Abstract
A single-nucleotide polymorphism (SNP) rs4236601 at the CAV1/CAV2 locus is associated with primary open-angle glaucoma (POAG). Rs4236601 is common in Caucasians but rare in East Asians. Here we conducted a haplotype-tagging SNP analysis followed by replication in a total of 848 POAG cases and 1574 controls drawn from 3 cities in China and 1 city in Japan. Two SNPs, rs4236601 (odds ratio [OR] = 6.25; P = 0.0086) and a tagging-SNP rs3801994 (OR = 1.32; P = 0.042), were associated with POAG in the Hong Kong Chinese cohort after age and gender adjustments. Rs4236601 was associated with POAG also in Shantou (OR = 6.09; P = 0.0037) and Beijing (OR = 3.92; P = 0.030) cohorts after age and gender adjustment, with a pooled-OR of 5.26 (P = 9.0 × 10−6) in Chinese; but it is non-polymorphic in the Osaka cohort. SNP rs3801994 showed a similar trend of effect in the Shantou and Beijing cohorts, with a pooled-OR of 1.23 (P = 0.022) and 1.20 (P = 0.063) in Chinese, prior to and after age and gender adjustment, respectively; but it showed a reverse effect in the Osaka cohort (OR = 0.58; P = 0.033) after the adjustments. We have thus confirmed the association of rs4236601 with POAG in different Chinese cohorts. Also, we found a common SNP rs3801994 of diverse associations with POAG between Chinese and Japanese.
Similar content being viewed by others
Introduction
Glaucoma is a group of optic neuropathies involving progressive loss of retinal ganglion cells and their axons, resulting in characteristic optic nerve head excavation and visual field defects1. It is a leading cause of irreversible blindness worldwide2. Primary open-angle glaucoma (POAG) is a major form of glaucoma. POAG has complex etiology, with major risk factors including a high intraocular pressure (IOP), older age, thin central corneal thickness, family history and genetic susceptibility3,4,5,6.
In a genome-wide association study (GWAS), a single-nucleotide polymorphism (SNP) rs4236601 at the CAV1/CAV2 locus on chromosome 7q31 was identified to be significantly associated with POAG in the Icelandic population and the association was replicated in European decedents and southern Chinese7. Notably, the minor allele frequencies (MAFs) and effect sizes of rs4236601 were different across populations. It is a common SNP in Caucasians, with the minor allele A presented in 20–28% of the control subjects, conferring an odds ratio (OR) of 1.1 to 1.38,9,10. In contrast, rs4236601 is rare in East Asians. For example in Chinese and Korean, the allele A presented in less than 1% of the population, but it conferred a higher OR of 2.9–5.5 for POAG7,11. Of note, in a Japanese study rs4236601 was found to be non-polymorphic12. In addition, the association of CAV1/CAV2 SNPs, rs42366019,10 and rs10529909,10,12, with POAG was reportedly stronger in female, suggesting a gender-specific association.
In view of the low MAF of SNP rs4236601 in East Asians, there could be other common SNPs at the CAV1/CAV2 locus that are associated with POAG. Therefore, we conducted the present study to investigate the association between common, haplotype-tagging SNPs in this locus and POAG in the Chinese and Japanese populations and assessed for gender-specific association.
Results
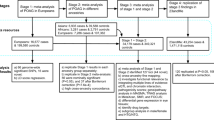
This study involved a total of 2422 study subjects, including 848 POAG patients and 1574 controls (Table 1). Among them, 890 (454 patients and 436 controls) were southern Chinese recruited from Hong Kong, 854 (123 patients and 731 controls) were southern Chinese from Shantou, 370 (170 patients and 200 controls) were northern Chinese from Beijing and 308 (101 patients and 207 controls) were Japanese from Osaka. All patients had high-tension POAG and the mean highest IOP was 29.0 ± 8.6 mmHg. The mean IOP of the control subjects was 15.0 ± 2.3 mmHg.
Association of CAV1/CAV2 with POAG
Two reported SNPs, rs4236601 and rs1052990 and 6 haplotype-tagging SNPs at the CAV1/CAV2 locus were genotyped in the Hong Kong cohort. All SNPs followed Hardy-Weinberg equilibrium (HWE) in the control group (P > 0.05; Supplementary Table 1). SNP rs4236601, which was identified in the GWAS7, conferred an increased risk of POAG (A allele, P = 0.0072, OR = 4.72; Table 2), with a population attributable risk (PAR) of 1.47%. A common SNP, rs3801994, showed a borderline association with POAG (A allele, P = 0.036, OR = 1.31; Table 2). This SNP presented in 18% of patients and 15% of controls, conferring a PAR of 4.33%. Logistic regression showed that when conditioned on each other these 2 SNPs were independently associated with POAG: rs4236601 (P = 0.0088, OR = 5.31) and rs3801994 (P = 0.038, OR = 1.22; Supplementary Tables 2 and 3). Moreover, the association of rs4236601 (P = 0.0086, OR = 6.25) and rs3801994 (P = 0.042, OR = 1.32) remained statistically significant after adjusting for age and gender. The other 6 SNPs did not show statistically significant associations with POAG (P > 0.05; Table 2).
Based on the findings in the Hong Kong cohort, we genotyped the SNPs rs4236601 and rs3801994 in the cohorts from Shantou, Beijing and Osaka. We also genotyped the SNP rs1052990 in the Osaka cohort for comparison with a previous study in Japanese12 as detailed in the methods. SNP rs4236601 was associated with POAG in the Shantou cohort prior to (P = 0.0079, OR = 4.23) and after adjusting for age and gender (P = 0.0037, OR = 6.09). It had a MAF of 0.49% in controls and 2.0% in patients. Also, rs4236601 showed a significant association with POAG in the Beijing cohort after adjusting for age and gender (P = 0.030, OR = 3.92) and a same trend of effect prior to adjustment (P = 0.057, OR = 2.86; Table 2). The MAF was higher in the Beijing cohort: 1.3% in controls and 3.6% in POAG patients. Of note, rs4236601 was non-polymorphic in our Osaka Japanese cohort. In contrast, rs3801994 was not significantly associated with POAG in the Shantou or Beijing cohort, but their ORs were toward the same direction with that in the Hong Kong cohort (OR = 1.14 in the Shantou cohort and 1.09 in the Beijing cohort; Table 2). Notably, rs3801994 showed an association with POAG in the Osaka cohort after adjusting for age and gender, with an OR toward the opposite direction (P = 0.033, OR = 0.58; Table 2). The MAF of this SNP in the Osaka controls was higher (33%) than that in Chinese (15–19%) and the SNP had a larger PAR 19.29% in the Osaka cohort than in the Hong Kong cohort (4.33%). The common SNP rs10529907,9 showed a significant association with POAG after adjusting for age and gender (allele G, P = 0.028, OR = 0.55; Table 2) in the Osaka cohort but not in the Hong Kong cohort, which again highlighted the ethnic difference in genetic associations. Linkage disequilibrium (LD) analysis showed that SNPs rs3801994 and rs1052990 were in weak LD (r2 = 0.26). Therefore, the association signals on these two SNPs should be independent.
By pooling the data of rs4236601 and rs3801994 from the 3 Chinese cohorts, we found that the SNP rs4236601 was strongly associated with POAG prior to (P = 1.1 × 10−4, OR = 3.80) and after adjusting for age and gender (P = 9.0 × 10−6, OR = 5.26), with no inter-cohort heterogeneity (I2 = 0; Fig. 1). SNP rs3801994 showed a borderline association with POAG (P = 0.022, OR = 1.23, I2 = 0) prior to age and gender adjustment and had the same trend of effect after adjustment (P = 0.063, OR = 1.20, I2 = 0; Fig. 2).
Gender-specific association with POAG
We assessed the association of the CAV1/CAV2 SNPs with POAG in different genders. SNP rs4236601 showed a significant association with POAG in females in the Hong Kong (P = 0.028, OR = 4.94) and Beijing (P = 0.025, OR = 5.76) cohorts, but in males in the Shantou cohort (P = 0.0071, OR = 11.72). The ORs of this SNP were not significantly different between males and females in the 3 cohorts (P > 0.05, Breslow-day test; Table 3). In the pooled samples from the 3 Chinese cohorts, rs4236601 had a significant association with POAG in female (P = 0.0011; OR = 4.84, 95% CI: 1.88–12.45; I2 = 0) and a borderline association in male (P = 0.052; OR = 2.83, 95% confidence interval [CI]: 0.99–8.10; I2 = 38%; Supplementary Fig. 1). The other 7 SNPs (i.e., rs1052990, rs6466579, rs7801950, rs3779512, rs3807989, rs3801994 and rs1049337) did not show a gender preference in the association with POAG (Table 3).
Discussion
In this study, we confirmed the association between the SNP rs4236601 at the CAV1/CAV2 locus and POAG in southern Chinese and, for the first time, in northern Chinese. The minor allele A increased the risk of POAG by over 4 fold in southern Chinese and nearly 3 fold in northern Chinese, but it is absent in Japanese. Moreover, we identified a common SNP rs3801994 for POAG, with an OR of 1.23 in Chinese and 0.58 in Japanese and a PAR of 4.33% in Chinese and 19.29% in Japanese. Another SNP, rs1052990 was found to be associated with POAG in the Osaka Japanese cohort (OR = 0.55) but not in the Hong Kong Chinese cohort (OR = 1.33). Thus, our data highlighted the ethnic diversities and independent roles of CAV1/CAV2 SNPs in the genetic susceptibility of POAG.
In Chinese, rs4236601 is an uncommon SNP, with a MAF of 0.43–1.3% in the control populations. But it confers a large risk effect on POAG (ORpooled = 5.26). This is in accordance with the findings in the GWAS7 and in a Korean cohort (MAFcontrol = 1.2%; OR = 2.9)11. Thus, the risk allele A of rs4236601 has a small PAR in Chinese and Koreans (about 1%), contributing to a small proportion of POAG. In this study, the A allele of rs4236601 was more common in the northern Chinese cohort (MAFcontrol 1.3% in northern Chinese compared with 0.43–0.49% in southern Chinese) and conferred a smaller risk effect (OR = 3.92 in the northern Chinese compared with 6.09–6.25 in southern Chinese). Such intra-ethnic diversity was also noted in other populations. For example, the MAFs and ORs of rs4236601 varied from 20.7% (OR = 1.33) to 28.1% (OR = 1.04) in Caucasian populations recruited from Sweden7, the United Kingdom7 and the United States9. Further studies in more Chinese populations are warranted to determine the allelic distribution and effects of rs4236601 across different geographic regions. Our study also revealed that rs4236601 is monomorphic in the Japanese cohort from Osaka, consistent with the findings in multiple regions of Japan12, suggesting that it is not a major gene marker for POAG in the Japanese population. The ethnic differences of rs4236601 in the allele frequency and effect size could be due to natural selection and population drift, which requires further investigation.
Apart from rs4236601, we identified a common, haplotype-tagging SNP, rs3801994, at the CAV1/CAV2 locus that conferred an independent effect on the risk of POAG. In the Hong Kong cohort, it showed a marginally significant association (P = 0.036, OR = 1.31; after adjustment, P = 0.042, OR = 1.32). Though this SNP did not show a statistically significant association with POAG in the Shantou and Beijing cohorts, the ORs were toward the same direction with that in the Hong Kong cohort. The meta-analysis showed that there was no heterogeneity across the 3 cohorts and the pooled P value was lowered (P = 0.022). Therefore, it is likely that rs3801994 is an associated SNP for POAG. The lack of statistical significance could be due to the limited statistical power in the Shantou (20.3%) and Beijing (16.2%) cohorts, assuming an OR of 1.31 and MAF of 0.15 according to the Hong Kong cohort. In contrast, rs3801994 is more common in Japanese, with a MAF of 0.33 in controls. Intriguingly, it conferred an OR towards the opposite direction in Japanese and had a PAR (19.29%) 4-fold higher than that in the Hong Kong cohort (PAR = 4.33%). This again highlights the ethnic differences in the genetic contribution of the CAV1/CAV2 locus to POAG. The difference in allelic frequencies between Chinese and Japanese found in this study may reflect differential occurrences of glaucoma and clinical features in the two populations, even though both are East Asians. In a previous study in Japanese, a SNP rs1052990 (P = 0.014, OR = 0.71, PAR = 8.79%)12 was found to be associated with normal tension glaucoma. Our study suggests that rs3801994 could be another common SNP associated with POAG in Japanese. Notably, however, the association of rs3801994 could not withstand the correction for multiple testing; therefore further studies in both Chinese and Japanese are warranted to confirm its role.
In this study, we have, for the first time, tested the SNP rs1052990 in Chinese POAG. However, in the Hong Kong Chinese cohort, we did not find a significant association. Of note, the minor allele of rs1052990 was G in our Hong Kong and Osaka cohorts, which in accordance with the data HapMap and another study in Japanese12. In contrast, allele T was the minor allele in Caucasians7,9. Moreover, the G allele was protective for normal-tension POAG in the Japanese cohorts in the study of Kato et al. (OR = 0.71) and for POAG in the Osaka Japanese cohort in our present study, being opposite to the risk effect observed in Caucasians in previous studies7,9 and the Hong Kong Chinese in the present study, highlighting the ethnic difference in the effects of rs1052990 on POAG. Of note in this study, a newly identified SNP, rs3801994 also showed a different effect (OR = 0.58) on POAG in the Osaka cohort as compared with the Chinese cohorts (pooled OR = 1.23). The MAF of this SNP in the Osaka controls was higher (33%) than that in Chinese (15–19%) and the SNP had a larger PAR 19.29% in the Osaka cohort than in the Hong Kong cohort (4.33%). Moreover, the SNPs rs3801994 and rs1052990 were in weak LD (r2 = 0.26), suggesting the association signals on these two SNPs are independent. Therefore, our study suggests that the effects of SNPs in the CAV1/CAV2 locus are different across different populations, even within East Asians. Further studies in more ethnic cohorts are warranted to confirm this finding.
Among the other 5 SNPs, only 2 SNPs (i.e., rs3779512 and rs3807989) have been reported in independent replication studies10,13,14, while rs6466579, rs7801950 and rs1049337 were tested in POAG for the first time. In this study, SNP rs3779512, which was associated with POAG in Caucasians (OR = 1.15)10, did not show a significant association (P = 0.68, OR = 0.92). Also, this SNP was not associated with POAG in an African population13, illustrating the ethnic diversity in the genetic contribution of this locus to POAG. Another SNP rs3807989 was not associated with POAG in our study or in African13,14.
Sex-specific association of rs4236601 has been reported in 2 studies in Caucasians9,10. These studies reported greater odds ratios and lower P values in females than in males. In our study cohorts, Breslow-day test did not support a significant difference in the ORs between females and males. This might be due to the decreased power after stratification. However, even with limited samples, rs4236601 remained significantly associated with POAG in Hong Kong and Beijing females and it showed a significant association in the pooled female samples (P = 0.0011, OR = 4.84). In contrast, we found rs4236601 to be associated with POAG in Shantou males (P = 0.0071, OR = 11.72) and have a borderline association in the pooled male subjects (P = 0.052, OR = 2.83). Thus, our data is consistent with the results in the Caucasian studies, in which CAV1/CAV2 SNPs are more significantly associated POAG in women than in men, suggesting the likely involvement of interactive effects among caveolins, eNOS and sex hormones9,10. Furthermore, we did not find gender preference in the association of rs1052990 with POAG, which could be due to the insufficient statistical power with the current sample size, confirmation of which requires further replication studies.
The SNP rs4236601 is located upstream to CAV1, while rs3801994 resides in the intronic region of CAV1. It has been shown that rs4236601 was not correlated with the expression of CAV1 and CAV2 in adipose tissue7 and in the retinas from normal donor eyes8. A recent study suggested an association between the allele G of rs17588172, a proxy SNP for rs105299011, with decreased CAV1 and CAV2 mRNA in skin and adipose15. In this study, the SNP rs7801950, which is a proxy SNP for rs1052990, did not show a significant association with POAG (P = 0.22).
Biologically, the involvement of caveolins in different biological pathways suggests their possible effects in the eye. In adult mouse brain, genetic ablation of caveolin-1, −2 or −3 increased neural stem cell (NSC) proliferation in the subventricular zone16. Adult NSC forms the three major cell lineages of the central nervous system16,17,18. In the mouse, cerebral ischemia may induce increases in endothelial caveolin-1 and -2 protein levels. Cerebral ischemia in the Cav-1 knockout mice resulted in an increased cerebral volume of infarction while in Cav-2 knockout mice reduced infarction volume19. These findings supported the involvement of caveolin proteins in the ischemia process. Of note, a marked pathological change in glaucoma is the progressive loss of RGCs along with supporting glia and vasculature1. Ischemia plays a central role in apoptotic RGC death and glaucoma patients can suffer from inadequate ocular blood flow20,21.
There were several limitations in this study. First, each of the study cohorts was of medium size, which may have limited the statistical power in identifying certain SNPs of mild effects, such as rs3801994, which showed a consistent trend of effect among the 3 Chinese cohorts but the P values in the Shantou and Beijing cohorts were not significant. The meta-analysis showed a marginally significant association between rs3801994 and POAG, with minimal inter-cohort heterogeneity (I2 = 0). Therefore, this association should be validated in more study cohorts. Second, only patients with high-tension glaucoma were enrolled in this study. Further studies are warranted to investigate the associations of the CAV1/CAV2 SNPs with normal-tension glaucoma, which will further enrich our understanding of the roles of the CAV1/CAV2 locus in POAG.
In summary, we confirmed the association of rs4236601 with POAG in the southern and northern Chinese and identified a common SNP at the CAV1/CAV2 locus, rs3801994, as a putative genetic biomarker for POAG in Chinese and Japanese, with diverse effects. Our findings enrich the understanding of the CAV1/CAV2 locus in POAG and provide new evidence on the ethnic diversities and allelic complexities in the genetic architecture of the disease.
Methods
Study subjects
This study included 4 cohorts from Hong Kong, Shantou (southern Chinese), Beijing (northern Chinese) and Osaka (Japanese). Part of the Chinese subjects from Hong Kong, Shantou and Beijing in the present study had been tested in our previous study22. All study subjects were unrelated. We recruited the Hong Kong subjects from the Hong Kong Eye Hospital and the Prince of Wales Hospital, Hong Kong. We recruited the Shantou subjects at the Joint Shantou International Eye Center of Shantou University and the Chinese University of Hong Kong, Shantou, China. The previous GWAS used part of the Hong Kong and Shantou subjects7. We recruited the Beijing cohort at the Beijing Tongren Eye Center, Beijing, China. We enrolled the Osaka cohort at the Department of Ophthalmology, Osaka University Hospital, Japan. We conducted this study in accordance with the tenets of the Declaration of Helsinki, under the ethics approvals from the ethics committees at respective collaborating institutions, such as the Chinese University of Hong Kong, the Joint Shantou International Eye Center of Shantou University, the Beijing Tongren Eye Center and Osaka University Hospital. We obtained written informed consent from each subject prior to the study.
All study subjects received complete ophthalmic examinations. The diagnostic criteria for POAG at all recruiting centers were: (1) evident glaucomatous retinal nerve fiber layer loss, characteristic optic disc changes (i.e. optic nerve head notching, hemorrhage, and/or increased vertical cup/disc ratio) and typical visual field loss by standard automated perimetry with the Glaucoma Hemifield Test (Humphrey Field Analyzer; Carl Zeiss Meditec, Inc., Dublin, CA); (2) normal gonioscopic appearance with Shaffer grade III or IV open iridocorneal angle; (3) exclusion of secondary causes, e.g., trauma, uveitis, steroid effects, or exfoliation glaucoma; and (4) the highest IOP measured by using Goldmann applanation tonometry, was greater than 22 mmHg. We enrolled Chinese control subjects with age more than 60 years from the 3 recruiting centers in China, whilst we recruited the Japanese control subjects from the staff, volunteers and patients with mild cataract or mild refractive errors from the recruiting center in Osaka. All control participants received complete ophthalmic examination and were free of glaucoma or other major eye diseases, except for mild cataract and/or mild refractive errors if any. They had IOPs lower than 21 mmHg and open iridocorneal angle.
SNP selection and genotyping
We investigated 8 SNPs in this study, including the SNP rs4236601 and rs1052990 that were reported in the previous GWAS7 and 6 haplotype-tagging SNPs covering the CAV1/CAV2 locus based on the Chinese Han Beijing population in HapMap project (namely rs6466579, rs7801950, rs3779512, rs3807989, rs3801994 and rs1049337) with a r2 cut-off of 0.8 and MAF cut-off of 0.1 (Supplementary Table 4).
We extracted genomic DNA from whole blood using the QIAamp Blood Kit (QIAGEN, Hilden, Germany). We conducted genotyping using TaqMan SNP genotyping assays (Applied Biosystems, Pleasanton, CA, USA) on a Roche LightCycler 480 Real-Time PCR System (Roche Diagnostics; Basel, Switzerland) according to the manufacturer’s instructions. We genotyped all of the 8 SNPs in the Hong Kong subjects and then genotyped 2 SNPs (rs4236601 and rs3801994), which showed an association (P < 0.05) with POAG in the Hong Kong cohort, in the Shantou, Beijing and Osaka subjects. Rs1052990 was also genotyped in the Osaka cohort as it was reported in GWAS7 and has been shown to be significant in a replication study in a Japanese population12.
Statistical analysis
We compared mean age and gender proportions between the case and control groups using the student t-test and χ2 test, respectively. We assessed the HWE for individual SNP in the control groups using the χ2 test in PLINK (version 1.07; http://pngu.mgh.harvard.edu/~purcell/plink/)23, with a P < 0.05 indicating deviation from HWE. We compared the differences in the MAFs of the SNPs between cases and controls using χ2 test or Fisher’s exact test using PLINK. We used logistic regression analysis to adjust the association for age and gender and condition on other SNPs to assess if an association was independent. We conducted subgroup analysis by gender to assess for gender-specific association. We performed the Breslow-day test for homogeneity of the ORs with the Tarone adjustment to determine if the ORs were significantly different between males and females24. We estimated population attributable risk of a SNP using the following equation, PAR% = 100 × p × (odds ratio−1) ÷ [p × (odds ratio−1) + 1], where p is the frequency of the associated alleles among control subjects25.
In association analysis, a P value of <0.05 suggested a potential association. To correct for multiple testing we used the SNPSpD program (Single Nucleotide Polymorphism Spectral Decomposition; http://gump.qimr.edu.au/general/daleN/SNPSpD/; accessed on 1 April, 2015)26. The SNPSpD takes into account inter-SNP correlations and generates the experiment-wide significance threshold that is required to keep the type I error rate of 5%. In this study, the effective number of independent markers was estimated to be 5, thus the significance threshold was 0.01 (0.05/5).
We pooled the data from the 3 Chinese cohorts by meta-analysis using the Review Manager (version 5.2.8; Copenhagen, The Nordic Cochrane Centre, The Cochrane Collaboration). We calculated the pooled-OR and 95% CI for each SNP using the fixed- or random-effect model based on inter-cohort heterogeneity. We assessed the heterogeneity by using the Q-statistic and the I2 27. An I2 value of <24%, 25% to 49%, 50% to 74% and ≥75% denoted none, low, moderate and high heterogeneity, respectively27,28. In this study, since the P value for the Q-statistic was >0.1 and the I2 < 50%, a fixed-effect model was adopted29.
Additional Information
How to cite this article: Rong, S. S. et al. Ethnic specific association of the CAV1/CAV2 locus with primary open-angle glaucoma. Sci. Rep. 6, 27837; doi: 10.1038/srep27837 (2016).
References
Kwon, Y. H., Fingert, J. H., Kuehn, M. H. & Alward, W. L. Primary open-angle glaucoma. N Engl J Med 360, 1113–1124 (2009).
Resnikoff, S. et al. Global data on visual impairment in the year 2002. Bull World Health Organ 82, 844–851 (2004).
Fan, B. J., Wang, D. Y., Lam, D. S. & Pang, C. P. Gene mapping for primary open angle glaucoma. Clin Biochem 39, 249–258 (2006).
Fan, B. J. & Wiggs, J. L. Glaucoma: genes, phenotypes and new directions for therapy. J Clin Invest 120, 3064–3072 (2010).
Janssen, S. F. et al. The vast complexity of primary open angle glaucoma: disease genes, risks, molecular mechanisms and pathobiology. Prog Retin Eye Res 37, 31–67 (2013).
Takamoto, M. & Araie, M. Genetics of primary open angle glaucoma. Jpn J Ophthalmol 58, 1–15 (2014).
Thorleifsson, G. et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet 42, 906–909 (2010).
Kuehn, M. H. et al. Chromosome 7q31 POAG locus: ocular expression of caveolins and lack of association with POAG in a US cohort. Mol Vis 17, 430–435 (2011).
Wiggs, J. L. et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum Mol Genet 20, 4707–4713 (2011).
Loomis, S. J. et al. Association of CAV1/CAV2 genomic variants with primary open-angle glaucoma overall and by gender and pattern of visual field loss. Ophthalmology 121, 508–516 (2014).
Kim, S. et al. Expression-associated polymorphisms of CAV1-CAV2 affect intraocular pressure and high-tension glaucoma risk. Mol Vis 21, 548–554 (2015).
Kato, T. et al. Association study of genetic variants on chromosome 7q31 with susceptibility to normal tension glaucoma in a Japanese population. Eye (Lond) 27, 979–983 (2013).
Liu, Y. et al. Investigation of known genetic risk factors for primary open angle glaucoma in two populations of African ancestry. Invest Ophthalmol Vis Sci 54, 6248–6254 (2013).
Williams, S. E., Carmichael, T. R., Allingham, R. R., Hauser, M. & Ramsay, M. The genetics of POAG in black South Africans: a candidate gene association study. Sci Rep 5, 8378 (2015).
Grundberg, E. et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet 44, 1084–1089 (2012).
Jasmin, J. F., Yang, M., Iacovitti, L. & Lisanti, M. P. Genetic ablation of caveolin-1 increases neural stem cell proliferation in the subventricular zone (SVZ) of the adult mouse brain. Cell Cycle 8, 3978–3983 (2009).
Eriksson, P. S. et al. Neurogenesis in the adult human hippocampus. Nat Med 4, 1313–1317 (1998).
Sanai, N. et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature 427, 740–744 (2004).
Jasmin, J. F. et al. Caveolin-1 deficiency increases cerebral ischemic injury. Circ Res 100, 721–729 (2007).
Caprioli, J. & Coleman, A. L. Blood pressure, perfusion pressure and glaucoma. Am J Ophthalmol 149, 704–712 (2010).
Harris, A., Chung, H. S., Ciulla, T. A. & Kagemann, L. Progress in measurement of ocular blood flow and relevance to our understanding of glaucoma and age-related macular degeneration. Prog Retin Eye Res 18, 669–687 (1999).
Chen, L. J. et al. SNP rs1533428 at 2p16.3 as a marker for late-onset primary open-angle glaucoma. Mol Vis 18, 1629–1639 (2012).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007).
Breslow, N. E. & Day, N. E. Statistical Methods in Cancer Research. Vol. 1 - The Analysis of Case-Control Studies (IARC Scientific Publications, 1980).
Zheng, S. L. et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med 358, 910–919 ((2008).
Nyholt, D. R. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74, 765–769 (2004).
Higgins, J. P. & Thompson, S. G. Quantifying heterogeneity in a meta-analysis. Stat Med 21, 1539–1558 (2002).
Higgins, J. P., Thompson, S. G., Deeks, J. J. & Altman, D. G. Measuring inconsistency in meta-analyses. BMJ 327, 557–560 (2003).
Kuritz, S. J., Landis, J. R. & Koch, G. G. A general overview of Mantel-Haenszel methods: applications and recent developments. Annu Rev Public Health 9, 123–160 (1988).
Acknowledgements
We express our gratitude to all participants in this study. The work in this paper was supported in part by the research grants 2140597 (C.K.S.L) and 14112514 (C.P.P.) from the General Research Fund, Hong Kong; and the Health and Medical Research Fund 01122236 (C.L.J.) & 11120801 (C.L.J.), Hong Kong. We express our sincere thanks to Dr Li Ma who has helped with part of the genotyping work in this study.
Author information
Authors and Affiliations
Contributions
S.S.R., C.P.P. and L.J.C. conceived the study idea and participated in the study design. C.K.S.L., A.M., K.M., M.T., L.J., N.H., M.Z., A.L.Y., N.W. and L.J.C. recruited the study subjects. S.S.R. and S.W.Y.C. conducted the genotyping work and carried out the statistical analysis. C.K.S.L., P.O.S.T., M.Z., N.W., K.N. and C.P.P. provided critical comments for the conduct of the study. S.S.R. and L.J.C. drafted the article. S.S.R., C.P.P. and L.J.C. revised the article. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Rong, S., Chen, L., Leung, C. et al. Ethnic specific association of the CAV1/CAV2 locus with primary open-angle glaucoma. Sci Rep 6, 27837 (2016). https://doi.org/10.1038/srep27837
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep27837
This article is cited by
-
Metabolic syndrome and risk of breast cancer mortality by menopause, obesity, and subtype
Breast Cancer Research and Treatment (2019)
-
Association of SIX1/SIX6 locus polymorphisms with regional circumpapillary retinal nerve fibre layer thickness: The Nagahama study
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.