Abstract
Novel factors involved in Mycobacteria antibiotics resistance are crucial for better targets to combat the ever-increasing drug resistant strains. Mycobacterium tuberculosis Rv1152, a novel GntR family transcriptional regulator and a promising vancomycin adjuvant target, was firstly characterized in our study. Overexpression of Rv1152 in Mycobacterium smegmatis decreased bacterial susceptibility to vancomycin. Moreover, a deficiency in MSMEG_5174, an Rv1152 homolog made M. smegmatis more sensitive to vancomycin, which was reverted by complementing the MSMEG_5174 deficiency with Rv1152 of M. tuberculosis. Rv1152 negatively regulated four vancomycin responsive genes, namely genes encoding the ribosome binding protein Hsp, small unit of sulfate adenylyltransferase CysD, L-lysine-epsilon aminotransferase Lat and protease HtpX. Taken together, Rv1152 controls the expression of genes required for the susceptibility to vancomycin. This is the first report that links the GntR family transcriptional factor with vancomycin susceptibility. Inhibitors of Rv1152 might be ideal vancomycin adjuvants for controlling multi-drug resistant Mycobacterial infections.
Similar content being viewed by others
Introduction
Tuberculosis, caused by Mycobacterium tuberculosis (M. tuberculosis) infection, remains the second highest pandemic disease with formidable rate of morbidity and mortality worldwide1, particularly in developing countries and HIV co-infected population despite decade’s implementation of TB control programs2. The emergence of multidrug-resistant strains of M. tuberculosis3, extensively drug resistance (XDR), even totally drug resistance (TDR) cases of TB4,5,6 is worsening the TB control. Globally, an estimated 480,000 cases of multidrug-resistant TB (MDR-TB) was reported in 2013. The genus Mycobacterium includes both pathogenic and saprophytic species that are able to survive under environmental stresses, including oxidative and genotoxic stress, hypoxia, nutrient starvation and multiple antibiotics7,8. Transcriptional regulation plays an important role in the bacterial response to environmental stresses. The GntR family of bacterial regulators is named after the Bacillus subtilis transcription regulator GntR, the first characterized transcriptional GntR-type repressor required for gluconate metabolism9,10. This holds true for the GntR family of transcriptional regulators, with around 2000 members in both bacterial and archaea genomes9,11. The proteins belong to GntR family share a characteristic conserved N-terminal domain with winged helix-turn-helix that is involved in DNA binding, which can be easily recognized by a Conserved Domain Database (CDD) search12. GntR consists of six subfamilies differing in C-terminal signaling domains involved in the effector binding11, namely FadR, HutC, MocR, YtrA, AraR and PlmA13,14. GntR regulators are defined as a part of specific subfamily15. The structures of FadR alone and in complex with its effector and operator DNA have been recently determined16,17,18,19, before no structural information is available for other three subfamilies of GntR-like regulators. GntR regulators in M. tuberculosis20 and M. smegmatis21 are just to only bioinformatically predicted, while lacking experimental evidences for the proposed functions.
In this study, we identified a novel M. tuberculosis GntR family regulator, Rv1152, which can alter cell wall permeability of M. smegmatis to acid and surface stress and play an important in vancomycin loss of susceptibility through negatively regulating the genes responsive to vancomycin. In brief, M. smegmatis overexpressed M. tuberculosis Rv1152 (MS_Rv1152) was more resistant to vancomycin than M. smegmatis harboring the vector only (MS_Vec), while the MSMEG_5174 (the homologous gene of Rv1152 in M. smegmatis) deletion mutant (△MSMEG_5174) was more sensitive to vancomycin than the wild type M. smegmatis. More importantly, the susceptibility phenotype of △MSMEG_5174 to vancomycin can be complemented by the Rv1152 (△MSMEG_5174::Rv1152). Several vancomycin responsive genes were down regulated in M. smegmatis overexpressed Rv1152 strain, while the expression of the same set of vancomycin responsive genes was up regulated in homologous gene MSMEG_5174 knock out strains. The genes regulated by Rv1152 are responsible for the sensitivity of M. smegmatis to vancomycin. These data suggest that Rv1152 involved in the loss of susceptibility to vancomycin through negatively regulating the expression of vancomycin responsive genes.
Material and Methods
Strains, Plasmids and Primers
M. smegmatis mc2155 strains were preserved by the Institute of Modern Biopharmaceuticals. Escherichia coli DH5α strain used for gene clone was grown at 37 °C in Luria-Bertani (LB) broth or on LB agar with appropriate antibiotics. M. smegmatis was grown at 37 °C in Middlebrook (MB) 7H9 liquid medium or on MB 7H10 agar supplemented with 0.2% (w/v) glucose, 0.5% (v/v) glycerol and 0.05% (v/v) Tween 80. Hygromycin (100 μg/ml) was added when required. All strains were stored with sterile 20% glycerol at −80 °C for further use. The genomic DNA of M. tuberculosis H37Rv was provided by Chongqing Pulmonary Hospital. The bacterial strains and plasmids used in this study are described in Table 1. All the PCR primers were synthesized by BGI (Shenzhen, Guangdong, China) and the sequences of primers are listed in Table 2.
Construction of overexpression strains
The full length of Rv1152 gene and several vancomycin responsive genes was amplified from M. tuberculosis H37Rv genome DNA using the specific gene primer pairs (Table 2). For Rv1152, The PCR product and the plasmid pALACE were digested with BamHI and ClaI to generate the recombinant pALACE-Rv1152. For the vancomycin responsive genes in M. tuberculosis and their homologous genes in M. smegmatis: including Rv0251c (MSMEG_0424), Rv1285 (MSMEG_4979), Rv0563 (MSMEG_1134) and Rv3290c (MSMEG_1764), the PCR products were ligated into the plasmid pALACE digested by BamH I and NdeI. All plasmids were electroporated into M. smegmatis, a non-pathogenic, fast-growing mycobacterium that, serves as a surrogate model organism to study genes functions of the virulent M. tuberculosis22,23. The electroporated recombinant M. smegmatis strains were plated on Middlebrook (MB) 7H10 agar containing 50 μg/ml hygromycin after in vitro growth in MB 7H9 liquid medium for 3 hour. The positive strains were further verified by Western blot.
Western blot and subcellular localization
Generally, the acetamide-induced recombinant MS_Rv1152 and MS_Vec were sonicated. The whole lysates were centrifuged at the speed of 3,000 × g for 5 min at 4 °C to remove un-lysed cells and cell debris. The supernatants were ultra-centrifuged at the speed of 27,000 × g for 40 min at 4 °C. After ultra-centrifugation, the pellets were considered the cell wall fraction and the supernatants were supposed to be cell membrane and cytosol fractions. The pellets were further suspended in 1 × PBS. Equal amounts of protein from pellet and supernatant fractions were subjected to Western blotting for analyzing the expression of Rv1152. Native M. smegmatis GroEL2, which contains a string of endogenous histidines24, was detected as a cytoplasmic control by an anti-His mouse primary antibody (TIANGEN, China).
Construction of the genetic deletion and complementation strains
An in-frame deletion of the gene MSMEG_5174 (the homolog of M. tuberculosis Rv1152), a GntR-family response regulator of M. smegmatis was constructed using Xer site-specific recombination25. Two DNA fragments, comprising 600 and 700 bp (including the first 50 bp and the last 50 bp of the gene, respectively), were amplified from the genome of M. tuberculosis using specific primers (Table 2) and cloned at the borders of the hygromycin excisable cassette. The resulting DNA fragment was purified and introduced by recombineering into M. smegmatis derivative containing pJV53, a replicative plasmid expressing two phage recombinases and conferring kanamycin resistance26,27. Two hygromycin-resistant colonies were isolated and tested by PCR forthe correct integration of the excisable cassette into the chromosome (not shown). Subsequently, they were grown for 10 generations in the absence of hygromycin and kanamycin to allow excision of the hygromycin cassette and the loss of pJV53. Hygromycin and kanamycin-sensitive colonies were finally recovered at the expected frequency. One of them was analyzed in parallel with a colony of the wild-type parental strain by PCR with primers flanking the region used for the recombination. BGI sequencing confirmed the successful construction of the knockout strain M. smegmatis (△MSMEG_5174).
For the complementation strains, the Rv1152 complemented M. smegmatis strains (△MSMEG_5174::Rv1152) were constructed by integrating M. tuberculosis homolog Rv1152 into the chromosomes of the respective deletion strains. Briefly, the Rv1152 gene was first cloned into a pALACE vector and the recombinant plasmid pALACE_Rv1152 was transformed into the respective M. smegmatis mutant strains. The complementation strain was selected on 7H10 medium (complemented with 0.2% glycerol) containing 50 μg/mL hygromycin and the hygromycin-resistant strains were selected and further confirmed by using Western blot.
In vitro growth of the bacteria under acid and surface stresses
Growth patterns of the two recombinant mycobacteria were examined according to previously described procedures28. Briefly, M. smegmatis was grown overnight in Middlebrook 7H9 medium (complemented with 0.05% Tween 80 and 0.2% glycerol). Recombinant MS_Vec and MS_Rv1152 were grown in presence of surface stress and acidic stress. For surface stress, acetamide-induced MS_Vec and MS_Rv1152 were treated with 0.05% SDS for 1, 2, 3 and 4 h. For acidic stress, HCl was added into the 7H9 medium and adjusted to pH = 4. MS_Rv1152 and MS_Vec were exposed for duration of 3, 6 and 9 h, respectively. After SDS and acidic treatment, the recombinant strains were diluted and plated onto MB 7H10 agar containing hygromycin, the bacteria were counted after 3 days of incubation.
The MIC determinations for antibiotics
Seven antibiotics including vancomycin (Van), norfloxacin (Nor), ciprofloxacin (Cip), ofloxacin (OFL), erythromycin (Ery), isoniazid (INH) and rifampicin (Rif) were used in this study. Growth patterns of the wild type M. smegmatis strain (WT), the gene deletion mutant (△MSMEG_5174) and complementation strains (△MSMEG_5174::Rv1152), the overexpression strain (MS_Rv1152) and the control strain (MS_Vec) were measured according to the procedures described previously with minor modification23,29. The MICs of antibiotics were determined by using serial two-fold dilution of the antibiotics in 7H9 medium as previously described30. Briefly, wells of a 96 well microtiter plate were filled with 100 μl of 7H9 medium. The required highest antibiotic concentration was prepared and 200 μl were added to the first vial. This was serially diluted to halve the concentration by mixing with equal volume of bacterial culture in the subsequent wells till second to the last well. Last vial was the control without antibiotics. The M. smegmatis strains were grown in replicates in 7H9 medium to an OD600 of 0.8, 1% of original bacteria was inoculated to 100 μl of the prepared culture with or without antibiotics. MIC values of each antibiotic were determined as drug concentration that inhibited bacterial growth by at least 99%.
Determination of bactericidal effect of antibiotics
To determine mycobacterial growth curves and the effect of antibiotics, M. smegmatis strains including WT, MS_Vec, MS_Rv1152, △MSMEG_5174 and △MSMEG_5174::Rv1152 were grown overnight in Middlebrook 7H9 broth (supplemented with 0.05% Tween80 and 0.2% glycerol). Hygromycin was not added in the 7H9 medium when assaying antibiotics resistance of all strains. When cells entered a stationary growth phase (OD600 between 0.8–1.0), each culture was 100-fold diluted in 100 μl of fresh 7H9 broth containing the indicated concentration of each antibiotic. The cultures were then allowed to grow further at 37 °C with shaking at 110 rpm. After 24 h treatment with these antibiotics with different concentration, the bacteria were diluted by 10-fold and plated into 7H10 agar medium. The bacterial numbers were counted after 3 days culture. The medium without any antibiotics serves as the control to make sure the normal growth of bacteria.
RT-PCR detection of the gene transcription
Isolation of mRNA and cDNA preparation were performed from wild type strains (WT), MSMEG_5174 deletion mutants (△MSMEG_5174), Rv1152 complementation strains (△MSMEG_5174::Rv1152), M. smegmatis harboring pALACE (MS_Vec) and Rv1152 overexpression M. smegmatis strains (MS_Rv1152). RT-PCR was used to compare the transcriptional levels of genes expression using gene specific primers and the real-time PCR analysis was subsequently carried out according to previously described procedures31. The reactions were performed in a RT-PCR machine (Bio-Rad IQ5) under the following thermocycling parameters: 95 °C for 5 min and 40 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. Amplification specificity was assessed using melting curve analysis. Gene expression levels were normalized to the levels of sigA gene transcription. Average relative expression levels and standard deviations were determined from three independent experiments. All the gene specific primers used for RT-PCR were listed in the Table 1S.
Statistical analysis
The experiments were performed in triplicate. Differences between groups were analyzed by using Prism 6 and Student’s t test. ***P < 0.001, **P < 0.01, *P < 0.05, means ± SEM from at least three biological replicates.
Results
M. smegmatis overexpressing Rv1152 showed altered response to stresses
To study the function of Rv1152, we constructed the recombinant M. smegmatis harboring pALACE_Rv1152 (MS_Rv1152) and vector only (MS_Vec). Rv1152 gene was successfully amplified from the M. tuberculosis genome by using gene specific primers (Fig. 1A). Western blot showed the presence of the expressed 14 kDa his-tagged in MS_Rv1152 but not in MS_Vec (Fig. 1B), suggesting that Rv1152 was successfully expressed in M. smegmatis. Rv1152 localized to both cell wall and cytoplasm of M. smegmatis: the target proteins were found in cell wall and cytoplasm of MS_Rv1152, but not in MS_Vec while the cytoplasm marker GroEL2 of M. smegmatis was detected in both MS_Rv1152 and MS_Vec (Fig. 1C).
The ability of M. tuberculosis to survive within the host requires resistance to various physiological and environmental stresses. Within granuloma, M. tuberculosis is able to persist for years even under severe stresses such as hypoxia, nutrient limitation, reactive oxygen and nitrogen intermediates, low pH, alveolar surfactants and free fatty acids32. However, bacterial factors that enhanced their survival under hash condition remain poorly understood. Mycobacterial responses to cell surface stress are of particular interest for understanding the pathogenesis of M. tuberculosis and its susceptibility to antibiotics33. The recombinant MS_Rv1152 showed more cell death than MS_Vec when the bacteria were exposed to acid stress (Fig. 2A). Although there was a rapid decrease in the bacterial numbers for all tested strains exposed to the detergent SDS, in comparison with MS_Vec, MS_Rv1152 was more tolerant to SDS: the percentage survival values were 5% for the MS_Rv1152 and 1.5% for the MS_Vec. The greater tolerant of the MS_Rv1152 to SDS was verified when bacterial survival was tested after 1 hour when incubation with SDS (Fig. 2B). These results suggest that the overexpression of Rv1152 in M. smegmatis modified the M. smegmatis response to surface and acid stress.
Effect of Rv1152 overexpression on acid and surface stress tolerance.
(A) Survival of recombinant MS_Vec and MS_Rv1152 after 0, 3, 6 and 9 h treatment with low pH (pH = 4). (B) Survival of recombinant MS_Vec and MS_Rv1152c after exposure to 0.05% sodium dodecyl sulfonate (SDS) for 0, 1, 2 and 4 hours.
Rv1152 confers M. smegmatis reduced susceptibility to vancomycin
Vancomycin (Van), the last-resort antibiotics against infections caused by meticillin-resistant Staphylococcus aureus (MRSA)34, is a glycopeptide blocking the transpeptidation and nascent peptidoglycan synthesis. Van, in combination with lipid biosynthesis targeting antibiotic, was recently found more effective in killing multidrug-resistant (MDR) and extensively-drug resistant (XDR) M. tuberculosis35. Interestingly, Rv1152 decreased susceptibility of M. smegmatis to vancomycin, as the MIC of MS_Rv1152 for vancomycin is 80 μg/ml while MS_Vec is 20 μg/ml, there is no significant difference in MIC when using Cip, OFL, Nor, Ery, INH and Rif (Table 3). In comparison with MS_Vec group, MS_Rv1152 is less susceptible to vancomycin even at the concentrations of 5 μg/ml. No bacteria of MS_Vec were detected on the plate supplemented with 20 μg/ml vancomycin (Fig. 3A). In addition, the obvious difference in survival of bacteria between MS_Rv1152 and MS_Vec were detected after treatment with vancomycin with various concentrations of for 24 h: there was about 1.3 × 105 (CFU/ml) of MS_Rv1152 survived while no colony of MS_Vec can be detected when 80 μg/ml vancomycin was used (Fig. 3B). These data suggest that overexpression of Rv1152 in M. smegmatis contributes to reduced susceptibility to vancomycin.
Overexpression of Rv1152 in M. smegmatis decreases vancomycoin susceptibility.
(A) Ten-fold serial dilutions of MS_Rv1152 and MS_Vec were plated onto 7H10 agar supplemented with 0, 5, 10 and 20 μg/ml vancomycin. The plate without any antibiotic served as the control to confirm the normal growth of bacteria. (B) MS_Rv1152 and MS_Vec were spotted on the 7H10 plate without any antibiotic after 24 hour treatment with 0, 5, 10, 20, 40, 80 μg/ml vancomycin, respectively. The bacteria numbers were counted after 3 days of incubation.
The isogenic deletion of MSMEG_5174 increased susceptibility to vancomycin
In order to further study the role of Rv1152 in vancomycin susceptibility, the homologous gene of Rv1152 in M. smegmatis, MSMEG_5174, was deleted (△MSMEG_5174). As shown in Fig. 4A, the sizes of the fragments amplified from the two colonies were consistent with expectations (300 bp for the mutant and 657 bp for the WT). The fragment amplified from the mutant bacteria was sequenced to verify the correctness of the excision: as expected, a copy of the DIF sequence flanked by two BglII restriction sites was found in the correct position, replacing the 247 central nucleotides of the target gene (Fig. 4B). Rv1152 was successfully expressed in △MSMEG_5174 after transformation Rv1152 gene into △MSMEG_5174 strain (Fig. 4C). The MSMEG_5174 gene knockout strain is more sensitive to vancomycin than the wild type M. smegmatis: most bacteria of △MSMEG_5174 were inhibited when using vancomycin at the concentration of 5 μg/ml and this sensitive phenotype was complemented in △MSMEG_5174::Rv1152 strain. No significant difference between WT and △MSMEG_5174 was detected when using other antibiotics (Table 4). The same result was reiterated when the strains were treated with different concentration of vancomycin (Fig. 5): with the increasing of the concentration of vancomycin, MSMEG_5174 deleted mutant are more sensitive than the WT strain, no bacteria of △MSMEG_5174 strains were detected, while there were obvious WT bacteria survived when using vancomycin at the concentration of 40 μg/ml.
Construction of the △MSMEG_5174 and △MSMEG_5174::Rv1152 strains.
(A) confirmation of MSMEG_5174 gene knockout by PCR amplification from WT and △MSMEG_5174 strains. M, Marker DL10000; 1, PCR band from △MSMEG_5174 strain; 2, PCR band from WT strain. (B) A map of the sequencing of PCR amplification products and DNAMAN alignment showing 28 bp DIF in △MSMEG_5174 strain. (C) Western blot showed that the Rv1152 gene was expressed in △MSMEG_5174 strain.
Rv1152 negatively regulates the expression of vancomycin responsive genes
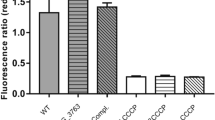
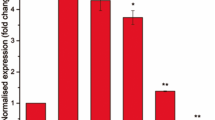
M. smegmatis has been widely used as a surrogate model bacterium to study the gene regulation and signal transduction mechanism of pathogenic mycobacteria23,36. The vancomycin responsive genes that transcriptionally regulated by Rv1152, together with their corresponding homologous genes in M. smegmatis were summarized in Table 5. The genome-wide regulator-DNA interaction network of M. tuberculosis H37Rv has shown that 68 genes are regulated by Rv115237. Global transcriptome has shown that several M. tuberculosis genes are differentially regulated upon exposure to vancomycin38. To define the molecular mechanisms of the role of Rv1152 in vancomycin susceptibility, we tested whether vancomycin responsive genes are regulated by Rv1152. In comparison with WT strains, four genes including heat stress induced ribosome binding protein (MSMEG_0424), L-lysine-epsilon aminotransferase (MSMEG_1764), small unit of sulfate adenylyltransferase (MSMEG_4979) and protease transmembrane protein heat shock protein (MSMEG_1134) were up-regulated in △MSMEG_5174 strain, such expression pattern was restored in the Rv1152 complementary strain (Fig. 6A). The relative expression of these four genes to sigA was also up regulated after the homologous gene MSMEG_5174 in M. smegmatis was knock out (Fig. 6B), suggesting that Rv1152 negatively regulated the expression of these four genes. There was no significant difference for other genes listed in the Table 5 among WT, △MSMEG_5174 and △MSMEG_5174::Rv1152 strains (data not shown). We further identified Rv1152 inhibited the expression of these four target genes in M. smegmatis (Fig. 6C). These data suggest that Rv1152 represses the expression of the four vancomycin responsive genes.
Transcription of genes responsive to vancomycin and regulated by Rv1152.
(A) Semi-quantitative RT-PCR. (B) qRT-PCR assay of the relative transcription of target genes in WT, △MSMEG_5174 and △MSMEG_5174::Rv1152. (C) qRT-PCR assay the relative transcription of target genes in MS_Vec and MS_Rv1152 strains.
Rv1152 regulated genes affect susceptibility to vancomycin
To determine whether Rv1152 regulated genes affect susceptibility to vancomycin, the recombinant strains expressing vancomycin responsive genes from M. tuberculosis and their homologous genes from M. smegmatis were constructed (Fig. 1S). The MIC for vancomycin of recombinant strains MS_Lat, MS_Rv1285, MS_Rv0251 and MS_Rv0563 was lower than MS_Vec, similar results were obtained from the M. smegmatis homologous gene recombinant strains (Table 2S). We have recently identified a lysine-epsilon aminotransferase (Lat) encoded by Rv3290c was involved in persister formation39. Interestingly, M. smegmatis overexpressing Lat exhibited more susceptibility to vancomycin than the wild type carrying the control vector –MS_Lat showed a greater bacterial cell death than MS_Vec (Fig. 7A). After 48 hour treatment with high concentration vancomycin, no bacteria was detectable (detection limit of 100 bacteria/ml) for MS_Lat while many colonies were recovered with MS_Vec control (Fig. 7B). These results demonstrate that overexpression of Lat in M. smegmatis make the host strain more sensitive to vancomycin. Overexpressing of another three vancomycin responsive genes (Rv_1285, Rv_0251 and Rv0563) from M. tuberculosis and their homologous genes (MSMEG_4979, MSMEG_0424 and MSMEG_1134) from M. smegmatis decreased susceptibility of host bacteria to vancomycin (Fig. 8). These data suggest that Rv1152 negatively regulated genes are responsive to vancomycin, resulting in decreased susceptibility of host bacteria to vancomycin.
Overexpression of Lat in M. smegmatis sensitizes vancomycin susceptibility.
(A) Log change in colony-forming units (CFU)/ml, from time zero, of MS_Vec (Log Cfu/ml = 7.2999) and MS_Lat (Log Cfu/ml = 7.4926) after treatment for 24 hours with different concentrations of vancomycin. (B) Killing curves of MS_Lat and MS_Vec upon 50 μg/ml vancomycin exposure for the indicated times. ♦untreated MS_Vec; ◾untreated MS_Lat; ▴antibiotic-treated MS_Vec; ⦁antibiotic-treated MS_Lat.
Overexpression of genes regulated by Rv1152 sensitizes vancomycin susceptibility.
(A) Ten-fold serial dilutions of MS_Vec, MS_Rv1285, MS_Rv0251, MS_Rv0563 plated onto 7H10 agar medium supplemented with 2.5 and 10 μg/ml vancomycin, respectively. (B) Ten-fold serial dilutions of MS_Vec, MS_MSMEG_0424, MS_MSMEG_4979 and MS_MSMEG_1134 plated onto 7H10 agar medium supplemented with 2.5 or 5 μg/ml vancomycin, respectively. The plate without any antibiotic served as the control to confirm the normal growth of bacteria.
Discussion
The unusual ability of M. tuberculosis to persist for years within human host and the requirement for lengthy antibiotic combination regimens to eliminate drug sensitive strains are hallmarks of this recalcitrant pathogen. Its prolonged chronic infection requires the expression of a complex array of genetic determinants, including those involved in secondary metabolism, cell wall processes, stress responses and signal transduction40. Transcriptional regulators are well-established key players for the mycobacteria adaptation, such as two-component regulatory systems, stress-responsive sigma factors41 including sigB, sigE, and sigH42,43,44. We expressed GntR-like family transcriptional regulator Rv1152 in M. smegmatis and found that Rv1152 localized to both cell wall and cytoplasm. The up-regulation of Rv1152 after 96 h of starvation prompted us to test whether recombinant MS_Rv1152 will affect the permeability to these antimicrobial factor, the growth characteristics of MS_Vec and MS_Rv1152 under acid and surface stress were analyzed, as the transcription of sigB depends on SigE under physiological conditions and following exposure to SDS41. Our data identified that overexpression of Rv1152 in M. smegmatis has no effect on the bacterial growth in standard culture conditions (Fig. 2S), while Rv1152 can modify the response of M. smegmatis to different stresses. Rv1152 enhanced tolerance of M. smegmatis to SDS, but it sensitized bacteria to acid stress.
Mycobacterial responses to stress are of particular interest to understand the pathogenesis of M. tuberculosis and its sensitivity and reaction to antibiotics33. The detailed pathways, signals, regulatory responses and molecular interactions are not yet well understood, despite of the existence of 190 in silico predicted transcription regulators in the M. tuberculosis genome45. The genus Mycobacterium includes both pathogenic and saprophytic species that are able to survive under environmental stresses, including oxidative and genotoxic stress, hypoxia, nutrient starvation and multiple antibiotics7,8. We suspected that Rv1152 might be involved in vancomycin susceptibility since the expression of Rv1152 was up-regulated under vancomycin exposure38. In congruent with our expectation, the expression of Rv1152 in M. smegmatis host bacteria loss of susceptibility to vancomycin. No difference was detected between MS_Vec and MS_Rv1152 in susceptibility to the other six antibiotics including norfloxacin, ciprofloxacin, ofloxacin, erythromycin, isoniazid and rifampicin. To elucidate the molecular mechanism underlying Rv1152 vancomycin loss of susceptibility. The MSMEG_5174 knock-out strain (△MSMEG_5174) was constructed, MSMEG_5174, the M. smegmatis homolog of Rv1152, bearing the signature of the YtrA subfamily member21. In comparison with the parental strain, MSMEG_5174 gene knock out has little effect on the growth of M. smegmatis (Fig. 3S). △MSMEG_5174 is more susceptible to vancomycin than the parental M. smegmatis, suggesting the involvement of MSMEG_5174 in the tolerance to vancomycin. In addition, the response of Rv1152 complemented △MSMEG_5174 strain to vancomycin was restored to the level of parental M. smegmatis. Taken together, our data indicate that Rv1152 plays an important role in vancomycin loss of susceptibility.
M. tuberculosis transcriptome alteration in response to vancomycin38 was mined for genes regulated by Rv1152 and involved in vancomycin loss of susceptibility. We found that the expression of four vancomycin responsive genes were down-regulated in Rv1152 overexpression M. smegmatis, including ribosome binding protein (MSMEG_0424), small unit of sulfate adenylyltransferase (MSMEG_4979), L-lysine-epsilon aminotransferase (MSMEG_1764) and protease HtpX (MSMEG_1134), respectively. Their expression was also increased in response to vancomycin treatment. The expression of these four genes was increased in △MSMEG_5174 and the complementary strains △MSMEG_5174::Rv1152 showed the restored levels of transcription for these four genes. We further found these four genes are responsible for susceptibility to vancomycin. In summary, our data demonstrate that M. tuberculosis Rv1152 plays a role in vancomycin loss of susceptibility via negatively regulating the expression of genes responsive to vancomycin. Vancomycin is a robust glycopeptide antibiotic against multiple drug resistant clinical strains. This is the first report of Mycobacteria GntR family transcriptional factor involved in vancomycin loss of susceptibility. Further discovery of inhibitors against Rv1152 may provide good adjuvants for vancomycin or other antibiotics targeting the cell wall biosynthesis.
Additional Information
How to cite this article: Zeng, J. et al. Mycobacterium tuberculosis Rv1152 is a Novel GntR Family Transcriptional Regulator Involved in Intrinsic Vancomycin Resistance and is a Potential Vancomycin Adjuvant Target. Sci. Rep. 6, 28002; doi: 10.1038/srep28002 (2016).
References
Dye, C., Williams, B. G., Espinal, M. A. & Raviglione, M. C. Erasing the world's slow stain: strategies to beat multidrug-resistant tuberculosis. Science 295, 2042–2046, doi: 10.1126/science1063814 (2002).
Corbett, E. L. et al. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med 163, 1009–1021, doi: 10.1001/archinte.163.9.1009 (2003).
Matteelli, A. et al. Multidrug-resistant and extensively drug-resistant Mycobacterium tuberculosis: epidemiology and control. Expert Rev Anti-infe 5, 857–871, doi: 10.1586/14787210.5.5.857 (2007).
Gandhi, N. R. et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368, 1575–1580, doi: 10.1016/S0140-6736(06)69573-1 (2006).
Velayati, A. A. et al. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in iran. Chest 136, 420–425, doi: 10.1378/chest.08-2427 (2009).
Shah, N. S. et al. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis 13, 380–387, doi: 10.3201/eid1303.061400 (2007).
Zahrt, T. C. & Deretic, V. Reactive nitrogen and oxygen intermediates and bacterial defenses: unusual adaptations in Mycobacterium tuberculosis. Antioxid Redox Sign 4, 141–159, doi: 10.1089/152308602753625924 (2002).
Hingley-Wilson, S. M., Sambandamurthy, V. K. & Jacobs, W. R., Jr. Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nat Immunol 4, 949–955, doi: 10.1038/ni981 (2003).
Haydon, D. J. & Guest, J. R. A new family of bacterial regulatory proteins. FEMS Microbiol Lett 63, 291–295 (1991).
Fujita, Y. & Fujita, T. The gluconate operon gnt of Bacillus subtilis encodes its own transcriptional negative regulator. P Natl Acad Sci USA 84, 4524–4528 (1987).
Rigali, S., Derouaux, A., Giannotta, F. & Dusart, J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR and YtrA subfamilies. J Biol Chem 277, 12507–12515, doi: 10.1074/jbc.M110968200 (2002).
Marchler-Bauer, A. et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res 33, D192–196, doi: 10.1093/nar/gki069 (2005).
Rigali, S. et al. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res 32, 3418–3426, doi: 10.1093/nar/gkh673 (2004).
Lee, M. H., Scherer, M., Rigali, S. & Golden, J. W. PlmA, a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J Bacteriol 185, 4315–4325 (2003).
Casali, N., White, A. M. & Riley, L. W. Regulation of the Mycobacterium tuberculosis mce1 operon. J Bacteriol 188, 441–449, doi: 10.1128/JB.188.2.441-449.2006 (2006).
van Aalten, D. M., DiRusso, C. C., Knudsen, J. & Wierenga, R. K. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J 19, 5167–5177, doi: 10.1093/emboj/19.19.5167 (2000).
Xu, Y., Heath, R. J., Li, Z., Rock, C. O. & White, S. W. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J Biol Chem 276, 17373–17379, doi: 10.1074/jbc.M100195200 (2001).
Yousuf, S., Angara, R., Vindal, V. & Ranjan, A. Rv0494 is a starvation-inducible, auto-regulatory FadR-like regulator from Mycobacterium tuberculosis. Microbiology 161, 463–476, doi: 10.1099/mic.0.000017 (2015).
Biswas, R. K. et al. Identification and characterization of Rv0494: a fatty acid-responsive protein of the GntR/FadR family from Mycobacterium tuberculosis. Microbiology 159, 913–923, doi: 10.1099/mic.0.066654-0 (2013).
Vindal, V., Ranjan, S. & Ranjan, A. In silico analysis and characterization of GntR family of regulators from Mycobacterium tuberculosis. Tuberculosis 87, 242–247, doi: 10.1016/j.tube.2006.11.002 (2007).
Vindal, V., Suma, K. & Ranjan, A. GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC genomics 8, 289, doi: 10.1186/1471-2164-8-289 (2007).
Goude, R. & Parish, T. Electroporation of mycobacteria. J Vis Exp, doi: 76110.3791/761 (2008).
Yang, M., Gao, C., Wang, Y., Zhang, H. & He, Z. G. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PloS one 5, e10672, doi: 10.1371/journal.pone.0010672 (2010).
Li, W. et al. Mycobacterium tuberculosis Rv3402c Enhances Mycobacterial Survival within Macrophages and Modulates the Host Pro-Inflammatory Cytokines Production via NF-Kappa B/ERK/p38 Signaling. PLoS One 9, e94418, doi: 10.1371/journal.pone.0094418 PONE-D-13-36639 (2014).
Cascioferro, A. et al. Xer site-specific recombination, an efficient tool to introduce unmarked deletions into mycobacteria. Appl Environ Microb 76, 5312–5316, doi: 10.1128/AEM.00382-10 (2010).
Bardarov, S. et al. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017 (2002).
van Kessel, J. C. & Hatfull, G. F. Recombineering in Mycobacterium tuberculosis. Nat Methods 4, 147–152, doi: 10.1038/nmeth996 (2007).
Zhang, L., Li, W. & He, Z. G. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem 288, 3085–3096, doi: 10.1074/jbc.M112.428110 (2013).
Zhang, H. et al. A novel marRAB operon contributes to the rifampicin resistance in Mycobacterium smegmatis. PloS one 9, e106016, doi: 10.1371/journal.pone.0106016 (2014).
Ribeiro, A. L. et al. Analogous mechanisms of resistance to benzothiazinones and dinitrobenzamides in Mycobacterium smegmatis. PloS one 6, e26675, doi: 10.1371/journal.pone.0026675 (2011).
Wang, Y., Huang, Y., Xue, C., He, Y. & He, Z. G. ClpR protein-like regulator specifically recognizes RecA protein-independent promoter motif and broadly regulates expression of DNA damage-inducible genes in mycobacteria. J Biol Chem 286, 31159–31167, doi: 10.1074/jbc.M111.241802 (2011).
Manganelli, R. et al. Sigma factors and global gene regulation in Mycobacterium tuberculosis. J Bacteriol 186, 895–902 (2004).
McKinney, J. D. In vivo veritas: the search for TB drug targets goes live. Nat Medicine 6, 1330–1333, doi: 10.1038/82142 (2000).
Hiramatsu, K. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect Dis 1, 147–155, doi: 10.1016/S1473-3099(01)00091-3 (2001).
Soetaert, K. et al. Increased vancomycin susceptibility in mycobacteria: a new approach to identify synergistic activity against multi-drug resistant mycobacteria. Antimicrob Agents Ch, doi: 10.1128/AAC.04856-14 (2015).
Li, Y., Zeng, J., Zhang, H. & He, Z. G. The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC microbiology 10, 242, doi: 10.1186/1471-2180-10-242 (2010).
Zeng, J., Cui, T. & He, Z. G. A genome-wide regulator-DNA interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J Proteome Res 11, 4682–4692, doi: 10.1021/pr3006233 (2012).
Provvedi, R., Boldrin, F., Falciani, F., Palu, G. & Manganelli, R. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155, 1093–1102, doi: 10.1099/mic.0.024802-0 (2009).
Duan, X. et al. Mycobacterium Lysine epsilon-aminotransferase is a novel alarmone metabolism related persister gene via dysregulating the intracellular amino acid level. Sci Rep 6, 19695, doi: 10.1038/srep19695 (2016).
Clark-Curtiss, J. E. & Haydel, S. E. Molecular genetics of Mycobacterium tuberculosis pathogenesis. Annu Rev Microbiol 57, 517–549, doi: 10.1146/annurev.micro.57.030502.090903 (2003).
Manganelli, R., Voskuil, M. I., Schoolnik, G. K. & Smith, I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 41, 423–437, doi: mmi2525 (2001).
Fisher, M. A., Plikaytis, B. B. & Shinnick, T. M. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol 184, 4025–4032 (2002).
He, H. & Zahrt, T. C. Identification and characterization of a regulatory sequence recognized by Mycobacterium tuberculosis persistence regulator MprA. J bacteriol 187, 202–212, doi: 10.1128/JB.187.1.202-212.2005 (2005).
Raman, S. et al. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J Bacteriol 183, 6119–6125, doi: 10.1128/JB.183.20.6119-6125.2001 (2001).
Bishai, W. The Mycobacterium tuberculosis genomic sequence: anatomy of a master adaptor. Trends Microbiol 6, 464–465 (1998).
Acknowledgements
This work was supported byNational Natural ScienceFoundation [grant numbers 81371851, 81511120001, 81071316, 81271882, 81301394], New Century Excellent Talents in Universities[grant number NCET-11-0703], National Megaprojects for KeyInfectious Diseases [grant numbers 2008ZX10003-006], Excellent Ph.D. thesis fellowship of Southwest University [grantnumbers kb2010017, ky2011003], the Fundamental Research Fundsfor the Central Universities [grant numbers XDJK2011D006, XDJK2012D011, XDJK2012D007, XDJK2013D003, XDJK2014D040], Graduate research and innovation project of graduate in Chongqing (CYS14044), The Chongqing Municipal Committee of Education for postgraduates excellence program [grant number YJG123104] and The undergraduates teaching reform program [grant number 2013JY201].
Author information
Authors and Affiliations
Contributions
The experiments were conceived and designed by J.Z., W.D. and J.X. Most experiments conducted by J.Z. and W.D., they contributed equally to this paper. W.Y., H.L., X.D., L.X., P.L., R.W., T.F. and A.E.A. performed several experiments. This paper was written by J.Z., W.D. and J.X. All authors have read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zeng, J., Deng, W., Yang, W. et al. Mycobacterium tuberculosis Rv1152 is a Novel GntR Family Transcriptional Regulator Involved in Intrinsic Vancomycin Resistance and is a Potential Vancomycin Adjuvant Target. Sci Rep 6, 28002 (2016). https://doi.org/10.1038/srep28002
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep28002
This article is cited by
-
Novel role of PE_PGRS47 in the alteration of mycobacterial cell wall integrity and drug resistance
Archives of Microbiology (2023)
-
Mycobacterium tuberculosis toxin Rv2872 is an RNase involved in vancomycin stress response and biofilm development
Applied Microbiology and Biotechnology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.