Abstract
Bacillus thuringiensis and Bacillus endophyticus both act as the companion bacteria, which cooperate with Ketogulonigenium vulgare in vitamin C two-step fermentation. Two Bacillus species have different morphologies, swarming motility and 2-keto-L-gulonic acid productivities when they co-culture with K. vulgare. Here, we report the complete genome sequencing of B. thuringiensis Bc601 and eight plasmids of B. endophyticus Hbe603, and carry out the comparative genomics analysis. Consequently, B. thuringiensis Bc601, with greater ability of response to the external environment, has been found more two-component system, sporulation coat and peptidoglycan biosynthesis related proteins than B. endophyticus Hbe603, and B. endophyticus Hbe603, with greater ability of nutrients biosynthesis, has been found more alpha-galactosidase, propanoate, glutathione and inositol phosphate metabolism, and amino acid degradation related proteins than B. thuringiensis Bc601. Different ability of swarming motility, response to the external environment and nutrients biosynthesis may reflect different companion mechanisms of two Bacillus species. Comparative genomic analysis of B. endophyticus and B. thuringiensis enables us to further understand the cooperative mechanism with K. vulgare, and facilitate the optimization of bacterial consortium.
Similar content being viewed by others
Introduction
The microbial communities have been deemed important organisms based on their metabolic capabilities and potential applications1. The co-cultured organisms can divide the labor reasonable and improve the robustness of the system2. For example, the ability to metabolize and degrade cellulose3, alkanes4, heavy metal toxins5 and natural products6. The multiple microbial species increased range of genes and metabolic capabilities in comparison to monocultures. Enumerating metabolic exchanges and the microbial diversity are still uncharacterized for complex metabolic needs and biosynthetic capabilities.
The microbial community of K. vulgare and Bacillus species have been widely used in the two-step vitamin C production7. In the community, K. vulgare is responsible for the bioconversion of sorbose to 2-keto-L-gulonic acid (2-KLG, the precursor of vitamin C). Mono-cultured K. vulgare grows poorly, even on rich natural media. Addition of amino acids8, vitamins9 and glutathione10 could enhance the growth of K. vulgare, implying the defects of oxidation metabolism, vitamin and amino acid synthesis11. The Bacillus species have been used to stimulate the growth of K. vulgare, including Bacillus megaterium, Bacilius cereus and B. endophyticus, and different 2-KLG productivities have been determined in the co-culture systems. Besides, mix cultured B. megaterium and B. cereus (1:3) could increase the 2-KLG yield compared to one-helper-strain co-culture system12. In our previous studies, the metabolomics approach has been demonstrated on the interactions between K. vulgare and Bacillus10,13. However, the different cooperative effects and mechanisms of companion bacteria are needed to interpret at molecular level.
Comparative genomics is a powerful approach for studying variation in physiological traits as well as the evolution and ecology of microorganisms14. Based on comparative genomics analysis, relationship of B. megaterium with other Bacillus species and numerous unique genetic traits were identified15. Through comparative genomics, the evolutionary relationships and major traits of Bacillus species were understood, including the diversity of sporulation and competence genes16. Whole-genome sequencing of B. thuringiensis 97-27 and B. cereus E33L was undertaken to identify shared and unique genes, the differences were revealed in terms of virulence, structural components and regulatory mechanisms17.
Identification the optimum companion bacterium has always been an important research topic in VC two-step fermentation. In present study, we observe the different phenotype of B. thuringiensis Bc601 and B. endophyticus Hbe603 and different 2- KLG productivities of K. vlugare in the co-culture system with different helper bacteria. In order to understand the genome evolution and the metabolic versatility, we have sequenced the complete genomes of B. thuringiensis Bc601 and eight plasmids of B. endophyticus Hbe603. Aided with comparative genomics analysis, characteristics of two species and cooperation mechanisms could be systematically understood, which are of great importance for consortium optimization.
Results
General genomic properties of two Bacillus species
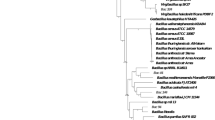
The complete genome of B. thuringiensis Bc601 consists of one circular chromosome and six plasmids (Fig. S1 and Table S1). B. anthracis, B. cereus and B. thuringiensis are closely-related members, which have been named as the B. cereus-group. Comparison of their 16S rRNA sequences places them within the same species18. B. thuringiensis and B. cereus mix together in the phylogenetic tree by blasting the whole genome sequences. B. thuringiensis Bc601 is most similar with B. thuringiensis serovar kurstaki HD73, which contains 15 contigs (Fig. S2). The genes in plasmids are mainly encodes transposases, transcriptional regulators, recombinases, type VII secretion proteins and cell surface proteins. Besides, B. thuringiensis Bc601 also encodes thiamine biosynthesis protein, 3-oxoacyl-ACP synthase, ATPase and flagellar biosynthesis protein on plasmids for nutrients synthesis.
The complete genome of B. endophyticus Hbe603 consists of one circular chromosome and eight plasmids (Table S2). Our previous research published the chromosome of B. endophyticus Hbe60319. Besides, it owns eight plasmids, which have the special sequence information and low similarity compared to diverse Bacillus spp. With the RAST analysis platform, we identify 468 genes in eight plasmids and almost half of them encode the hypothetical proteins or unknown enzymes. Six plasmids contain phage proteins which may derive from horizontal transfer. The genes in plasmids are mainly about sporulation, germination, regulation and transportation. The sporulation cycle is complete in B. endophyticus and plasmids also harbor some sporulation-related proteins, including AbrB family transcription regulatory proteins, spores coat protein and germination proteins GerKA, GerKC and GerKB. B. endophyticus has abundant transcriptional regulatory proteins in response to changes of external environment, and the plasmids harbor sigma-W, sigma-B and two-component signal transduction system for the regulation of cell density. In addition, it contains a large number of other transcription factors, covering MerR, ArsR, MarR, AbrB, GntR, AraC and LacI family. Besides, B. endophyticus also encodes five cell division proteins and 36 mobile element proteins on plasmids for cell division and proliferation.
The different morphologies and companion effects of two Bacillus species
By microscopic observation, we found the obvious differences in the morphology of two Bacillus species. B. thuringiensis has a long rod-shaped with no obvious changes before and after sporulation (Fig. 1A,B). B. endophyticus keeps the long chain distribution characteristics in the vegetative state, and becomes elliptic and reduces half of its volume after sporulation (Fig. 1C,D). We speculate that its cell structure is changeable. The different morphologies after sporulation may be connected with the characteristics of two species, particularly the cell structure related proteins. The highest cell density of single-cultured B. thuringiensis was 1.59 times of single-cultured B. endophyticus, and the cell density decreased significantly after 20 hours due to the sporulation (Fig. 2A). B. thuringiensis may absorb more nutrients for its strong growth ability compared to B. endophyticus in the mixed bacteria system. In the co-culture system, the 2-KLG productivity of K. vulgare with B. endophyticus was higher than that with B. thuringiensis after 36 hours and tended to be stable in 72 hours (Fig. 2B). Swarming motility is a multi-cellular behavior which can help us to better understand the metabolic interaction and the cooperative mechanism between the two species. We orthogonal cultured B. thuringiensis, B. endophyticus and K. vulgare on an agar plate. The horizontal cultivation was K. vulgare, and vertical cultivation was B. thuringiensis Bc601 and B. endophyticus Hbe603. Swarming motility of B. thuringiensis could be induced when it was co-cultured with K. vulgare, while B. endophyticus didn’t swarm to K. vulgare (Fig. 1E).
The vegetative state (A) and sporulation (B) of B. thuringiensis Bc601 and the vegetative state (C) and sporulation (D) of B. endophyticus Hbe603. Swarming pattern of the ecosystem via chemotaxis of species (E). The photographs show monocultures of B. thuringiensis, B. endophyticus and K. vulgare and coculture after 96 hours of growth at 30 °C on soft agar.
Comparative analysis of the versatile metabolism in two Bacillus species
To gain further insight into the relationship between B. thuringiensis Bc601 and B. endophyticus Hbe603, the common and unique genes were calculated using the CD-HIT rapid clustering20. B. thuringiensis Bc601 and B. endophyticus Hbe603 have 1524 common genes, and the number of specific genes is 4189 and 3698, respectively. The distribution of COG classification was compared to facilitate the difference of gene function in two species (Table S3). In B. thuringiensis Bc601, the number of genes related to carbohydrate transport and metabolism (G), Energy production and conversion (C) and lipid metabolism (I) is less than that in B. endophyticus Hbe603, respectively. We speculate that B. endophyticus Hbe603 may have greater promotion for K. vulgare due to greater capacity of nutritional supplements. From our observation, the cell wall and membrane structure of B. thuringiensis Bc601 are significantly thicker than B. endophyticus Hbe603, and the ratio of peptidoglycan biosynthesis related gene number is 29:21. Only B. thuringiensis Bc601 harbors femX, femA, femB, murN, mtgA, pbpC and pbpB. Besides, D-alanyl-D-alanine carboxypeptidases, collagenases, ATPases, spore coat and two-component system related proteins in B. thuringiensis Bc601 are more than those in B. endophyticus Hbe603. We speculate these factors may affect B. endophyticus Hbe603’s ability to communicate with the external environment. Furthermore, B. endophyticus Hbe603 has more PTS related transporters and ferredoxins than B. thuringiensis Bc601 (Table 1).
Based on the transcriptome analysis of B. thuringiensis sporulation process, 1646 genes were differentially expressed and most of them were connected with transport, transcriptional regulation, cell motility and DNA repair21. In the process of sporulation, the companion Bacillus bacterium further releases abundant nutrients for the growth and 2-KLG production of K. vulgare22. In order to analyze the different metabolic capacities in B. thuringiensis Bc601 and B. endophyticus Hbe603, metabolic network of two species was obtained, including the central carbon, amino acid and cofactor metabolism (Fig. 3).
In the central carbon metabolism, we identified the complete glycolysis, citrate cycle (TCA cycle) and pentose phosphate pathway in the two species. Besides, B. thuringiensis Bc601 owns glyceraldehyde-3-phosphate dehydrogenases and 2- oxoglutarate ferredoxin oxidoreductase, which converses glyceraldehyde-3-phosphate to glycerate-3-phosphate, and converts 2-oxoglutarate into succinyl-CoA, respectively. In B. thuringiensis Bc601, the alpha-galactosidases, pentose and glucuronate conversion related proteins, which can utilize D-galacturonate, D-altronate, D-mannonate, xylitol, xylose and galactose for carton implying, are less than that in B. endophyticus Hbe603. In the propanoate metabolism, only B. endophyticus Hbe603 harbors a complete pathway of six steps to converse 2-oxobutanoate to succinyl-CoA, thus implies for the citrate cycle. The absence of propionyl-CoA carboxylase and methylmalonyl-CoA mutase in B. thuringiensis Bc601 significantly impedes the metabolic flux to citrate cycle. Particularly, three methylmalonyl-CoA mutases are identified in B. endophyticus Hbe603, which have only been found in B. megaterium, Geobacillus kaustophilus and Bacillus halodurans.
In the amino acid metabolism, B. endophyticus Hbe603 has more lysine degradation related genes than B. thuringiensis Bc601, the ratio of gene number is 26:18. In the tyrosine metabolism, the two species are both lack of adequate genes. B. thuringiensis Bc601 uses 4- hydroxyphenylpyruvate dioxygenase and homogentisate 1, 2-dioxygenase to converse tyrosine to 4-maleyl-acetoacetate, and phenylalanine-4-hydroxylase to converse phenylalanine to tyrosine. B. endophyticus Hbe603 uses tyrosinase (EC 1.14.18.1) to converse tyrosine to pheomelanin and eunelanin. In the tryptophan metabolism, the two species are both lack of the degradation pathway. They use tryptophan 2, 3- dioxygenase and kynureninase to converse tryptophan to formyl-anthranilate. The two species both have the complete phenylalanine, tyrosine and tryptophan biosynthesis pathway while only B. thuringiensis Bc601 has the phenylalanine-4-hydroxylase to converse the phenylalanine to tyrosine. The glutathione metabolism in B. thuringiensis Bc601 is not complete and B. endophyticus Hbe603 contains five gamma-glutamyl-transpeptidases to converse glutathione to glycine, cysteine and glutamate.
In the cofactor and vitamin metabolism, the two species both have the complete biosynthesis pathway of folate, protoheme, pantothenate and CoA, while the lipoic acid and biotin biosythensis pathway are defect. In the inositol phosphate metabolism, only B. endophyticus Hbe603 contains a complete pathway of thirteen key enzymes, including myo-inositol 2-dehydrogenase, inosose dehydratase, glucuronate isomerases, 5-dehydro-2-deoxygluconokinase, 6-phospho-5-dehydro-2-deoxy-D-gluconate aldolase, methylmalonate-semialdehyde dehydrogenase and triosephosphate isomerase. That pathway converses inositol to acetyl-CoA and glyceraldehyde-3-phosphate, which participates in TCA cycle and glycolysis, respectively. In the nicotinate and nicotinamide metabolism, the two species both can transfer L-aspartate to nicotinate, but only B. thuringiensis Bc601 contains three nucleotidases that can transfer L-aspartate to nicotinamide.
Discussion
Microorganisms can often utilize and secrete a large number of metabolites. This plastic network is readily adapted and regulated in response to nutrients. Swarming motility is a multi-cellular behavior which can help us to better understand the metabolic interaction and cooperative mechanism23,24. Swarming motility of B. thuringiensis could be induced when it was co-cultured with K. vulgare, while B. endophyticus didn’t swarm to K. vulgare. The metabolic interaction and companion mechanism of the two Bacillus species are completely different in bacterial consortium (Fig. 4).
For the ability of response to the external environment, B. thuringiensis Bc601 has more collagenases, ATPases, two-component system and peptidoglycan biosynthesis related proteins than B. endophyticus Hbe603 (marked in blue words). For the ability of nutrients biosynthesis, B. endophyticus Hbe603 has more PTSsystem, propanoate, glutathione and inositol phosphate metabolism, and amino acid degradation related proteins than B. thuringiensis Bc601 (marked in red words).
By the means of metabolomics, the metabolites changes were identified by B. thuringiensis, K. vlugare and consortium10. The contents of nutritional compounds in the medium surrounding K. vulgare were fairly higher than in fresh medium. Erythrose, erythritol, guanine and inositol accumulated around B. thuringiensis were consumed by K. vulgare, and the oxidization products of K. vulgare were sharply increased. For the ability of response to the external environment, B. thuringiensis Bc601 has more two-component system, sporulation coat and peptidoglycan biosynthesis related proteins than B. endophyticus Hbe603. B. thuringiensis is capable of crawling along K. vulgare, indicating that it has a stronger ability to communicate to the external environment and respond to the nutrients surrounding K. vulgare.
Our previous research showed that the sub-cultivated B. thuringiensis and K. vulgare significantly increased the productivity of 2-keto-L-gulonic acid25. By culturing the B. thuringiensis and K. vulgare orthogonally on agar plates, the swarming distance of B. thuringiensis along the trace of K. vulgare decreased after 150 days’ sub-cultivation26. Metabolomic analysis showed that the ability of nutrients searching and intaking was increasing in the evolved B. thuringiensis and it provided more nutrients to K. vulgare. For the ability of nutrients biosynthesis, B. endophyticus Hbe603 has more alpha-galactosidases, propanoate, glutathione and inositol phosphate metabolism, and amino acid degradation related proteins than B. thuringiensis Bc601. B. endophyticus didn’t swarm to K. vulgare, probably due to its adequate metabolic capacity in consortium. The high production of 2-KLG is also connected with the abundant nutrition that B. endophyticus provided.
Although thousands of B. thuringiensis strains were isolated, less than 30 complete genomes were obtained. The complete genome sequencing of B. thuringiensis Bc601 adds a new member in genome library of B. thuringiensis family, and the genomic analysis will give us the opportunity to investigate the diversity and evolution among B. thuringiensis-B. cereus family. B. endophyticus Hbe603 is the first in that species with complete genome, which will provide the important genetic background and molecular information for gene modification. All in all, comparative genomic analysis of B. endophyticus and B. thuringiensis enables us to identify the unique genes for each species and understand the companion mechanism for system optimization.
Methods
Bacteria and cultivation conditions
The growth medium was composed of 2% L-sorbose, 0.3% corn-steep liquor (CSL), 1% peptone, 0.3% yeast extract, 0.3% beef extract, 0.1% urea, 0.1% KH2PO4, 0.02% MgSO4·7H2O and 0.1% CaCO3. The fermentation medium contained 8% L-sorbose, 2% corn-steep liquor (CSL), 1.2% urea, 0.1% KH2PO4, 0.05% MgSO4.7H2O and 0.1% CaCO3. The seed cultures of K. vulgare and Bacillus species were cultivated in 250 mL flasks with 50 mL growth medium (30 °C, 250 rpm) for 24 hours. Subsequently, the two species were co-inoculated into 250 mL flasks with 50 mL fermentation medium at 250 rpm, 30 °C for 96 hours. Swarming motility was observed by culturing the two Bacillus species with K. vulgare orthogonally on agar plates. Bacillus species and K. vulgare were cultivated for 12 hours and 36 hours, respectively, and then inoculated separately in solid sorbose-CSL medium containing 1.5% agar and cultivated at 30 °C for 96 hours.
Analyses of 2-KLG and biomass
The concentration of extracellular 2-KLG were determined by the High Performance Liquid Chromatography (HPLC) (Waters Corp., Massachusetts, USA), equipped with an Aminex HPX-87H column (Bio-Rad, CA) and a refractive index detector. The mobile phase used in the HPLC system was 5 mM H2SO4 at 65 °C with a flow rate of 0.6 mL/min. The cell density was measured as optical density at 600 nm (OD600) with a spectrophotometer after dissolving CaCO3 in 100 mM HCl.
DNA extraction, genome sequencing and assembly
Isolation of genomic DNA was carried out using SDS method. Total DNA obtained was subjected to quality control by agarose gel electrophoresis and quantified by Qubit. DNA was used to construct a 10 kb SMRTbell library, and the genome was sequenced by Single Molecule, Real-Time (SMRT) technology. Sequencing was performed at the Beijing Novogene Bioinformatics Technology Co., Ltd. SMRT Analysis 2.3.0 were used to filter low quality reads and the filtered reads were assembled by SOAPdenovo (http://soap.genomics.org.cn/soapdenovo.html) to generate the complete genome, which has been confirmed by PCR amplification.
Gene prediction, annotation and protein classification
Gene prediction was performed on the genome assembly by GeneMarkS27. Transfer RNAs (tRNAs) were predicted with tRNAscan-SE28, ribosome RNAs (rRNAs) were predicted with rRNAmmer29 and sRNAs were predicted by BLAST against Rfam database30. PHAST31 was used for prophage prediction and CRISPRFinder32 was used for CRISPR identification. A whole genome Blast33 search was performed against KEGG database (Kyoto Encyclopedia of Genes and Genomes)34, COG database (Clusters of Orthologous Groups)35, NR database (Non-Redundant Protein Database) and Swiss-Prot database36. The origin of replication (oriC) and putative DnaA boxes were identified by Ori-Finder37. CVTree was performed for the phylogenetic analysis38 and the phylogenetic tree was generated using the MEGA program39. The GC-Profile was used to compute the GC content variation in DNA sequences and to predict the horizontal gene transfer40. CGView Server was used for the visualization of circular genomes41 and the metabolic network was constructed by KEGG automatic annotation server KAAS42.
Nucleotide sequence accession numbers
The genome sequence of the B. thuringiensis Bc601 has been deposited at DDBJ/EMBL/GenBank under the accession numbers CP015150 (chromosome), CP015151 to CP015156 (six plasmids, respectively). The genome sequence of the B. endophyticus HBe603 has been deposited at DDBJ/EMBL/GenBank under the accession numbers CP011974 (chromosome), CP015323 to CP015330 (eight plasmids, respectively).
Additional Information
How to cite this article: Jia, N. et al. Comparative genomics analysis of the companion mechanisms of Bacillus thuringiensis Bc601 and Bacillus endophyticus Hbe603 in bacterial consortium. Sci. Rep. 6, 28794; doi: 10.1038/srep28794 (2016).
References
Ponomarova, O. & Patil, K. R. Metabolic interactions in microbial communities: untangling the Gordian knot. Curr. Opin. Microbiol. 27, 37–44 (2015).
Hays, S. G., Patrick, W. G., Ziesack, M., Oxman, N. & Silver, P. A. Better together: engineering and application of microbial symbioses. Curr. Opin. Biotechnol. 36, 40–49 (2015).
Jiménez, D. J., Korenblum, E. & van Elsas, J. D. Novel multispecies microbial consortia involved in lignocellulose and 5-hydroxymethylfurfural bioconversion. Appl. Microbiol. Biotechnol. 98, 2789–2803 (2014).
Embree, M., Nagarajan, H., Movahedi, N., Chitsaz, H. & Zengler, K. Single-cell genome and metatranscriptome sequencing reveal metabolic interactions of an alkane-degrading methanogenic community. ISME J. 8, 757–767 (2014).
Maleke, M. et al. Optimization of a bioremediation system of soluble uranium based on the biostimulation of an indigenous bacterial community. Environ. Sci. Pollut. Res. 22, 8442–8450 (2015).
Zhou, K., Qiao, K., Edgar, S. & Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol. 33, 377–383 (2015).
Takagi, Y., Sugisawa, T. & Hoshino, T. Continuous 2-Keto-L-gulonic acid fermentation by mixed culture of Ketogulonicigenium vulgare DSM 4025 and Bacillus megaterium or Xanthomonas maltophilia . Appl. Microbiol. Biotechnol. 86, 469–480 (2010).
Liu, L., Chen, K., Zhang, J., Liu, J. & Chen, J. Gelatin enhances 2-keto-l-gulonic acid production based on Ketogulonigenium vulgare genome annotation. J. Biotechnol. 156, 182–187 (2011).
Fan, S. et al. Development of a minimal chemically defined medium for Ketogulonicigenium vulgare WSH001 based on its genome-scale metabolic model. J. Biotechnol. 169, 15–22 (2014).
Zhou, J. et al. Metabolome profiling reveals metabolic cooperation between Bacillus megaterium and Ketogulonicigenium vulgare during induced swarm motility. Applied. Environ. Microbiol. 77, 7023–7030 (2011).
Jia, N. et al. Insights into mutualism mechanism and versatile metabolism of Ketogulonicigenium vulgare Hbe602 based on comparative genomics and metabolomics studies. Sci. Rep. 6, 23068 (2016).
Yang, W., Han, L., Wang, Z. & Xu, H. Two-helper-strain co-culture system: a novel method for enhancement of 2-keto-l-gulonic acid production. Biotechnol. Lett. 35, 1853–1857 (2013).
Du, J., Zhou, J., Xue, J., Song, H. & Yuan, Y. Metabolomic profiling elucidates community dynamics of the Ketogulonicigenium vulgare–Bacillus megaterium consortium. Metabolomics. 8, 960–973 (2012).
Thompson, D., Regev, A. & Roy, S. Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu. Rev. Cell. Dev. Biol. 31, 399–428 (2015).
Eppinger, M. et al. Genome sequences of the biotechnologically important Bacillus megaterium strains QM B1551 and DSM319. J. Bacteriol. 193, 4199–4213 (2011).
Alcaraz, L. D. et al. Understanding the evolutionary relationships and major traits of Bacillus through comparative genomics. BMC genomics. 11, 332 (2010).
Ivanova, N. et al. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis . Nat. 423, 87–91 (2003).
Ash, C., Farrow, J. A., Dorsch, M., Stackebrandt, E. & Collins, M. D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int. J. Syst. Evol. Microbiol. 41, 343–346 (1991).
Jia, N., Du, J., Ding, M.-Z., Gao, F. & Yuan, Y.-J. Genome Sequence of Bacillus endophyticus and analysis of its companion mechanism in the Ketogulonigenium vulgare-Bacillus strain consortium. PloS one. 10, e0135104 (2015).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 22, 1658–1659 (2006).
Bassi, D. et al. Transcriptome analysis of Bacillus thuringiensis spore life, germination and cell outgrowth in a vegetable-based food model. Food microbiol. 55, 73–85 (2016).
Ma, Q. et al. Integrated proteomic and metabolomic analysis of an artificial microbial community for two-step production of vitamin C. PloS one. 6, e26108 (2011).
Gao, S., Wu, H., Yu, X., Qian, L. & Gao, X. Swarming motility plays the major role in migration during tomato root colonization by Bacillus subtilis SWR01. Biol. Control. 98, 11–17 (2016).
Strehmel, J. et al. Sensor kinase PA4398 modulates swarming motility and biofilm formation in Pseudomonas aeruginosa PA14. Appl. Environ. Microbiol. 81, 1274–1285 (2015).
Zou, Y. et al. Enhancement of 2-keto-gulonic acid yield by serial subcultivation of co-cultures of Bacillus cereus and Ketogulonigenium vulgare . Bioresource Technol. 132, 370–373 (2013).
Ding, M.-Z., Zou, Y., Song, H. & Yuan, Y.-J. Metabolomic analysis of cooperative adaptation between co-cultured Bacillus cereus and Ketogulonicigenium vulgare . PloS one. 9, e94889 (2014).
Besemer, J., L., A. & Borodovsky, M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618 (2001).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 0955–964 (1997).
Lagesen, K. et al. RNammer: consistent annotation of rRNA genes in genomic sequences. Nucleic Acids Res. 35, 3100–3108 (2007).
Gardner, P. P. et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 37, D136–D140 (2009).
Zhou, Y., Liang, Y., Lynch, K. H., Dennis, J. J. & Wishart, D. S. PHAST: a fast phage search tool. Nucleic Acids Res. gkr485 (2011).
Grissa, I., Vergnaud, G. & Pourcel, C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57 (2007).
Altschul, S. F. & Gish, W. Local alignment statistics. Method. Enzymol. 266, 460–480 (1996).
Kanehisa, M. et al. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34, D354–D357 (2006).
Tatusov, R. L. et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 4, 41 (2003).
Magrane, M. & Consortium, U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011, bar009 (2011).
Gao, F. & Zhang, C.-T. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC bioinformatics. 9, 79 (2008).
Xu, Z. & Hao, B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 37, W174–W178 (2009).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Gao, F. & Zhang, C.-T. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 34, W686–W691 (2006).
Grant, J. R. & Stothard, P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, W181–W184 (2008).
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007).
Acknowledgements
This work was funded by the Ministry of Science and Technology of China (“973” Program: 2014CB745102, 2015AA020101), and the National Natural Science Foundation of China (Major Program: 21390203, general program: 31571358). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
F.G., M.-Z.D. and Y.-J.Y. designed the project and experiments; N.J. performed the experiments; F.G. and Y.-J.Y. contributed reagents/materials/analysis tools; N.J., M.-Z.D. and F.G. analyzed the final data and wrote the manuscript. All the authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jia, N., Ding, MZ., Gao, F. et al. Comparative genomics analysis of the companion mechanisms of Bacillus thuringiensis Bc601 and Bacillus endophyticus Hbe603 in bacterial consortium. Sci Rep 6, 28794 (2016). https://doi.org/10.1038/srep28794
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28794
This article is cited by
-
Genomic–proteomic analysis of a novel Bacillus thuringiensis strain: toxicity against two lepidopteran pests, abundance of Cry1Ac5 toxin, and presence of InhA1 virulence factor
Archives of Microbiology (2023)
-
Enhanced 2-keto-l-gulonic acid production by a mixed culture of Ketogulonicigenium vulgare and Bacillus megaterium using three-stage temperature control strategy
Brazilian Journal of Microbiology (2021)
-
Comparative genomic analysis and mosquito larvicidal activity of four Bacillus thuringiensis serovar israelensis strains
Scientific Reports (2020)
-
Enhanced 2-keto-l-gulonic acid production by applying l-sorbose-tolerant helper strain in the co-culture system
AMB Express (2018)
-
Biodegradation of gentamicin by bacterial consortia AMQD4 in synthetic medium and raw gentamicin sewage
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.