Abstract
Under consecutive monoculture, the biomass and quality of Pseudostellaria heterophylla declines significantly. In this study, a three-year field experiment was conducted to identify typical growth inhibition effects caused by extended monoculturing of P. heterophylla. Deep pyrosequencing was used to examine changes in the structure and composition of soil fungal community along a three-year gradient of monoculture. The results revealed a distinct separation between the newly planted plot and the two-year, three-year monocultured plots. The Shannon and Simpson diversity indices were significantly higher in the two-year and three-year monoculture soils than in the newly planted soil. Consecutive monoculture of this plant led to a significant increase in relative abundance of Fusarium, Trichocladium and Myrothecium and Simplicillium, etc., but a significant decrease in the relative abundance of Penicillium. Quantitative PCR analysis confirmed a significant increase in Fusarium oxysporum, an agent known to cause wilt and rot disease of P. heterophylla. Furthermore, phenolic acid mixture at a ratio similar to that found in the rhizosphere could promote mycelial growth of pathogenic F. oxysporum. Overall, this study demonstrated that consecutive monoculture of P. heterophylla can alter the fungal community in the rhizosphere, including enrichment of host-specific pathogenic fungi at the expense of plant-beneficial fungi.
Similar content being viewed by others
Introduction
Pseudostellaria heterophylla, which is a perennial herbaceous plant that belongs to family Caryophyllaceae, is highly valued in traditional Chinese medicine. P. heterophylla contains various active constituents, including saponins, polysaccharides, amino acids, cyclopeptides and sapogenins. This plant provides treatments for spleen deficiency, anorexia, hyperirritability, palpitation and lassitude ailments1. It is mainly planted in the so-called geo-authentic production region, which has the most suitable soil and climate conditions; however, consecutive monoculture of this medicinal plant in the same land leads to a significant decline in the yield and quality of the underground tubers because of increasing disease pressure, which is known as soil sickness or consecutive monoculture problem2. Accordingly, farmers and Chinese traditional medicine industry have made understanding the mechanism of consecutive monoculture problems exhibited in P. heterophylla a priority.
Many factors have been reported to cause the decline in crop yield and quality in a monoculture regime, such as soil nutrients deficiency, root exudates’ autotoxicity and imbalances in soil microbial community3,4. The autotoxicity of root exudates has been considered as one of the main reasons for consecutive monoculture problems3. However, previous studies frequently use the filter paper bioassays enriched with a single chemical to determine the negative effects of exuded metabolites on target plant growth, which was considered as insufficient and controversial evidence since the influences of soil chemical properties and microbial communities were excluded5. Therefore, the shifts of rhizospheric microbial community under monoculture have been recently attracting considerable attention6. As is well-known, soil ecosystem functioning is largely governed by rhizospheric microbial dynamics since microbial composition and diversity affect geochemical cycles, humus formation and degradation, soil structure and biological interactions7,8. Soil fungal communities, in terms of abundance, composition and diversity play crucial roles in the betterment of ecosystem to guarantee soil quality and crop health9,10. Wu et al.11 found that Rehmannia glutinosa monoculture can alter fungal community in the rhizosphere, leading to an increase in pathogenic Fusarium oxysporum. Zhao et al.2 successfully isolated F. oxysporum from infected P. heterophylla plants and found that the amount of F. oxysporum increased significantly in the rhizosphere of this plant under consecutive monoculture. However, the responses of the rhizospheric fungal community and their functional significance to consecutive monoculture of P. heterophylla have not yet been fully elucidated. Therefore, this study was conducted to evaluate how the abundance and composition of the soil fungal community change with the increasing years of monoculture and which soil properties contribute to shape fungal community structure.
Results
The morphology and yield of P. heterophylla under consecutive monoculture
When compared with the newly planted plants (FY), the two-year (SY) and three-year (TY) monocultured plants displayed poorer growth with more adventitious fibrous roots and less aboveground biomass (Fig. 1a). Additionally, the dry weight of P. heterophylla tuber roots, the part most useful for traditional Chinese medicine, was significantly (P < 0.05) higher in the newly planted plots (FY) than in the two-year (SY) and three-year (TY) monocultured plots (Fig. 1b).
Soil chemical properties of different treatments
Compared with the newly planted soil, available nitrogen (AN) and available potassium (AK) were significantly higher in the two-year monoculture soil, and all available nutrients including AN, AK and available phosphorus (AP) were significantly higher in the three-year monoculture soil. However, the newly planted soil had significantly higher total nitrogen (TN) than the two-year and three-year monoculture soils. Moreover, total potassium (TK), TP and AP were significantly higher in the newly planted soil than in the two-year monoculture soil (Table 1).
OTU cluster and species annotation
ITS2 deep pyrosequencing was applied to assess the effects of consecutive monoculture of P. heterophylla on soil fungal community. In total, 756,116 ITS2 effective tags with species annotation were obtained from 12 soil samples, with each providing an average of 63,010 effective tags (Fig. S1). Rarefaction analyses showed that the observed species number tended to plateau at 50,000 sequences (Fig. 2). Sequences from 12 soil samples were assigned to 6,242 OTUs at the 97% similarity cut-off level. There were 628, 442, 457 and 554 OTUs in CK, FY, SY and TY, respectively (Fig. S1). On average, about 99.6% of the effective sequences could be grouped to the phylum level, and more than 94% were grouped to the species level (Fig. S2).
Alpha diversity indices
The richness and diversity indices of the fungal community in different soil samples were all calculated based on 52,585 sequences. Shannon and Simpson diversity indices were significantly higher in SY and TY than in FY (P < 0.05). The observed species and Chao1 were higher in SY and TY than in FY, but the difference was not significant (P > 0.05) (Table 2).
Beta diversity indices
The weighted Unifrac and unweighted Unifrac distances between FY and SY were 0.614 and 0.413, respectively, while they were 0.897 and 0.431 between FY and TY, respectively (Fig. 3). In comparison with FY, both weighted and unweighted Unifrac distances increased with the extended monoculture.
PCoA and UPGMA clustering
Both PCoA analysis and hierarchical cluster analysis revealed distinct differences in fungal community structure among different treatments and similar patterns for the same treatments when analyzed in triplicate (Fig. 4). Furthermore, PCoA analysis showed the fungal communities in FY and SY were separated from that in TY by principle component 1, and the community in FY was separated from that in SY by principle component 2 (Fig. 4b).
Principal coordinate analysis (PCoA) (a) and hierarchical clustering (b) of fungal communities based on weighted Unifrac algorithm for four different soil samples. CK, FY, SY and TY represent the control with no P. heterophylla cultivation, the newly planted, two-year monocultured and three-year monocultured plots, respectively.
Shifts in soil fungal community composition under consecutive monoculture
The fungal OTUs were comprised mainly of six phyla, Ascomycota, Zygomycota, Basidiomycota, Glomeromycota, Chytridiomycota and Rozellomycota. Ascomycota was the dominant microbial taxa, accounting for 91.6%, 94.6%, 96.4% and 70.7% of the total population in CK, FY, SY and TY, respectively (Fig. S3).
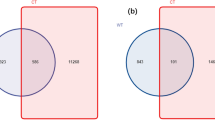
The percentage of species-level taxa shared in FY, SY and TY was 44.7% (438 species). Most species belongs to the phylum Ascomycota (88%) (Fig. 5a). The number of OTUs exclusively found in FY samples was 63 (6.4%), and these were mainly assigned to the phyla Ascomycota (50%) and Basidiomycota (44%) (Fig. 5b). The number of exclusive OTUs shared between SY and TY was 103 (10.5%), showing the similar community structure in SY and TY (Fig. 5c,d).
Venn diagram of exclusive and shared species-level taxa among the newly planted (FY), two-year monocultured (SY) and three-year monocultured (TY) soils. (a) Pie chart of shared OTUs (438) among 3 time-scale soil samples based on the average abundance of each phylum in FY, SY and TY. (b) Pie chart of exclusive OTUs (63) in FY; (c) Pie chart of exclusive OTUs (103) shared between SY and TY based on the relative abundance of each phylum in SY. (d) Pie chart of exclusive OTUs (103) shared between SY and TY based on the relative abundance of each phylum in TY.
Furthermore, at fungal genus level, consecutive monoculture of this plant led to a significant increase in the relative abundances of Fusarium, Clonostachys, Mortierella, Trichocladium and Myrothecium, but a significant decrease in the relative abundance of Penicillium (Fig. 6). Pathogenic F. oxysporum was frequently isolated from consecutively monocultured soil and infected P. heterophylla (Fig. S4). A F. oxysporum strain isolated from the infected plant parts could rapidly cause wilt disease on the seedlings of P. heterophylla in both tissue culture vessels under sterile conditions and in pots with sterilized soils (Fig. S4). Compared with NP, the genus Trichoderma increased in SY but decreased in TY.
Heat map analysis of the top 35 most abundant genera within a hierarchical cluster showed obvious variations in fungal community structure across the three time-scale replanted soils. Moreover, when compared with FY, the difference in community structure increased with increasing years of monoculture, indicating a gradual shift in the rhizosphere fungal community following extended monoculture (Fig. 7). When SY and TY were combined as one sample and termed “consecutively monocultured soil (CM)”, SIMPER analysis (relative abundance of genera, logarithmic transformation and standardization) showed that the pairwise dissimilarity in the fungal community between FY and CM was 52.5%. The top genera with 40% cumulative contribution to the dissimilarity between FY and CM are listed in Table 3, and these included Penicillium, Fusarium, Isaria, Myrothecium and Simplicillium, etc. (Table 3).
Effects of soil chemical properties on dominant genera
Redundancy analysis (RDA) of the dominant genera data and soil chemical properties revealed remarkable variations in fungal community structure in the three time-scale replanted soils. The first two RDA components (RDA1 and RDA2) could explain 65.4% and 23.2% of the total variance, respectively. The first RDA component (RDA1) separated the newly planted soils (NP) from the two-year (SY) and three-year (TY) monocultured soils. The newly planted soil was positively related to the higher relative abundance of Penicillium, and the higher contents of total N (TN), but negatively related to Fusarium, as well as higher contents of available N (AN) and available K (AK) (Fig. 8). A forward selection of environmental variables was carried out by a Monte-Carlo permutation test. Variables including total N (P = 0.02), available N (P = 0.04), total K (P = 0.02) and available K (P = 0.02) were significantly correlated with variations in the fungal community structure. The relative abundance of Penicillium was positively correlated with soil total N and negatively correlated with soil available N and available K. In contrast, the relative abundance of Fusarium was positively correlated with soil available N and available K and negatively correlated with soil total N (Fig. 8).
Redundancy analysis (RDA) of dominant genera and soil chemical properties for individual sample from four different treatments. CK, FY, SY and TY represent the control with no P. heterophylla cultivation, the newly planted, two-year monocultured and three-year monocultured plots, respectively. TN and AN represent total and available nitrogen, respectively. TK and AK represent total available potassium, respectively.
Abundance of F. oxysporum by quantitative PCR
The amount of F. oxysporum was significantly greater in consecutive monoculture soils (SY and TY) than in newly planted soils (FY) (Fig. 9a). The result was consistent with the deep pyrosequencing analysis except for the control. Moreover, a phenolic acid mixture at the same ratio found in the P. heterophylla rhizosphere could significantly promote the mycelial growth of isolated F. oxysporum. The mycelium diameter increased as the concentration increased until reaching a plateau at 120 μmol/L concentration (Fig. 9b).
Discussion
Consecutive monoculture problem, also known as replant disease or soil sickness, is common to Chinese medicinal herbs in all growing regions. About 70% of medicinal plant species with tuber roots have various degrees of consecutive monoculture problems, including P. heterophylla, Rehmannia glutinosa, and Panax notoginseng, etc12. Our three-year field experiment revealed typical growth inhibition effects caused by consecutive monoculture of P. heterophylla, with poor plant performance and insufficient resistance for disease and pests (Fig. 1). Studies of consecutive monoculture problems have centered on nutrient availability and the autotoxicity of allelochemicals released by roots3,4. However, increasing studies have shown that soil available nutrients did not decrease under consecutive monoculture of medicinal plants and fertilization was not effective for eliminating replant disease13,14. In the current work, we also found that some available nutrients including AN, AK and AP were significantly higher in the consecutively monocultured soils than in the newly planted soil (Table 1). Besides, many researchers have begun to doubt that allelochemicals are present in sufficient concentrations in soil to directly affect the growth of neighboring plants or the host plant5,15,16. If they could have these effects, the observation that replant disease can endure for many years after harvest would imply that allelochemicals are extraordinarily resilient to the degradation by soil microbes17. In our previous study, we found that most phenolic acids in rhizosphere soil of P. heterophylla did not continuously accumulate with increasing years of monoculture18. Moreover, mixtures of these phenolic acids at the same ratio found in the rhizosphere soil did not show direct autotoxicity toward tissue culture seedlings of P. heterophylla18.
Recently, the roles of belowground microbial community in aboveground plant performance have been of increasing interest6,19,20. Microbes are the unseen majority in soil and act as the most important drivers of plant health and productivity21,22. Fungi, an important group of microbes in the soil ecosystem, are crucial for soil functions and plant health23,24,25. In this study, pyrosequencing of ITS2 amplicons showed great shifts in fungal community composition and diversity in the rhizosphere after consecutive monoculture of P. heterophylla. The Shannon and Simpson diversity indices were significantly higher in the two-year and three-year monoculture soils than in the newly planted soil (Table 2). Xiong et al.26 found that soil fungal diversity index including the Chao1, ACE and Shannon index increased significantly after monoculture of vanilla (Vanilla planifolia). Zhou et al.27 found that exogenously applied p-hydroxybenzoic acid, an autotoxin of cucumber, increased the Shannon-Wiener index of the fungal community in the rhizosphere. Furthermore, we found that consecutive monoculture of this plant led to a significant increase in the relative abundances of Fusarium, Myrothecium and Simplicillium, etc., but a significant decrease in the relative abundance of Penicillium (Table 3). Among them, Fusarium spp. are well-known pathogens for many important crops28. The pathogenic fungi F. oxysporum, which is an agent of wilt and rot disease of P. heterophylla, was frequently isolated from infected tuber roots of this plant2. Quantification analysis by qPCR in this study further confirmed a significant increase in F. oxysporum in rhizosphere soil after P. heterophylla monoculture (Fig. 9). Moreover, Myrothecium spp. and Simplicillium spp., which have been reported to be pathogens of a number of plants29,30, exhibited a higher relative abundance in SY or TY than in FY (Table 3). However, the relative abundance of genus Penicillium decreased from 66.47% in FY to 2.63% in SY and 2.10% in TY (Table 3). Although some Penicillium species can cause plant diseases31, many of them are reportedly plant beneficial fungi that could enhance protection against pathogens by induced systemic resistance (ISR) and promote plant growth32,33. Hossain et al.34 demonstrated that the plant-growth-promoting-fungus Penicillium sp. GP16-2 could induce systemic resistance against Pseudomonas syringae in Arabidopsis thaliana. In this study, the decrease of genus Penicillium from FY to SY and TY was mainly attributed to the decrease of Penicillium sp. P_33. The total abundance of other Penicillium species (including Penicillium guanacastense, Penicillium oxalicum, Penicillium sp. MI 31 and Penicillium sumatrense) accounted for less than 2% of the total in the four soils. Penicillium sp. P_33 (NCBI Taxonomy ID: 472018, http://www.ncbi.nlm.nih.gov/nuccore/157419957) has been reported to possess antagonistic activity against plant pathogen Phytophthora citricola. These results suggest that the replant disease of P. heterophylla might be attributed to the rapid proliferation of potential pathogens at the expense of plant-growth-promoting-fungi in replanted soil. Li et al.28 found that fungal pathogens accumulated at the expense of plant-beneficial fungi under consecutive monoculture of peanut (Arachis hypogaea L.). In addition, many studies reported that the consecutive monoculture problems of plants resulted from shifts in the soil microbial community induced by root exudates rather than direct allelopathic autotoxicity11,35,36. Increasing studies have shown that root exudates could select microorganisms in the rhizosphere, and that these plant-associated microorganisms could then influence plant growth and health37,38,39. In this study, a phenolic acid mixture at the same ratio detected in the P. heterophylla rhizosphere could significantly promote the mycelial growth of pathogenic F. oxysporum. Zhou et al.40 found p-coumaric acid detected in root exudates of cucumber could significantly increase the densities of pathogenic F. oxysporum f.sp. cucumerinum Owen in soil. Moreover, Li et al.35 found peanut root exudates could selectively inhibit or stimulate certain microorganisms, particularly, the relative abundance of F. oxysporum. However, it should be noted that the positive effects of root-released phenolic acids on F. oxysporum may not be limited to the promotion of mycelial growth, but also include promotion of spore germination, toxin production and the epigenetic regulation of gene expression. Moreover, other root exudates such as soluble sugar, organic acids, and amino acids might also play important roles in the growth of F. oxysporum. All of these factors may contribute to the proliferation of pathogens. Accordingly, more researches in these aspects are needed and are currently underway.
In conclusion, consecutive monoculture of P. heterophylla can alter the fungal communities in the rhizosphere, resulting in fewer plant-beneficial fungi and more pathogenic fungi, which may be a contributing factor to soil sickness of this plant. However, further work is needed to explore the underlying mechanisms of the rhizosphere interactions between pathogenic and beneficial microorganisms and the roles of root exudates in the rhizosphere. The isolation of specific microorganisms (i.e., Penicillium spp. and Fusarium spp.) and their relationships with replant disease of this plant would also help understand this phenomenon.
Materials and Methods
Field experiment and soil sampling
P. heterophylla cultivar ‘Zheshen 2’, which is common in the main production region, was used as the test plant material. P. heterophylla was planted on November 20 and harvested by June 30 of the following year. After harvest, fields were kept fallow from July 1 to November 19. The experiment was conducted at Fuding City, Fujian Province (27°26′N, 120°04′E), which has a subtropical oceanic monsoon climate, annual mean temperature of 18.4 °C and an annual mean precipitation of 1668.3 mm. A field previously cultivated with Oryza sativa was used for this experiment with four treatments: i) the newly planted (FY), ii) two-year consecutive monoculture (SY), iii) three-year consecutive monoculture (TY) and iv) control with no P. heterophylla cultivation (CK). The soil chemical properties before the experiment were as follows: total nitrogen 1.83 g · kg−1, total phosphorus 0.47 g · kg−1, total potassium 8.46 g · kg−1, available nitrogen 26.23 mg · kg−1, available phosphorus 96.34 mg · kg−1, available potassium 365.21 mg · kg−1, pH 5.32. All treatments were organized within a single field site to keep the same soil and climatic conditions and subjected to the same fertilization and field management during the experimental period. Each treatment had three replicate plots and the study plots were completely randomized.
On April 15, 2014 we collected soil samples from 5 random locations within each plot because of the significant difference in growth status between different treatments on this date. The rhizosphere soil that clung to roots and rhizomes of P. heterophylla was brushed off and collected. Soil samples were sieved through 2 mm mesh to immediately extract total soil DNA. The rest of the soil samples were air-dried and used to determine soil physical–chemical properties13.
DNA extraction and PCR amplification
Total DNA were extracted from soil samples using SoilGen DNA kit (CWBIO, Beijing, China) following the manufacturer’s instructions. DNA concentration was determined by using Nanodrop 2000C Spectrophotometer (Thermo Scientific, USA) and then DNA was diluted to 1 ng/μL using sterile water. Internal transcribed spacer (ITS2) was amplified by using specific primer ITS2F (GCATCGATGAAGAACGCAGC) and ITS2R (TCCTC CGCTTATTGATATGC) with the barcode. All PCR reactions were carried out with Phusion High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, USA).
PCR purification and pyrosequencing
PCR products were monitored on 2% agarose gel and samples with bright main strip between 400–450 bp were chosen for further analysis. PCR products were mixed at an equal ratio and purified with Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany). Sequencing libraries were generated using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, USA) following the manufacturer’s instructions. Pyrosequencing was performed on an Illumina HiSeq2500 platform and 250 bp paired-end reads were generated.
OTU (Operational Taxonomic Unit)-based sequence analysis
The sequence reads were assigned to each sample based on their unique barcode and truncated by cutting off the barcode and primer sequences. Paired-end reads were merged using FLASH (V1.2.7)41. Through quality filtering and chimera removal, the retained sequences called as effective tags were used to perform OTU cluster and species annotation. Sequences with ≥97% similarity were assigned to the same OTU through Uparse software (Uparse v7.0.1001)42. Species annotation was carried out via the Unite Database (https://unite.ut.ee/)43 based on Blast algorithm which was calculated by QIIME software (Version 1.7.0).
Quantitative PCR for F. oxysporum
Quantitative PCR (qPCR) was carried out to quantify F. oxysporum in different soil samples by using the primers ITS1-F (CTTGGTCATTTAGAGGAAGTAA) and AFP308R (CGAATTAACGCGAGTCCCAA)44. Quantitative PCR was performed in 15 μl reaction mixture containing 7.5 μl 2× SYBR green I SuperReal Premix (TIANGEN, Beijing, China), 0.5 μl of each primer (10 μM) and template DNA (20 ng of total soil DNA or a serial dilution of plasmid DNA for standard curves). Four independent quantitative PCR assays were performed for each treatment.
The effects of phenolic acids on the growth of isolated F. oxysporum
Based on the HPLC results of phenolic acids in the P. heterophylla rhizosphere18, a phenolic acid mixture at the same ratio as detected in soil including gallic acid, coumaric acid, p-hydroxybenzoic acid, vanillic acid, syringic acid, vanillin, ferulic acid and benzoic acid was used to assess the effects on the growth of isolated F. oxysporum. Specifically, the isolated F. oxysporum was inoculated onto the center of a 9 cm diameter Petri dish filled with a 10-fold dilution of soil-extract agar medium (SEM) containing different final concentrations of phenolic acid mixtures (30, 60, 120, 240 and 480 μmol/L). Each treatment had three replicates. The mycelium diameter was measured after eight days of incubation at 28 °C in 0:24 under a 0:24 light: dark cycle.
Statistical analyses
The abundances of OTUs were normalized, after which alpha and beta diversity analyses were performed based on the normalized data. Alpha diversity was applied to analyze species complexity for a sample through six indices, observed-species, community richness indices (chao1, ACE) and diversity indices (Shannon’s, Simpson’s). Beta diversity analysis was used to evaluate differences in species complexity between samples, including principal coordinate analysis (PCoA) and unweighted pair-group method with arithmetic means (UPGMA) clustering.
For all parameters, one way analysis of variance (ANOVA) followed by Tukey’s test (P < 0.05) was used for multiple comparisons through DPS software version 7.51. Similarity percentage analysis (SIMPER) for assessing the relative contribution (%) of each microbial taxa to the dissimilarity between samples was performed with the PRIMER V5 software package (PRIMER-E Ltd, Plymouth, UK)45.
Additional Information
How to cite this article: Wu, L. et al. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 6, 26601; doi: 10.1038/srep26601 (2016).
References
Zhao, W. O., Pang, L., Dong, N. & Yang, S. LC-ESI-MS/MS analysis and pharmacokinetics of heterophyllin B, a cyclic octapeptide from Pseudostellaria heterophylla in rat plasma. Biomed. Chromatogr. 29, 1693–1699 (2015).
Zhao, Y. P. et al. Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f.sp. heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. 5, 8197 (2015).
Huang, L. F. et al. Plant-soil feedbacks and soil sickness: from mechanisms to application in agriculture. J. Chem. Ecol. 39, 232–242 (2013).
Bennett, A. J., Bending, G. D., Chandler, D., Hilton, S. & Mills, P. Meeting the demand for crop production: the challenge of yield decline in crops grown in short rotations. Biol. Rev. 87, 52–71 (2012).
Kaur, H., Kaur, R., Kaur, S. & Baldwin, I. T & Inderjit. Taking ecological function seriously: soil microbial communities can obviate allelopathic effects of released metabolites. Plos ONE 4, e4700 (2009).
Berendsen, R. L., Pieterse, C. M. & Bakker, P. A. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012).
Huang, L. F. et al. Plant-soil feedbacks and soil sickness: from mechanisms to application in agriculture. J. Chem. Ecol. 39, 232–242 (2013).
Bennett, A. J., Bending, G. D., Chandler, D., Hilton, S. & Mills, P. Meeting the demand for crop production: the challenge of yield decline in crops grown in short rotations. Biol. Rev. 87, 52–71 (2012).
Manici, L. M. & Caputo, F. Fungal community diversity and soil health in intensive potato cropping systems of the east Po valley, northern Italy. Ann. Appl. Biol. 155, 245–258 (2009).
Xu, L., Ravnskov, S., Larsen, J., Nilsson, R. H. & Nicolaisen, M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 46, 26–32 (2012).
Wu, L. K. et al. Plant-microbe rhizosphere interactions mediated by Rehmannia glutinosa root exudates under consecutive monoculture. Sci. Rep. 5, 15871 (2015).
Zhang, Z. Y. & Lin, W. X. Continuous cropping obstacle and allelopathic autotoxicity of medicinal plants. Chin. J. Eco-Agric. 17, 189–196 (2009).
Wu, L. K. et al. Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture. Appl. Soil. Ecol. 67, 1–9 (2013).
Bramley, R. G. V., Ellis, N., Nable, R. O. & Garside, A. L. Changes in soil chemical properties under long-term sugar cane monoculture and their possible role in sugar yield decline. Soil Res. 34, 967–984 (1996).
Weidenhamer, J. D., Li, M., Allman, J., Bergosh, R. G. & Posner, M. Evidence does not support a role for gallic acid in Phragmites australis invasion success. J. Chem. Ecol. 39, 323–332 (2013).
Ehlers, B. K. Soil microorganisms alleviate the allelochemical effects of a thyme monoterpeneon the performance of an associated grass species. Plos ONE 6, e26321 (2011).
Mazzola, M. & Manici, L. M. Apple replant disease: role of microbial ecology in cause and control. Annu. Rev. Phytopathol. 50, 45–65 (2012).
Wu, H. M. et al. Mixed phenolic acids mediated proliferation of pathogens Talaromyces helicus and Kosakonia sacchari in continuously monocultured Radix pseudostellariae rhizosphere soil. Front. Microbiol. 7, 335 (2016).
Wardle, D. A. et al. Ecological linkages between aboveground and belowground biota. Science 304, 1629–1633 (2004).
Haney, C. H. & Ausubel, F. M. Plant microbiome blueprints. Science 349, 788–789 (2015).
Van Der Heijden, M. G., Bardgett, R. D. & Van Straalen, N. M. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008).
Santhanam, R., Weinhold, A., Goldberg, J., Oh, Y. & Baldwin, I. T. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. P. Natl. Acad. Sci. USA 112, E5013–E5020 (2015).
Xu, L., Ravnskov, S., Larsen, J., Nilsson, R. H. & Nicolaisen, M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 46, 26–32 (2012).
Liu, X. et al. Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. Plos ONE 9, e86610 (2014).
Zhou, X. & Wu, F. Dynamics of the diversity of fungal and Fusarium communities during continuous cropping of cucumber in the greenhouse. FEMS Microbiol. Ecol. 80, 469–478 (2012).
Xiong, W. et al. Different continuous cropping spans significantly affect microbial community membership and structure in a vanilla-grown soil as revealed by deep pyrosequencing. Microbial Ecol. 70, 209–218 (2015).
Zhou, X., Yu, G. & Wu, F. Responses of soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.) to exogenously applied p-hydroxybenzoic acid. J. Chem. Ecol. 38, 975–983 (2012).
Li, X. G., Ding, C. F., Zhang, T. L. & Wang, X. X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 72, 11–18 (2014).
Moreira, F. G., Reis, S. D., Costa, M. A. F., Souza, C. G. M. D. & Peralta, R. M. Production of hydrolytic enzymes by the plant pathogenic fungus Myrothecium verrucaria in submerged cultures. Braz. J. Microbiol. 36, 07–11 (2005).
Ward, N. A., Schneider, R. W. & Aime, M. C. Colonization of soybean rust sori by Simplicillium lanosoniveum . Fungal Ecol. 4, 303–308 (2011).
Leelasuphakul, W., Hemmanee, P. & Chuenchitt, S. Growth inhibitory properties of Bacillus subtilis strains and their metabolites against the green mold pathogen (Penicillium digitatum Sacc.) of citrus fruit. Postharvest Biol. Tec. 48, 113–121 (2008).
Van Wees, S. C., Van der Ent, S. & Pieterse, C. M. Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448 (2008).
Gulden, R. H. & Vessey, J. K. Penicillium bilaii inoculation increases root-hair production in field pea. C. J. Plant Sci. 80, 801–804 (2000).
Hossain, M. M., Sultana, F., Kubota, M. & Hyakumachi, M. Differential inducible defense mechanisms against bacterial speck pathogen in Arabidopsis thaliana by plant-growth-promoting-fungus Penicillium sp. GP16-2 and its cell free filtrate. Plant Soil 304, 227–239 (2008).
Li, X. G. et al. Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 78, 149–159 (2014).
Zhou, X., Yu, G. & Wu, F. Soil phenolics in a continuously mono-cropped cucumber (Cucumis sativus L.) system and their effects on cucumber seedling growth and soil microbial communities. Eur. J. Soil Sci. 63, 332–340 (2012).
Haichar, F. Z. et al. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230 (2008).
Paterson, E., Gebbing, T., Abel, C., Sim, A. & Telfer, G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 173, 600–610 (2007).
Huang, X. F. et al. Rhizosphere interactions: root exudates, microbes, and microbial communities 1. Botany 92, 267–275 (2014).
Zhou, X. & Wu, F. p-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f.sp. cucumerinum Owen. Plos ONE 7, e48288 (2012).
Magoč, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Kõljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277 (2013).
Lievens, B. et al. Quantitative assessment of phytopathogenic fungi in various substrates using a DNA macroarray. Environ. Microbiol. 7, 1698–1710 (2005).
Rees, G. N., Baldwin, D. S., Watson, G. O., Perryman, S. & Nielsen, D. L. Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Anton. Leeuw. Int. J. G. 86, 339–347 (2004).
Acknowledgements
This work was supported by grants from the National Science Foundation of China (Grant nos 81303170, U1205021, 81573530 and 31401950) and the Outstanding Youth Scientific Fund of Fujian Agriculture and Forestry University (Grant no. XJQ201501).
Author information
Authors and Affiliations
Contributions
W.X.L. and L.K.W. conceived the study; L.K.W., W.X.L., J.C. and M.U.K. wrote the paper. L.K.W., J.C., H.M.W. and J.Y.W. performed experiments; L.K.W., J.C. and S.L. performed the statistical analyses; Z.Y.Z., Y.H.W. and J.C. are involved in field management and soil sampling. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wu, L., Chen, J., Wu, H. et al. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci Rep 6, 26601 (2016). https://doi.org/10.1038/srep26601
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26601
This article is cited by
-
Impacts of continuous and rotational cropping practices on soil chemical properties and microbial communities during peanut cultivation
Scientific Reports (2022)
-
Potato tillage method is associated with soil microbial communities, soil chemical properties, and potato yield
Journal of Microbiology (2022)
-
Effects of continuous cropping of Pinellia ternata (Thunb.) Breit. on soil physicochemical properties, enzyme activities, microbial communities and functional genes
Chemical and Biological Technologies in Agriculture (2021)
-
Leaching alleviates phenol-mediated root rot in Panax notoginseng by modifying the soil microbiota
Plant and Soil (2021)
-
Long-term continuously monocropped peanut significantly disturbed the balance of soil fungal communities
Journal of Microbiology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.