomes were checked for misassembly using REAPR 1.0.1762. Scaffolding was performed in the following way. First, we mapped mate-pair reads using CLC Assembly Cell 4.2 ( www.clcbio.com) by requiring at least 80% of the read length to map with identity of at least 99%. We did not use paired-end reads for scaffolding, because due to their short insert sizes they are less capable of resolving repeats. We then visualized mate pair links between the contigs using Circos 0.6763. Additionally, to identify possibly overlapping sequences between the ends of the contigs, we performed a BLASTN alignment of the plastid contig set to itself. The result was inspected visually by plot construction in Circos. To assess for misassembled repeat regions, we mapped all reads by CLC Assembly Cell to the contigs and visually searched for areas of increased coverage. Combined analyses of mate-pair links and repeats allowed us to determine the order of the contigs in the plastid genome. We manually joined the contigs into a scaffold and filled the gaps using GapFiller 1.1064.
It is difficult to precisely assemble regions abundant in repeats, which are present in the P. rotundifolia and M. uniflora plastomes. Thus, we performed three steps to assemble them as strictly as possible:
(1) We mapped mate-pair reads to the region containing the repeats.
(2) We calculated an average insert size of mate-pairs in which the left read is located to the left of the repeat-containing region and the right read to the right of this region.
(3) If the average insert length is shorter than the average insert length at regions of a genome that do not contain repeats, then the number of copies of tandem repeats is underestimated; however, if it is larger, then the number of copies of tandem repeats is overestimated. In the first case, we added one copy of a tandem repeat unit and in the second case, we removed one. These procedures were iteratively performed until the average insert size in the tandem repeat region became equal to the average insert size in non-repeat regions.
The copies of repeats in a tandem repeat regions can be divergent. To account for this, we mapped mate-pair reads by CLC Assembly Cell, demanding that 80% of a reads’ length map with an identity of at least 98%; we then replaced nucleotides at positions in which a consensus of mapped reads suggested another nucleotide than that present in a current version of the sequence. Then, we remapped the reads. This operation was iteratively performed until no discrepancies between the mapping and the contig sequence remained.
To verify the assembled plastid genomes, we mapped all reads by CLC Assembly Cell, demanding that at least 80% of the read length map with an identity of at least 99% and ensuring no sites with decreased coverage. Additionally, we mapped the mate-pair reads and estimated the average insert size over all positions of the genomes and a number of mate-pair inserts (“coverage by inserts”) that span each genome position (see Supplementary Fig. 4). The uniformity of insert size distribution suggests no unnoticed artificially made insertions and deletions in the sequence and the absence of sharp declines in the “coverage by inserts” indicate no incorrect contig connections.
Annotation
The initial annotation was performed using the on-line service DOGMA65. The annotation was performed with relaxed parameters: an e-value < 10−5 and a sequence similarity to reference genes >25%. The annotations were then checked manually. To verify borders of intronless genes, we calculated ORFs in the plastid genomes using CLC Genomics Workbench 7.0 by considering sequences that start with any of 7 possible plastid start codons (ATG, TTG, GTG, CTG, ATA, ATT, ATC), end with a stop codon, have no internal stop codons and are longer than 90 bp. To refine the predictions of tRNAs, we scanned the entire plastid genomes with an online version of tRNAscan-SE 1.2.1166 at http://selab.janelia.org/tRNAscan-SE/ using both the general and organellar tRNA modes of the program. Additionally, we verified the gene prediction made by DOGMA by generating BLASTN and TBLASTN alignments of the genes and proteins of Camellia sinensis (as one of the most studied plants in Ericales) with a maximum e-value of 10−5 and by inspecting the alignments visually. Verification of borders of genes with introns was made similarly, by aligning genes that have introns in C. sinensis to assembled genomes by BLASTN and TBLASTN, but more sensitively - with a maximum e-value of 10 and word sizes of 7 for BLASTN and 2 for TBLASTN. Finally, we performed a multiple alignment of our three plastid genomes using mVista67 to ascertain the presence of any unusually conserved regions in the genomes, regions that could correspond to unnoticed genes. The maps of the plastid genomes (Fig. 1) were built using the on-line tool OGDRAW68. Plastome sequences are deposited in NCBI Genbank under accession numbers KU878156 (Hypopitys monotropa), KX228067 (Monotropa uniflora) and KU833271 (Pyrola rotundifolia).
Analysis of the genomes
The domain structures of ORF357, accD and clpP were analysed using the InterPro server69 at http://www.ebi.ac.uk/interpro/.
A computational simulation was performed to evaluate whether the transmembrane domains of ORF357 are an artefact of domain prediction. Utilizing random sequences of 999 bp each, we removed stop codons from the respective 333 codons and translated the remaining codons into an amino acid sequence. Potential transmembrane helices in this randomly generated amino acid sequence were predicted by TMHMM 2.070. This operation was performed for sequences with all possible GC-contents from 0 to 1 with a step of 0.05. For each GC-content, 1000 random sequences were generated and then we calculated for each GC-content the average fraction of sequence length that potentially forms a transmembrane helix and a standard deviation of this value. To compare individually the percent of sequence of ORF357 potentially forming transmembrane helices with that parameter of random ORFs, we also generated 1000 random ORFs with GC-contents as in ORF357 of Pyrola and as in Orthilia and calculated the percentile in the random ORFs above which that value falls.
Repeats in the genomes were tabulated using BLASTN alignment of the genomes to themselves with a word size of 9 bp. For Fig. 1, we compiled all repeats with a minimum length of 20 bp and a minimum sequence similarity of 95%, irrespective of the e-value. Circos was then used to generate diagrams of the repeats. The same set of parameters was used to calculate repeats for Supplementary Table 1. For dot plot diagrams (Supplementary Figs 1 and 2), BLASTN self-alignment in NCBI Genome Workbench 2.9.5 ( http://www.ncbi.nlm.nih.gov/tools/gbench/ was performed with a word size of 9 bp and a maximum e-value of 10−5. The low-complexity filter (“-dust no” option) was switched off for all BLAST alignments.
To build a phylogenetic tree (Fig. 2), we used sequences of plastid protein-coding genes, plastid rDNAs, nuclear 18s rDNAs and the nuclear internal transcribed spacer (ITS) of M. uniflora, H. monotropa, A. unedo, V. macrocarpon, P. rotundifolia and C. sinensis. Nuclear 18S rDNA and the ITS sequences for our species were found among the contigs from the de novo assembly by BLASTN alignment of the respective sequences from close relatives deposited in GenBank. We verified the found sequences by making their BLASTN alignment to NCBI NT database. We used consensus sequences, generated by SPAdes and hence did not account for possible polymorphisms between genomic copies of the sequences. The 5.8S rDNA situated between ITS1 and ITS2 was removed. Multiple alignments of the sequences from these four sets were then performed separately. The common protein-coding genes of the aforementioned species were aligned using a combination of Muscle71 and TranslatorX72 with default options, which allows alignment of protein-coding genes by taking into account a respective amino acid alignment. Poorly aligned regions were removed using Gblocks73 in the codon mode with the most liberal settings for poorly aligned region removal available on the Gblocks server ( http://molevol.cmima.csic.es/castresana/Gblocks_server.html). The plastid rDNA, nuclear 18S rDNA and ITS sequences were aligned separately using Muscle and poorly aligned regions were removed by Gblocks in a nucleotide mode, also with the most liberal settings. The four respective alignments were concatenated and a phylogenetic tree was built using RAxML 8.1.2074. We used the GTR+Gamma model and 5 partitions, with RAxML calculating its own parameters of the GTR+Gamma model for each. The partitions were as follows: (1) the first and second bases in each codon of protein-coding genes; (2) the third base in each codon of protein-coding genes (they are often synonymous and, therefore, accumulate mutations faster); (3) plastid rDNAs; (4) nuclear 18S rDNAs; (5) nuclear ITS. RAxML builds 100 starting trees and after topology estimation, it performs 1000 iterations of fast bootstrap analysis.
To estimate the selective pressure acting on protein-coding plastid genes, a multiple alignment of the concatenated sequences of those genes was performed using a combination of Muscle and TranslatorX with default parameters on TranslatorX server. The dN/dS (the ratio of rates of non-synonymous and synonymous substitutions) was then calculated using the branch model in PAML 4.775 with the topology estimated as indicated above. Statistical significance of difference between substitution accumulation rates of the species was estimated by Tajima’s relative rate test in MEGA 676. We performed pairwise comparisons between all pairs of the five studied Ericaceae (ten pairwise comparisons), using Camellia as an outgroup. To calculate pairwise dN/dS values between the sequences of ORF357 from Pyrola and Orthilia, we aligned them using Muscle and TranslatorX and then calculated dN/dS in PAML in the pairwise mode. To test whether these values are significantly different from 1, we performed the same computations, while fixing dN/dS to 1 and then comparing likelihoods between the models with fixed and floating dN/dS by the likelihood ratio test.To estimate the selective pressure acting on the genes clpP and accD in Ericaceae, we generated multiple alignments of these sequences using the webPRANK server77 ( http://www.ebi.ac.uk/goldman-srv/webprank/) in the codon alignment mode with default parameters. For the analyses of both clpP and accD we used all species of Ericaceae where they are intact, i.e. Uniflora, Hypopitys, Vaccinium and Pyrola for clpP and Uniflora, Hypopitys, Arbutus and Pyrola for accD. We chose PRANK for alignments of clpP and accD because it has been noted that for highly divergent sequences, dN/dS analysis performed on alignments generated by PRANK provides more precise results than other alignment tools54. Then, we ran PAML in site models M1a (which allows for negative selection and neutral evolution) and M2a (which allows for negative selection, neutral evolution and positive selection). To calculate the p-value of the hypothesis that positive selection exists, a likelihood ratio test was performed for the likelihoods of these two models. In all calculations performed by PAML, the Gy94_3 × 4codon model was used and the transition/transversion ratio was estimated by PAML with a starting value of 2.0. As a model for codon substitutions we used the “basic” model, which is a simplified version of the model of Goldman and Yang78. In the alignments, the columns with gaps were not analysed (cleandata = 1 option). To estimate the effect of the alignment tool on selection estimation, we also performed multiple alignments of accD and clpP genes by combinations of TranslatorX and, separately, PRANK, MUSCLE, MAFFT79, ClustalW80 and T-Coffee81. The alignments were made on the TranslatorX server with default parameters.
To calculate the percentage of identity among protein-coding genes and proteins, we aligned them using Muscle+TranslatorX and Muscle, respectively (in both cases with default parameters) and calculated the percent identity in BioEdit 7.2.582. Alignments generated by PRANK usually contain excessive amounts of short gaps, which distort the results of sequence identity calculations. The codon usage analysis was performed in CodonW 1.4.283.
Additional Information
How to cite this article: Logacheva, M. D. et al. Comparative analysis of plastid genomes of non-photosynthetic Ericaceae and their photosynthetic relatives. Sci. Rep. 6, 30042; doi: 10.1038/srep30042 (2016).
References
Kron, K. A. et al. Phylogenetic classification of Ericaceae: molecular and morphological evidence. Bot. Rev. 68, 335–423 (2002).
Read, D. J. & Stribley, D. P. Effect of mycorrhizal infection on nitrogen and phosphorus nutrition of ericaceous plants. Nature. New Biol. 244, 81–82 (1973).
Fajardo, D. et al. Complete plastid genome sequence of Vaccinium macrocarpon: structure, gene content and rearrangements revealed by next generation sequencing. Tree Genet. Genomes 9, 489–498 (2013).
Martínez-Alberola, F. et al. Balanced gene losses, duplications and intensive rearrangements led to an unusual regularly sized genome in Arbutus unedo chloroplasts. PLos One 8, e79685 (2013).
Tedersoo, L., Pellet, P., Kõljalg, U. & Selosse, M.-A. Parallel evolutionary paths to mycoheterotrophy in understorey Ericaceae and Orchidaceae: ecological evidence for mixotrophy in Pyroleae. Oecologia 151, 206–217 (2007).
Barrett, C. F. et al. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids and implications for heterotrophic angiosperms. Mol. Biol. Evol. 31, 3095–3112 (2014).
Schelkunov, M. I. et al. Exploring the limits for reduction of plastid genomes: a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol. Evol. 7, 1179–1191 (2015).
Braukmann, T. & Stefanović, S. Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol. 79, 5–20 (2012).
Delannoy, E., Fujii, S., des Francs-Small, C. C., Brundrett, M. & Small, I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28, 2077–2086 (2011).
Barrett, C. F. & Davis, J. I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 99, 1513–1523 (2012).
Nickrent, D. L., Ouyang, Y. & Duff, R. J. et al. Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Mol. Biol. 34, 717–729 (1997).
Molina, J. et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 31, 793–803 (2014).
Allen, J. F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 112, 10231–10238 (2015).
Barbrook, A. C., Howe, C. J. & Purton, S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 11, 101–108 (2006).
Peng, L., Yamamoto, H. & Shikanai, T. Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta BBA - Bioenerg. 1807, 945–953 (2011).
Martín, M. & Sabater, B. Plastid ndh genes in plant evolution. Plant Physiol. Biochem. PPB Société Fr. Physiol. Végétale 48, 636–645 (2010).
de Vries, J., Stanton, A., Archibald, J. M. & Gould, S. B. Streptophyte terrestrialization in light of plastid evolution. Trends Plant Sci., doi: 10.1016/j.tplants.2016.01.021 (2016).
Ruhlman, T. A. et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 15, 100 (2015).
Burrows, P. A., Sazanov, L. A., Svab, Z., Maliga, P. & Nixon, P. J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 17, 868–876 (1998).
Horváth, E. M. et al. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123, 1337–1350 (2000).
Martín, M., Funk, H. T., Serrot, P. H., Poltnigg, P. & Sabater, B. Functional characterization of the thylakoid Ndh complex phosphorylation by site-directed mutations in the ndhF gene. Biochim. Biophys. Acta BBA - Bioenerg. 1787, 920–928 (2009).
Peredo, E. L., King, U. M. & Les, D. H. The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. Plos One 8, e68591 (2013).
Drescher, A., Ruf, S., Calsa, T., Carrer, H. & Bock, R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J. 22, 97–104 (2000).
Saski, C. et al. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera and comparative analyses with other grass genomes. Theor. Appl. Genet. 115, 571–590 (2007).
Kikuchi, S. et al. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574 (2013).
de Vries, J., Sousa, F. L., Bölter, B., Soll, J. & Gould, S. B. YCF1: a green TIC? Plant Cell 27, 1827–1833 (2015).
Nakai, M. YCF1: a green TIC: response to the de Vries et al. Commentary. Plant Cell 27, 1834–1838 (2015).
Wickett, N. J. et al. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol. Biol. Evol. 25, 393–401 (2008).
Börner, T., Aleynikova, A. Y., Zubo, Y. O. & Kusnetsov, V. V. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta 1847, 761–769 (2015).
Krause, K., Berg, S. & Krupinska, K. Plastid transcription in the holoparasitic plant genus Cuscuta: parallel loss of the rrn16 PEP-promoter and of the rpoA and rpoB genes coding for the plastid-encoded RNA polymerase. Planta 216, 815–823 (2003).
Logacheva, M. D., Schelkunov, M. I., Nuraliev, M. S., Samigullin, T. H. & Penin, A. A. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol. Evol. 6, 238–246 (2014).
Samigullin, T. H., Logacheva, M. D., Penin, A. A. & Vallejo-Roman, C. M. Complete plastid genome of the recent holoparasite Lathraea squamaria reveals earliest stages of plastome reduction in Orobanchaceae. PloS One 11, e0150718 (2016).
Wicke, S. et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the Broomrape Family. Plant Cell 25, 3711–3725 (2013).
Kamikawa, R. et al. Proposal of a twin arginine translocator system-mediated constraint against loss of ATP synthase genes from nonphotosynthetic plastid genomes. Mol. Biol. Evol. 32, 2598–2604 (2015).
Tiller, N. & Bock, R. The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120 (2014).
Ueda, M. et al. Substitution of the gene for chloroplast RPS16 was assisted by generation of a dual targeting signal. Mol. Biol. Evol. 25, 1566–1575 (2008).
Lerbs-Mache, S. Regulation of rDNA transcription in plastids of higher plants. Biochimie 82, 525–535 (2000).
Courtois, F., Merendino, L., Demarsy, E., Mache, R. & Lerbs-Mache, S. Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis. PLANT Physiol. 145, 712–721 (2007).
Gruzdev, E. V. et al. The complete chloroplast genome of parasitic flowering plant Monotropa hypopitys: extensive gene losses and size reduction. Mitochondrial DNA in press, (2016).
Kim, J. S., Kim, H. T. & Kim, J.-H. The largest plastid genome of monocots: a novel genome type containing AT residue repeats in the slipper orchid Cypripedium japonicum. Plant Mol. Biol. Report. 33, 1210–1220 (2015).
Lam, V. K. Y., Soto Gomez, M. & Graham, S. W. The Highly Reduced Plastome of Mycoheterotrophic Sciaphila (Triuridaceae) Is Colinear with Its Green Relatives and Is under Strong Purifying Selection. Genome Biol. Evol. 7, 2220–2236 (2015).
Bellot, S. & Renner, S. S. The plastomes of two species in the endoparasite genus Pilostyles (Apodanthaceae) each retain just five or six possibly functional genes. Genome Biol. Evol. evv251, doi: 10.1093/gbe/evv251 (2015).
Naumann, J. et al. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol. Evol. 8, 345–363 (2016).
Wicke, S., Schneeweiss, G. M., dePamphilis, C. W., Müller, K. F. & Quandt, D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297 (2011).
Chumley, T. W. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol. Biol. Evol. 23, 2175–2190 (2006).
Guisinger, M. M., Kuehl, J. V., Boore, J. L. & Jansen, R. K. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats and codon usage. Mol. Biol. Evol. 28, 583–600 (2011).
Haberle, R. C., Fourcade, H. M., Boore, J. L. & Jansen, R. K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66, 350–361 (2008).
Rocha, E. P. C. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 13, 1123–1132 (2003).
Rousseau-Gueutin, M. et al. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 161, 1918–1929 (2013).
Olinares, P. D. B., Kim, J. & van Wijk, K. J. The Clp protease system; a central component of the chloroplast protease network. Biochim. Biophys. Acta BBA - Bioenerg. 1807, 999–1011 (2011).
Yao, X. et al. The First Complete Chloroplast Genome Sequences in Actinidiaceae: Genome Structure and Comparative Analysis. PLos One 10, e0129347 (2015).
Jansen, R. K., Wojciechowski, M. F., Sanniyasi, E., Lee, S.-B. & Daniell, H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (Leguminosae). Mol. Phylogenet. Evol. 48, 1204–1217 (2008).
Dugas, D. V. et al. Mimosoid legume plastome evolution: IR expansion, tandem repeat expansions and accelerated rate of evolution in clpP. Sci. Rep. 5, 16958 (2015).
Fletcher, W. & Yang, Z. The effect of insertions, deletions and alignment errors on the branch-site test of positive selection. Mol. Biol. Evol. 27, 2257–2267 (2010).
Matasci, N. et al. Data access for the 1,000 Plants (1KP) project. GigaScience 3, 17 (2014).
Zhang, J. et al. Coevolution between nuclear-encoded DNA replication, recombination and repair genes and plastid genome complexity. Genome Biol. Evol. 8, 622–634 (2016).
Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19, 11–15 (1987).
Leggett, R. M., Clavijo, B. J., Clissold, L., Clark, M. D. & Caccamo, M. NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics 30, 566–568 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Hunt, M. et al. REAPR: a universal tool for genome assembly evaluation. Genome Biol. 14, R47 (2013).
Krzywinski, M. et al. Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Boetzer, M. & Pirovano, W. Toward almost closed genomes with GapFiller. Genome Biol. 13, R56 (2012).
Wyman, S. K., Jansen, R. K. & Boore, J. L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252–3255 (2004).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. & Dubchak, I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32, W273–279 (2004).
Lohse, M., Drechsel, O. & Bock, R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 52, 267–274 (2007).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Abascal, F., Zardoya, R. & Telford, M. J. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38, W7–W13 (2010).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Yang, Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Löytynoja, A. & Goldman, N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11, 579 (2010).
Goldman, N. & Yang, Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736 (1994).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Larkin, M. A. et al. Clustal W and Clustal X version 2.0. Bioinforma. Oxf. Engl. 23, 2947–2948 (2007).
Notredame, C., Higgins, D. G. & Heringa, J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000).
Hall, T. A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98 (1999).
Peden, J. F. Analysis of codon usage. (University of Nottingham, 1999).
Acknowledgements
We are grateful to Alexey S. Kondrashov for providing plants samples, to American Journal Experts company for editing the manuscript and to two anonymous reviewers for helpful comments on the initial version of manuscript. The study was supported by the Russian Foundation for Basic Research (project #11-04-02031, initial sequencing of Monotropa uniflora and Hypopitys monotropa plastomes) and Russian Science Foundation (project # 14-50-00029, sequencing of Pyrola rotundifolia plastome, assembly and analysis).
Author information
Authors and Affiliations
Contributions
M.D.L. designed and coordinated the study, performed sequencing and drafted the manuscript, M.I.S. performed data analyses and contributed to writing, V.Y.S. participated in annotation and edited the manuscript, M.V.M. participated in assembly, A.A.P. participated in sequencing.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Logacheva, M., Schelkunov, M., Shtratnikova, V. et al. Comparative analysis of plastid genomes of non-photosynthetic Ericaceae and their photosynthetic relatives. Sci Rep 6, 30042 (2016). https://doi.org/10.1038/srep30042
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep30042
This article is cited by
-
Distinctive plastome evolution in carnivorous angiosperms
BMC Plant Biology (2023)
-
A comparative study across the parasitic plants of Cuscuta subgenus Grammica (Convolvulaceae) reveals a possible loss of the plastid genome in its section Subulatae
Planta (2023)
-
Chloroplast genome assemblies and comparative analyses of commercially important Vaccinium berry crops
Scientific Reports (2022)
-
Higher level constructive neutral evolution
Biology & Philosophy (2022)
-
GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes
Genome Biology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.