Abstract
Bacillus subtilis ATCC 6051a is an undomesticated strain used in the industrial production of enzymes. Because it is poorly transformable, genetic manipulation in this strain requires a highly efficient genome editing method. In this study, a Streptococcus pyogenes CRISPR/Cas9 system consisting of an all-in-one knockout plasmid containing a target-specific guide RNA, cas9 and a homologous repair template was established for highly efficient gene disruption in B. subtilis ATCC 6051a. With an efficiency of 33% to 53%, this system was used to disrupt the srfC, spoIIAC, nprE, aprE and amyE genes of B. subtilis ATCC 6051a, which hamper its use in industrial fermentation. Compared with B. subtilis ATCC 6051a, the final mutant, BS5 (ΔsrfC, ΔspoIIAC, ΔnprE, ΔaprE, ΔamyE), produces much less foam during fermentation, displays greater resistant to spore formation and secretes 2.5-fold more β-cyclodextrin glycosyltransferase into the fermentation medium. Thus, the CRISPR/Cas9 system proved to be a powerful tool for targeted genome editing in an industrially relevant, poorly transformable strain.
Similar content being viewed by others
Introduction
Bacillus subtilis, a well-characterized gram-positive bacterium, has been widely used for the production of heterologous proteins. This species and some of its close relatives have excellent protein secretory capability and are generally recognized as safe (GRAS), making them important hosts for the production of antibiotics, medicinal proteins and industrial enzymes. B. subtilis 168 is a model laboratory strain that carries many mutations that have occurred during its modification via irradiation and selection1. These modifications have made the organism tryptophan-deficient and improved its transformablility. The B. subtilis strains commonly used for recombinant protein production, such as WB600 and WB800, were constructed on the basis of B. subtilis 1682.
Because the recombinant protein productivity of B. subtilis ATCC 6051a is superior to that of B. subtilis 168, it has been widely applied to the production of industrial enzymes3,4. However, B. subtilis ATCC 6051a has some undomesticated properties that hamper the extracellular production of recombinant proteins. In particular, it can produce large amounts of foam, highly resistant spores, multiple types of extracellular protease and high level of amylase during fermentation, which related to srfC5, spoIIAC6, nprE7, aprE8 and amyE9, respectively. To improve the usefulness of this important strain, we sought to modify these properties by inactivating the five genes. B. subtilis ATCC 6051a is poorly transformable, compared with laboratory strains, because it harbours an 84-kb endogenous plasmid pBS32, which encodes a single-pass trans-membrane protein ComI that inhibits the competence of DNA uptake10. Due to its poor competence, the genetic manipulation of B. subtilis ATCC 6051a is difficult and require a highly efficient genome editing method. The genome sequence of B. subtilis ATCC 6051a was recently determined by Jeong et al.11, which facilitates genetic manipulation.
Clustered regularly interspaced short palindromic repeat (CRISPR) systems, which are composed of CRISPR RNAs (crRNA), trans-activating CRISPR RNAs (tracrRNAs) and CRISPR-associated (Cas) proteins constitute an immune system in bacteria and archaea that efficiently cleaves foreign DNA entering the cell, including phages and plasmids12. Some CRISPR/Cas systems require multiple proteins13, whereas the type II CRISPR/Cas system requires a single nuclease; Cas protein 9 (Cas9). In the widely used Streptococcus pyogenes type II CRISPR/Cas system14, the 20-bp complementary region (N20) within the crRNA guide Cas9 nuclease to its specific target, a roughly 20-nt sequence known as the protospacer, which contains a specific protospacer-adjacent motif (PAM) at its 3′ end15. The PAM sequence leads cas9 to create a double-strand break at protospacer (target) sequence14,16 and it was repaired through homologous recombination using a repair template that is supplied along with the CRISPR/Cas9 system17. Recently, a single chimeric guide RNA (sgRNA) containing features of both crRNA and tracrRNA has been developed18, which simplify the genome editing design. As an efficient genome editing technology, the type II CRISPR/Cas9 system has been proved to be feasible in point mutation, single gene deletion/insertion and large-size gene cluster deletion19. And until recently, it has been widely applied in various organisms including, but not restricted to, Escherichia coli14, Streptococcus pneumonia14, Saccharomyces cerevisiae20, Lactobacillus reuteri21, Bombyx mori22, Drosophila23, mice24 and humans cell lines17.
Although there have been several successful implementations of the CRISPR/Cas9 system in microbial systems, there have not been any reports of genome editing in B. subtilis using a CRISPR/Cas9 system. This study describes the establishment and optimization of a CRISPR/Cas9 system in B. subtilis ATCC 6051a. We used this system to disrupt five genes in the B. subtilis ATCC 6051a genome (srfC, spoIIAC, nprE, aprE and amyE) that hamper its use during industrial fermentation. Compared with B. subtilis ATCC 6051a, this mutant strain, named BS5, produces much less foam at the level of control, exhibits great resistance to spore formation and secretes 2.5 times more β-cyclodextrin glycosyltransferase (β-CGTase) into the fermentation medium. β-CGTase is widely used in the production of β-cyclodextrin and mainly produced from wild strains and E. coli25, which exist low secretion and food safety problem, respectively. The increase of β-CGTase secretion from Bacillus subtilis can largely reduce the cost of β-cyclodextrin production, which shows the high industrial value of BS5.
Results
Construction of the CRISPR/Cas9 system all in one plasmid
We assembled a complete CRISPR/Cas9 system in a single knockout plasmid that could be used for highly efficiently genome editing in B. subtilis (Fig. 1a). The six knockout plasmids (pHYcas9dsrf1, pHYcas9dsrf2, pHYcas9dspo, pHYcas9dnpr, pHYcas9dapr and pHYcas9damy), which originated from plasmid pHY300PLK-β-CGTase, consist of the cas9 gene amplified from plasmid pwtcas9-bacterial, a sgRNA and its promoter P43, the temperature-sensitive replicon PE194 and a homologous repair template. The synthesis of Cas9 protein in vivo was driven by the α-amylase promoter (PamyQ), which originates from B. amyloliquefaciens. The P43 promoter is a constitutively expressed promoter26 that can drive strong transcription of the sgRNA in B. subtilis. The homologous repair template was obtained through overlap extension PCR of regions upstream and downstream of the target locus. The length of the upstream and downstream regions ranged from 450 to 550 bp and they introduced an Xho I site into the target locus. The knockout plasmids and plasmid pHY300PLK-β-CGTase encode ampicillin resistance in E. coli and tetracycline resistance in both E. coli and B. subtilis.
CRISPR/Cas9 system plasmids, homologous repair template and editing procedure.
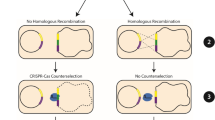
(a) Relevant features of pHY300PLK-β-CGTase and the knockout plasmids. p15A ori, E. coli replication origin; repB, B. subtilis replication origin; PE194, B. subtilis temperature-sensitive replication origin; AmpR, ampicillin-resistance marker; TcR, tetracycline resistance marker; PamyR, promoter of ampicillin-resistance marker; PamyQ, α-amylase promoter from B. amyloliquefaciens; β-CGTase, β-CGTase encoding gene; cas9, Cas9 encoding gene; sgRNA, target-specific guide RNA; N20, 20-bp complementary region; repair template, homologous repair template obtained by overlap PCR. (b) Diagram showing the design of a 934 bp repair template for the specific target within srfC. The 6 bp deletion includes 1 bp of the PAM and the last 5 bp of the guide sequence. The 11 bp insertion includes an Xho I restriction site and 5 bp of random sequence. (c) Detailed diagram of CRISPR/Cas9 system mediated continual genome editing in B. subtilis ATCC 6051a.
Disruption of the srfC gene using the CRISPR/Cas9 system
We used B. subtilis ATCC 6051a as the initial strain in which to perform genetic manipulations. Fermentation of B. subtilis ATCC 6051a in a 3 L fermenter in our laboratory produced a massive amount of foam that requires large quantities of antifoam agent (Fig. 2). Accumulation of the amphiphilic molecule surfactin promotes foam production and the gene srfC is crucial to the regulation of surfactin production5. The CRISPR/Cas9 system developed in this study was first tested using srfC as the target (Fig. 1c). We used the knockout plasmid pHYcas9dsrf1, which produces both a sgRNA specific to srfC and the Cas9 protein, to transform the initial strain, B. subtilis ATCC 6051a. Because the temperature-sensitive knockout plasmid is maintained at low copy in transformants at 37 °C, the tetracycline-resistant transformants were confirmed by colony PCR of cas9. As for the commonly same genotype of transformation colonies, the target cleavage by Cas9 and homology-directed repair are commonly performed during the liquid incubation of transformation. The resulting mutants had an Xho I site within the repair locus and this disruption genotype passed to the next generation through passaging in liquid media or LB plate. To test the effectiveness of the CRISPR/Cas9 system, the regions upstream and downstream of the repair locus were amplified by PCR and then the PCR products were digested with Xho I (Fig. 3). There are 66 ± 14 transformants after transformation of knockout plasmid pHYcas9dsrf1. 30 transformants were screened and 13 ± 2 colonies yielded a recombinant genotype with a disruption efficiency of 43% ± 6%. The results, combined with DNA sequencing of the homologous regions of the mutants (Supplementary Fig. S1a) demonstrated that the editing system described above worked efficiently. Curing the knockout plasmid pHYcas9dsrf1 through overnight incubation at 51 °C produced the desired mutant, named BS1. Colonies of BS1 were used as the initial strain for next gene disruption. There are 20 ± 4 transformants after transformation of initial knockout plasmid pHYcas9d and no srfC gene mutant was found among the transformants. Though DNA nonhomologous end joining exist in B. subtilis27, the low survival rate was probably due to the double-strand breaks in chromosome created by cas9 protein. To test foam production by B. subtilis ATCC 6051a and mutant BS1, cells were grown in a 3 L fermenter for 80 h and the growth rate of B. subtilis ATCC 6051a and BS1 has no significant difference. During the whole fermentation process, B. subtilis ATCC 6051a produced much foam and required to add antifoam (490 ul) continuously; BS1 produced much less foam at a controllable level that needed 60 ul antifoam. The foam height of B. subtilis ATCC 6051a and BS1 was similar after 42 h, while the foam of BS1 shows great sensitive to antifoam (Fig. 2).
The foam height and cell growth of B. subtilis ATCC 6051a and BS1 in 3 L fermenter.
During the fermentation, the foam height of B. subtilis ATCC 6051a (Δ) and BS1 (▲) and the dry cell weight (DCW) of B. subtilis ATCC 6051a (□) and BS1 (■) was measured. The antifoam was added to B. subtilis ATCC 6051a (dotted arrow) and BS1 (solid arrow) when foam height reach the limit.
Confirmation of the srfC disruption, spoIIAC disruption, nprE disruption, aprE disruption and amyE disruption.
Digesting the PCR product of upstream and downstream regions with Xho I. The digestion products were analysed by agarose gel electrophoresis. Lane M: DNA marker; lane WT: digestion of overlap PCR product using B. subtilis ATCC 6051a genomic DNA as the template; lane MT: digestion of overlap PCR product using the indicated gene disruption mutant genomic DNA as the template.
In addition to disruption described above, which deleted a relatively small section of the genome, we also designed and attempted a larger deletion of the srfC gene. An 1100 bp homologous repair template was designed to have a 500 bp upstream region and 600 bp downstream region flanking the Cas9 cleavage site. Using knockout plasmid pHYcas9dsrf2 that containing this repair template to delete a 284 bp region, which includes 44 bp upstream and 240 bp downstream of the PAM sequence (Supplementary Fig. S2). The efficiency of the 284 bp deletion is 9.1% in B. subtilis ATCC 6051a, which is lower than the efficiency of disruption gene. The low efficiency may related to the short length of homologous repair template that result in low homologous recombination efficiency28.
Disruption of spoIIAC gene using the CRISPR/Cas9 system
In order to cope with limiting nutrient sources and high cell density, B. subtilis can form highly resistant spores29 that will germinate and grow in favourable living conditions. These spores drastically hamper B. subtilis fermentations and limit their application in the food industry30. The SpoIIAC gene encodes sigma factor F, which permits cells to proceed through stage II of sporulation6,31,32,33. Using knockout plasmid pHYcas9dspo, we disrupted spoIIAC gene using the method described above. The resulting mutant, named BS2, was screened by amplifying the upstream and downstream regions and digesting the PCR products with Xho I (Fig. 3). The homologous regions of mutant BS2 were also subjected to DNA sequencing (Supplementary Fig. S1b) to confirm the result. The disruption efficiency of SpoIIAC gene was 36% ± 3%. After being cultured at 40 °C for 48 h in a culture medium that favours spore formation, the sporulation efficiency of BS1 was 28.44% (262 colonies/921 colonies), while the sporulation efficiency of BS2 was 0% (0 colonies/182 colonies), which shows that mutant BS2 has great resistant to spore formation.
Disruption of the nprE and aprE genes using the CRISPR/Cas9 system
Strains that lack several extracellular protease genes generally show superior extracellular protein productivity7,34. The nprE and aprE genes encode alkaline and neutral extracellular proteases, respectively, in B. subtilis. We constructed the knockout plasmids pHYcas9dnpr and pHYcas9dapr and used them to sequentially disrupt nprE and aprE gene of mutant BS2 using the CRISPR/Cas9 method described above. Amplification of the upstream and downstream regions and then digesting the PCR products with Xho I allowed us to select the appropriate mutant, named BS3 with disruption efficiency of 53% ± 6% (Fig. 3). The nprE and aprE double gene mutant, named BS4 with disruption efficiency of 33% ± 3%. The homologous regions of mutant BS3 and BS4 were subjected to DNA sequencing (Supplementary Fig. S1c,d) to confirm the disruption. When cultured on specify agar plates containing 5% non-fat powdered milk, strain BS3 forms a protein clearance zone smaller than the one formed by its parent, mutant BS2, demonstrating that BS3 has reduced protease activity. And strain BS4 shows substantial inhibition of protein degradation on a 5% non-fat powdered milk plate (Fig. 4a).
Detection of protease activity and α-amylase activity.
(a) Transparent rings formed on 5% non-fat powdered milk medium to detect the protease activity of mutants BS2, BS3 and BS4; (b) The mutants BS4 and BS5 were grown on LB plate containing 1% soluble starch, then the colonies were wiped; and (c) the plate was stained with iodine.
Disruption of the amyE gene using the CRISPR/Cas9 system
B. subtilis releases many extracellular enzymes during the post-exponential growth phase and α-amylase is one of the major proteins released9,35. These proteins hamper the purification of industrial products. We constructed the knockout plasmid pHYcas9damy and used it to disrupt the amyE gene of mutant BS4 as described above. After amplifying the sequences upstream and downstream of the target region, the PCR products were digested with Xho I to select the amyE deletion mutant, which was named BS5 with disruption of 53% ± 6% (Fig. 3). The homologous regions of mutant BS5 were also subjected to DNA sequencing (Supplementary Fig. S1d) to confirm the disruption. The results of a starch-plate assay demonstrate that BS5 fails to release α-amylase activity (Fig. 4b,c).
Extracellular expression of β-CGTase using B. subtilis ATCC 6051a and mutant BS5
To evaluate the utility of the strain BS5, in which five genes (srfC, spoIIAC, nprE, aprE and amyE) have been disrupted, as a host for extracellular recombinant protein expression, the ability of mutant BS5 to produce β-CGTase was compared with that of B. subtilis ATCC 6051a. Both expression plasmid pHY300PLK-β-CGTase and empty expression plasmid pHY300PLK were transferred into B. subtilis ATCC 6051a and mutant BS5 and the resulting strains were used in an expression study. After 48 h of cultivation in TB medium, the culture supernatants of B. subtilis ATCC 6051a and BS5 that harbouring empty expression vector PHY300PLK show no β-CGTase activity. After 80 h of cultivation in 3 L fermenter, the highest β-CGTase activity of B. subtilis ATCC 6051a that harbours expression plasmid pHY300PLK-β-CGTase was 110.8 U/ml in 56 h and the highest dry cell weight (DCW) was 68.8 g/L in 66 h; the highest β-CGTase activity of BS5 that harbours expression plasmid pHY300PLK-β-CGTase was 277.8 U/ml in 70 h, which was 2.5 times of B. subtilis ATCC 6051a and the highest DCW was 70.3 g/L in 75 h (Fig. 5).
Discussion
In this study, we established a CRISPR/Cas9 system that can disrupt target genes in B. subtilis ATCC 6051a (Fig. 1c) and then used this system to construct a mutant strain with improved fermentation characteristics. All of the elements required by the CRISPR system are present in a single plasmid that contains a constitutively expressed cas9, a strongly transcribed sgRNA and a homologous repair template. The sgRNA recognizes a specific site on the B. subtilis genome (Supplementary Table S1) and guides the Cas9 protein to the target genome locus, where it creates a double-stranded break. This is followed by homology-directed repair that utilizes a homologous repair template provided by the knockout plasmid. Since the knockout plasmid contains the PE194 temperature-sensitive replicon, it can be easily cured after the mutation step by incubating the mutants at 51 °C overnight. The mutant colonies cured of the knockout plasmid can be used as the parent strain for additional genetic modification. With an efficiency of 33% to 53%, the operate procedure of CRISPR system was simple and time-saving compared with the currently existing Bacillus subtilis genome editing methods (Table 1).
The transformation efficiency in B. subtilis 168 and B. subtilis ATCC 6051a are 1553 ± 213 and 60 ± 13 transformants/μg of the knockout plasmid, respectively. Its poor competence makes genetic manipulation of B. subtilis ATCC 6051a inconvenient. To increase the transformability of B. subtilis ATCC 6051a, we considered disrupting the gene encoding ComI. However, we were unable to find a target-specific sgRNA target within the 93 bp comI gene. The original knockout plasmid (pHYcas9d) did not contain a homologous repair template; therefore, a homologous repair template was transferred into B. subtilis ATCC 6051a in the form of PCR fragment, along with the knockout plasmid. This attempt did not meet the efficiency required for genetic editing, perhaps because B. subtilis ATCC 6051a was unable to simultaneously take up the knockout plasmid and the homologous repair PCR fragment.
The target recognition of the sgRNA mainly depends on the last 12 bp of the guide sequence; thus, the existence of highly homologous regions in chromosomal DNA may result in off-target effects36. These off-target effects have been reported in eukaryotic cells37, whereas little attention has been paid to this problem in prokaryotes. Although off-target effects may be less common in bacteria because of their relatively small genome size, bacterial genomes contain some high-homology clusters38. There are methods that can be used to reduce the off-target efficiency; for example, designing two sgRNAs to guide the Cas9 protein to cleave the genome at the adjacent sites, using a Cas9 nickase mutant and making sure the last 12 bp of the guide sequence is highly specific39.
Like many Bacillus strains, B. subtilis produces surfactin, which contains a peptide moiety and a β-hydroxy fatty acid side chain40. Because surfactin is an amphiphilic molecule, its accumulation at gas-liquid interfaces can lead to foam production40,41. The biosynthesis of surfactin is controlled by a non-ribosomal peptide synthase enzyme (SrfC)42 and a thioesterase/acyltransferase (SrfD)43. The internal thoiesterase domain of srfC, which is a one-module enzyme, controls the conversion of a linear lipoheptapeptide to its cyclic form, as well as the release of surfactin41. B. subtilis ATCC 6051a produces a large amount of foam during fermentation, which has an extreme affect on fermentation process control and may lead to contamination. Compared with its parent strain, the mutant BS1 produces much less foam and at the level of control. This shows that surfactin may be a major mediator of foam formation and that there is more work to be done to thoroughly inhibit foam production.
Spore development in B. subtilis is governed by multiple RNA polymerase sigma factors32. Sigma F, encoded by SpoIIAC gene, controls the forespore, sigma E controls the early stage of sporulation and then sigmas G and K control later stages32,33. SpoIIAC nonsense mutations can block the processing of sigma E precursor protein P31 to sigma E and prevent transcription of spoIID31. The transcription product, SpoIID, is a membrane-anchored enzyme essential for sporulation6,44. The observation that mutant BS2 shows great resistant to spore formation demonstrates that inactivation of sigma F can largely block the sporulation of B. subtilis.
Bacillus species produce many different types of extracellular protease to degrade heterologous extracellular proteins45 and many protease-deficient strains display favorable heterologous protein production7,34. B. subtilis WB600, which is deficient in six extracellular proteases due to knockouts of the nprB, nprE, aprE, mpr, bpr and epr genes. This strain displays a level of recombinant protein secretion higher than that of its parent strain46. Recently, a B. amyloliquefaciens strain lacking six extracellular protease genes was constructed. This strain displays improved production of levan and α-amylase34. B. subtilis releases multiple extracellular enzymes during the post-exponential period; α-amylase is among the major proteins released35. Secretion of a large amount of α-amylase by B. subtilis makes the isolation and purification of recombinant proteins difficult on an industrial scale. In addition, the massive expression of endogenous α-amylase increases secretion stress and influences the production of recombinant proteins47.
In summary, we established a CRISPR/Cas9 system in the poorly transformable strain B. subtilis ATCC 6051a, which is an undomesticated strain with favorable growth characteristics. To improve the usefulness of B. subtilis ATCC 6051a as an industrial expression host, we disrupted the srfC, spoIIAC, nprE, aprE and amyE genes with an efficiency of 33% to 53%. Compared with B. subtilis ATCC 6051a, the final mutant (BS5) forms less foam during fermentation, displays greater resistant to spore formation and secretes 2.5 times more β-CGTase. Thus, the CRISPR system developed here can be used to modify industrially relevant strains with high efficiency and mutant BS5 can be applied as a superior expression host.
Materials and Methods
Strains and plasmids
All bacterial strains and plasmids used in this study are described in Table 2. Escherichia coli JM109 was used for plasmid construction. Plasmid pHY300PLK-β-CGTase was previously constructed in our laboratory by inserting the β-CGTase gene of Bacillus circulans 251 into the B. subtilis-E. coli shuttle expression vector pHY300PLK (Takara, Dalian, China)48. Plasmid pwtcas9-bacterial was purchased from Addgene (Addgene plasmid # 44250)49. B. subtilis ATCC 6051a was purchased from the American Type Culture Collection (ATCC).
Reagents and enzymes
PrimeSTAR polymerase, restriction enzymes, calf intestinal alkaline phosphatase, Dpn I, T4 DNA ligase, In-Fusion HD Cloning Plus kit and vector (pMD18-T) were purchased from Takara (Dalian, China). The plasmid mini-prep kit, PCR purification kit and the agarose gel DNA purification kit were purchased from Tiangen Co. Ltd (Beijing, China). DNA sequencing and DNA primer synthesis were performed by Shanghai RuiDi Biological Technology Co. Ltd. (Shanghai, China). Tryptone and yeast extract were obtained from Oxoid (Hampshire, UK). β-cyclodextrin was purchased from Sigma-Aldrich (Milwaukee, WI, USA). Non-fat powdered milk was purchased from BBI Life Sciences (Shanghai, China). Other chemicals were purchased from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China).
Media and growth conditions
For routine construction of plasmids and B. subtilis mutants, E. coli, B. subtilis and B. subtilis derivatives were cultured in LB medium (10 g/L tryptone, 5 g/L yeast extract and 10 g/L NaCl) at 37 °C with shaking at 200 rpm. To evaluate spore formation, B. subtilis mutants BS1 and BS2 were cultured at 40 °C with shaking at 200 rpm for 48 h in a medium containing 30 g/L dextrin, 30 g/L bean peptone, 1 g/L CaCl2 and 0.5 g/L NaCl. Ampicillin (100 mg/L) and tetracycline (20 mg/L) were added as needed.
Fermentation Cultivate condition
Seed culture was obtained by inoculating 100 ul frozen glycerol stock (stored at −80 °C) into 100 ml LB medium and then incubating 12 h at 37 °C with shaking at 200 rpm. As for Shake-flask cultivation, a portion of 5% (v/v) seed culture (2.5 ml) was used to inoculate 50 ml of TB medium (Supplementary material and method), which was then incubated at 30 °C with shaking at 200 rpm for 48 h. While for the fermentation in 3 L fermenter (BioFlo 110, New Brunswick Scientific Co., Edison, NJ) that containing 0.86 L fermentation medium (Supplementary material and method), before a 10% (v/v) seed culture (100 ml) was inoculated, its pH was adjusted to 7.0 with 20% (v/v) H3PO4 and NH4OH, its temperature was adjusted to 37 °C and it was added with 30 ml 166.7 g/L glucose. After 2 h of inoculation, the inducer of 30 ml 333.3 g/L lactose was added into fermentation culture as needed. The feed solution (Supplementary material and method) was fed at rate 0 to 15 g glucose/h to confirm the glucose concentration was maintained 0.2 to 0.5 g/L. During the fermentation process, the batch cultivation was carried out at 37 °C, pH 7.0 and 30% dissolved oxygen that were maintained by automatically adjusting stirrer speed (300 rpm-900 rpm) and flow rate of air (1.5–4.0 L/min). Tetracycline (20 mg/L) was added as needed every 24 h. The foam height was measured as needed and antifoam was added manually. For both cultivation, the samples collected at certain time intervals were centrifuged at 12,000 × g for 10 min at 4 °C. For DCW determination, the pellet was resuspended with 0.9% (w/v) NaCl and centrifuged at 12000 × g for 10 min and dried to a constant weight at 105 °C. As for the host strains that harbour expression plasmid pHY300PLK-β-CGTase, the culture supernatant contains β-CGTase.
Plasmids construction
The sequences of all of the primers used in this study are listed in Table 3. The initial CRISPR system knockout plasmid pHYcas9d was assembled from four fragments (One, Two, Three and Four), each of which overlaps its two adjacent fragments by 15 bp, using the In-Fusion HD Cloning Plus kit50. Fragment One, which encodes the Cas9 protein, was amplified from plasmid pwtcas9-bacterial using the primer pair P03/P04. Fragment Two, which includes a P43 promoter for sgRNA expression, a target-specific 20-nt guide sequence specific for srfC fused with the sgRNA sequence and a temperature-sensitive replicon PE194 for plasmid curing, was synthesized and ligated into the cloning vector pMD18-T. This fragment was amplified using the primer pair P07/P08. Fragment Three includes an E. coli replication origin (p15A ori), an ampicillin-resistance marker (ampR) and the α-amylase promoter (PamyQ) from B. amyloliquefaciens. This fragment was amplified from plasmid pHY300PLK-β-CGTase using primers P02 and P05. Fragment Four includes a tetracycline resistance marker (TcR) and a terminator from B. amyloliquefaciens. This fragment was amplified from plasmid pHY300PLK-β-CGTase using primers P01and P06. We amplified the upstream and downstream regions of the target locus using B. subtilis ATCC 6051A genomic DNA as a template and two primer pairs: P09/P10 and P11/P12. The amplified fragments were ligated together using overlap extension PCR, forming homologous repair template. To form the final knockout plasmid, pHYcas9dsrf1, the appropriate homologous repair template was inserted into the Xba I site of pHYcas9d (Fig. 1a). During double-strand repair, this repair template removes 6 bp of native sequence and inserts an Xho I restriction site and 5 bp of random sequence (Fig. 1b).
The other five knockout plasmids (pHYcas9dsrf2, pHYcas9dspo, pHYcas9dnpr, pHYcas9dapr and pHYcas9damy) were constructed from pHYcas9dsrf1 by changing the 20-nt guide sequence (except pHYcas9dsrf2) and replacing the homologous repair template. The 20-nt guide sequence was changed using inverse PCR with the four primer pairs P17/P18 (for pHYcas9dspo), P23/P24 (for pHYcas9dnpr), P29/P30 (for pHYcas9dapr) and P35/P36 (for pHYcas9damy), which hanging the new modified 20-nt guide sequence at the 5′ end. The homologous repair templates were created by overlap extension PCR of sequences upstream and downstream of the 20-nt guide sequence. These sequences were amplified from the B. subtilis ATCC 6051a genome using primer pairs P13/P14 and P15/P16 (for pHYcas9dsrf2), P19/P20 and P21/P22 (for pHYcas9dspo), P25/P26 and P27/P28 (for pHYcas9dnpr), P31/P32 and P33/P34 (for pHYcas9dapr) and P37/P38 and P39/P40 (for pHYcas9damy). Then the appropriate homologous repair template was inserted into the modified pHYcas9dsrf1 by digesting the PCR product and the modified plasmid with Xba I and ligating the appropriate fragments.
Genome editing
B. subtilis competent cells were made by the method of Anagnostopoulos and Spizizen51. Tetracycline-resistant transformants were confirmed by colony PCR of the cas9 gene. Using transformant genomic DNA as the template, verification PCR reactions were carried out using specific primers (Supplementary Table S2) that anneal outside the homologous repair template. PCR products from disruption mutants were identified by digestion with Xho I and the homologous repair regions of the mutants were subsequently verified by DNA sequencing.
Plasmid curing
To cure the mutants of the knockout plasmid, edited colonies harbouring knockout plasmid were used to inoculate 10 mL of LB medium containing tetracycline (20 mg/L). The culture was incubated at 37 °C and then streaked onto LB plates that were subsequently incubated overnight at 51 °C52. The colonies cured of knockout plasmid were confirmed by streaking them onto LB plates containing tetracycline (20 mg/L); colonies cured of plasmid fail to grow at 37 °C. These colonies were used in the next round of genome editing.
Calculation of sporulation efficiency
After cultivation in a medium conducive to spore formation at 40 °C for 48 h, Cells of mutants BS1 and BS2 were diluted and divided into two same amount of portions, respectively, one of which was heat at 75 °C for 20 min53, then streak them onto LB plates that were subsequently incubated overnight at 37 °C. The cell ratio of two portions was calculated as sporulation efficiency.
Detection of protease activity
To detect the protease activity of mutants BS2, BS3 and BS4, strains were dropped onto a 5% non-fat powdered milk plate at 37 °C and incubated for 36 h54. Under these conditions, secreted proteases form a clear ring around the secreting colony.
Detection of α-amylase activity
The α-amylase expression of mutants BS4 and BS5 was detected by dropping the strains onto LB plates containing 1% soluble starch at 37 °C for 24 h, then staining the plates with iodine55. Under these conditions, secreted α-amylases create a colourless ring around the secreting colony.
Enzyme assay of β-CGTase
The β-cyclodextrin-forming activity was determined using the observation that β-cyclodextrin forms a stable, colourless inclusion complex with phenolphthalein56. A small sample (0.1 ml) of appropriately diluted culture supernatant was incubated with 2 ml of 1% (w/v) soluble starch in 25 mM phosphate buffer (pH 5.5) at 50 °C for 10 min. The amount of β-cyclodextrin formed was determined by titrating the sample with a standard phenolphthalein solution. One unit of activity was defined as the amount of enzyme that produce 1 μmol of β-cyclodextrin per min57.
Additional Information
How to cite this article: Zhang, K. et al. Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas9 system. Sci. Rep. 6, 27943; doi: 10.1038/srep27943 (2016).
References
Zeigler, D. R. et al. The origins of 168, W23 and other Bacillus subtilis legacy strains. J Bacteriol. 190, 6983–6995 (2008).
Wu, S. C. et al. Functional production and characterization of a fibrin-specific single-chain antibody fragment from Bacillus subtilis: effects of molecular chaperones and a wall-bound protease on antibody fragment production. Appl Environ Microbiol. 68, 3261–3269 (2002).
Skolpap, W., Scharer, J. M., Douglas, P. L. & Moo-Young, M. Fed-batch optimization of alpha-amylase and protease-producing Bacillus subtilis using Markov chain methods. Biotechnol Bioeng. 86, 706–717 (2004).
Widner, B. et al. Development of marker-free strains of Bacillus subtilis capable of secreting high levels of industrial enzymes. J Ind Microbiol Biot. 25, 204–212 (2000).
Coutte, F. et al. Effect of pps disruption and constitutive expression of srfA on surfactin productivity, spreading and antagonistic properties of Bacillus subtilis 168 derivatives. J Appl Microbiol. 109, 480–491 (2010).
Clarke, S. & Mandelstam, J. Regulation of stage II of sporulation in Bacillus subtilis. J Gen Appl Microbiol. 133, 2371–2380 (1987).
Kawabata, Y., Kimura, K. & Funane, K. Extracellular production of cycloisomaltooligosaccharide glucanotransferase and cyclodextran by a protease-deficient Bacillus subtilis host-vector system. Appl Microbiol Biot. 93, 1877–1884 (2012).
Wu, X. C., Lee, W., Tran, L. & Wong, S. L. Engineering a Bacillus subtilis expression-system with a strain deficient in 6 extracellular proteases J Bacteriol. 173, 4952–4958 (1991).
Gupta, M. & Rao, K. K. Phosphorylation of DegU is essential for activation of amyE expression in Bacillus subtilis. J Biosci. 39, 747–752 (2014).
Konkol, M. A., Blair, K. M. & Kearns, D. B. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J Bacteriol. 195, 4085–4093 (2013).
Jeong, H., Sim, Y. M., Park, S.-H. & Choi, S.-K. Complete genome sequence of Bacillus subtilis strain ATCC 6051a, a potential host for high-level secretion of industrial enzymes. Genome announcements. 3, 10.1128/genomeA.00532-15 (2015).
Sorek, R., Lawrence, C. M. & Wiedenheft, B. in Annual Review of Biochemistry Vol. 82 (ed R. D. Kornberg ) 237–266 (Annual Reviews, 2013).
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 321, 960–964 (2008).
Jiang, W., Bikard, D., Cox, D., Zhang, F. & Marraffini, L. A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat biotechnol. 31, 233–239 (2013).
Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 471, 602–607 (2011).
Jinek, M. et al. A Programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas Systems. Science. 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via cas9. Science. 339, 823–826 (2013).
Huang, H., Zheng, G., Jiang, W., Hu, H. & Lu, Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Bioch Bioph Sin. 47, 231–243 (2015).
DiCarlo, J. E. et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 41, 4336–4343 (2013).
Oh, J.-H. & van Pijkeren, J.-P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 42, 10.1093/nar/gku623 (2014).
Wang, Y. et al. The CRISPR/Cas System mediates efficient genome engineering in Bombyx mori. Cell Res. 23, 1414–1416 (2013).
Yu, Z. et al. Highly efficient genome modifications mediated by CRISPR/Cas9 in Drosophila. Genetics. 195, 289–291 (2013).
Shen, B. et al. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 23, 720–723 (2013).
Ong, R. M. et al. Cloning, extracellular expression and characterization of a predominant beta-CGTase from Bacillus sp G1 in E-coli. J Ind Microbiol Biot. 35, 1705–1714 (2008).
Ye, R. Q. et al. High-level secretory production of intact, biologically active staphylokinase from Bacillus subtilis. Biotechnol Bioeng. 62, 87–96 (1999).
Weller, G. R. et al. Identification of a DNA nonhomologous end-joining complex in bacteria. Science. 297, 1686–1689 (2002).
Yan, X., Yu, H.-J., Hong, Q. & Li, S.-P. Cre/lox system and PCR-based genome engineering in Bacillus subtilis. Appl Environ Micro. 74, 5556–5562 (2008).
de Jong, I. G., Veening, J. W. & Kuipers, O. P. Heterochronic phosphorelay gene expression as a source of heterogeneity in Bacillus subtilis spore formation. J Bacteriol. 192, 2053–2067 (2010).
Pandey, R. et al. Live cell Imaging of germination and outgrowth of individual Bacillus subtilis spores; the effect of heat stress quantitatively analyzed with spore tracker. Plos One. 8, 10.1371/journal.pone.0058972 (2013).
Jonas, R. M. & Haldenwang, W. G. Influence of spo mutations on sigma E synthesis in Bacillus subtilis. J Bacteriol. 171, 5226–5228 (1989).
Pereira, F. C. et al. The spore differentiation pathway in the enteric pathogen Clostridium difficile. Plos Genet. 9, 10.1371/journal.pgen.1003782 (2013).
Pishdadian, K., Fimlaid, K. A. & Shen, A. SpoIIID-mediated regulation of sigmaK function during Clostridium difficile sporulation. Mol Microbiol. 95, 189–208 (2015).
Feng, J. et al. Recruiting a new strategy to improve levan production in Bacillus amyloliquefaciens. Sci Rep. 5, 13814, 10.1038/srep13814 (2015).
Pretorius, I. S., de Kock, M. J., Britz, T. J., Potgieter, H. J. & Lategan, P. M. Numerical taxonomy of alpha-amylase producing Bacillus species. J Appl Microbiol. 60, 351–360 (1986).
Anders, C., Niewoehner, O., Duerst, A. & Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 513, 569–573 (2014).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat biotechnol. 31, 822–826 (2013).
Morton, J. T., Freed, S. D., Lee, S. W. & Friedberg, I. A large scale prediction of bacteriocin gene blocks suggests a wide functional spectrum for bacteriocins. BMC bioinformatics. 16, 10.1186/s12859-015-0792-9 (2015).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154, 1380–1389 (2013).
Willenbacher, J., Rau, J.-T., Rogalla, J., Syldatk, C. & Hausmann, R. Foam-free production of surfactin via anaerobic fermentation of Bacillus subtilis DSM 10(T). Bba-Mol Cell Res. 5, 10.1186/s13568-015-0107-6 (2015).
Vater, J., Wilde, C. & Kell, H. In situ detection of the intermediates in the biosynthesis of surfactin, a lipoheptapeptide from Bacillus subtilis OKB 105, by whole-cell cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in combination with mutant analysis. Rapid Commun Mass Sp. 23, 1493–1498 (2009).
Ullrich, C., Kluge, B., Palacz, Z. & Vater, J. Cell-free biosynthesis of surfactin, a cyclic lipopeptide produced by Bacillus subtilis. Biochemistry. 30, 6503–6508 (1991).
Steller, S. et al. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry. 43, 11331–11343 (2004).
Gutierrez, J., Smith, R. & Pogliano, K. SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation. J Bacteriol. 192, 3174–3186 (2010).
Westers, L., Westers, H. & Quax, W. J. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Bba-Mol Cell Res. 1694, 299–310 (2004).
Wu, X. C., Ng, S. C., Near, R. I. & Wong & S. L. Efficient production of a functional single-chain antidigoxin antibody via an engineered Bacillus subtilis expression-secretion system. Bio-Technology. 11, 71–76 (1993).
Waldeck, J. et al. Targeted deletion of genes encoding extracellular enzymes in Bacillus licheniformis and the impact on the secretion capability. J Biotechnol. 130, 124–132 (2007).
Knegtel, R. M. K. et al. Crystallographic studies of cyclodextrin glycosyltransferase from Bacillus-circulans strain-251 with natural substrates and products. J Biol Chem. 270, 29256–29264 (1995).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 152, 1173–1183 (2013).
Sleight, S. C. & Sauro, H. M. BioBrick assembly using the in-fusion PCR cloning kit. Methods Mol Biol. 1073, 19–30 (2013).
Anagnostopoulos, C. & Spizizen, J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 81, 741–746 (1961).
Dempsey, L. A. & Dubnau, D. A. Localization of the replication origin of plasmid pE194. J Bacteriol. 171, 2866–2869 (1989).
Ochi, K., Kandala, J. C. & Freese, E. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J Biol Chem. 256, 6866–6875 (1981).
Kasana, R. C. & Yadav, S. K. Isolation of a psychrotrophic Exiguobacterium sp SKPB5 (MTCC 7803) and characterization of its alkaline protease. Curr Microbiol. 54, 224–229 (2007).
Zhang, X.-Z., Yan, X., Cui, Z.-L., Hong, Q. & Li, S.-P. MazF, a novel counter-selectable marker for unmarked chromosomal manipulation in Bacillus subtilis. Nucleic Acids Res. 34, 10.1093/nar/gkl358 (2006).
Makela, M., Korpela, T. & Laakso, S. Colorimetric determination of beta-cyclodextrin: two assay modifications based on molecular complexation of phenolphtalein. J Biochem Bioph Meth. 14, 85–92 (1987).
Penninga, D. et al. The raw starch binding domain of cyclodextrin glycosyltransferase from Bacillus circulans strain 251. J Biol Chem. 271, 32777–32784 (1996).
Biswas, I., Gruss, A., Ehrlich, S. D. & Maguin, E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 175, 3628–3635 (1993).
Reyrat, J. M., Pelicic, V., Gicquel, B. & Rappuoli, R. Counterselectable markers: Untapped tools for bacterial genetics and pathogenesis. Infection and Immunity. 66, 4011–4017 (1998).
Acknowledgements
This work was funded by grants from the National Science Fund for Distinguished Young Scholars (31425020), the National Natural Science Foundation of China (31271813, 31401636), the project of outstanding scientific and technological innovation group of Jiangsu Province, the Natural Science Foundation of Jiangsu Province (BK20140142), the 111 Project (No. 111-2-06), the China Postdoctoral Science Foundation Funded Project (2015M580390) and the Production and Research Prospective Joint Research Project of Jiangsu Province (BY2015019-18).
Author information
Authors and Affiliations
Contributions
K.Z. and X.G.D. led the design and performance of the experiments, analysis of the data and writing of the paper. J.W. participated in experimental design, analysis and editing the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, K., Duan, X. & Wu, J. Multigene disruption in undomesticated Bacillus subtilis ATCC 6051a using the CRISPR/Cas9 system. Sci Rep 6, 27943 (2016). https://doi.org/10.1038/srep27943
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27943
This article is cited by
-
Identification and optimization of genes potentially related to protein expression for enhancing α-amylase production in Bacillus subtilis
Systems Microbiology and Biomanufacturing (2024)
-
Development and application of a rapid all-in-one plasmid CRISPR-Cas9 system for iterative genome editing in Bacillus subtilis
Microbial Cell Factories (2022)
-
A facile and robust T7-promoter-based high-expression of heterologous proteins in Bacillus subtilis
Bioresources and Bioprocessing (2022)
-
Development and application of a fast and efficient CRISPR-based genetic toolkit in Bacillus amyloliquefaciens LB1ba02
Microbial Cell Factories (2022)
-
Enhancing Extracellular Pullulanase Production in Bacillus subtilis Through dltB Disruption and Signal Peptide Optimization
Applied Biochemistry and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.