Abstract
This study assessed the prevalence and zoonotic potential of Shiga toxin-producing Escherichia coli (STEC) sampled from 104 dairy units in the central region of Zambia and compared these with isolates from patients presenting with diarrhoea in the same region. A subset of 297 E. coli strains were sequenced allowing in silico analyses of phylo- and sero-groups. The majority of the bovine strains clustered in the B1 ‘commensal’ phylogroup (67%) and included a diverse array of serogroups. 11% (41/371) of the isolates from Zambian dairy cattle contained Shiga toxin genes (stx) while none (0/73) of the human isolates were positive. While the toxicity of a subset of these isolates was demonstrated, none of the randomly selected STEC belonged to key serogroups associated with human disease and none encoded a type 3 secretion system synonymous with typical enterohaemorrhagic strains. Positive selection for E. coli O157:H7 across the farms identified only one positive isolate again indicating this serotype is rare in these animals. In summary, while Stx-encoding E. coli strains are common in this dairy population, the majority of these strains are unlikely to cause disease in humans. However, the threat remains of the emergence of strains virulent to humans from this reservoir.
Similar content being viewed by others
Introduction
Shiga toxigenic Escherichia coli (STEC) are emerging pathogens of public health concern worldwide, including in Europe, North and South America and Asia1,2. Ruminants, in particular cattle, have been identified as the predominant reservoir of STEC3,4, indicating that the bacteriophage-encoded Shiga toxins (Stx) are likely to confer an advantage to E. coli in these host animals. In Africa there is little information on the epidemiology of STEC in livestock systems and their impact on human health1. It is evident that only a subset of STEC are a serious threat to human health, these enterohaemorrhagic E. coli (EHEC) are associated with specific serogroups in particular the seven that have been defined as adulterants in beef production in the USA, O157, O26, O111, O45, O145, O103, O1215,6. Similar serotypes, especially O157 & O26 are also an issue in Europe. Typical EHEC strains can be further characterised by possession of a type 3 secretion system (T3SS) that enables colonisation of the gastrointestinal tract7. EHEC infections in humans are associated with diarrhoea and bloody diarrhoea, with the more serious sequelae of kidney and brain damage due to activity of Stx on the microvasculature in these organs4,8.
The cost of whole genome sequencing (WGS) has drastically reduced and it is now possible to sequence large numbers of isolates and use bioinformatics approaches to extract strain relatedness and gene carriage data. For E. coli, strains have classically been assigned into phylogroups that provide a good correlation with commensal versus pathogenic strains9. The phylogroups are based on particular combinations of specific genes and can be assigned from the WGS. Whole genome core SNP analysis to define strain relatedness is now commonly used and provides greater resolution than MLST10. In addition the serogroup, the O-chain of LPS, can also be inferred from their genetic determinants using WGS. The WGS of any strain collection is therefore a valuable resource allowing relatively rapid comparison of phylogeny and pathogenic potential.
Both small holding and large-scale dairy farming is important to the economic survival of communities in many developing nations, including Zambia11,12. As such, it is important to understand if practices on these units and their products may represent a threat to human health and where such risks exist suggest possible mitigation measures. A study has recently been carried out to sample E. coli strains from cattle across small, medium and large-scale (commercial) dairy farms in central Zambia, with the primary aim of understanding antibiotic use and antimicrobial resistance patterns in this sector13.
These isolates have now been further analysed in the present study for stx prevalence and any association with the farming system. In addition, E. coli isolates from patients with diarrhoea were also screened and sequenced to determine evidence of relationship to the bovine strains.
Results
Detection of Shiga toxin alleles (stx1 and stx2) in isolates from Zambian dairy cattle and humans
Eleven percent (41/371) of the bovine E. coli isolates were positive for the presence of Shiga toxin genes as defined by detection of appropriately sized PCR amplification products using an established stx multiplex assay14. Based on this, both stx1 and stx2 were detected in 54% (22/41) of the STEC, while 37% (15/41) had stx2 only and 10% (4/41) had stx1 only. Using this data, the overall adjusted prevalence of STEC across the different farming scales for the central Zambian study area can be estimated at 6% (95% CI: 2.5–10.2). The adjusted prevalence per farming scale was higher in medium-scale 17.1% (95% CI: 5.9–28.2) and small-scale 10.6% (95% CI: 6.6–14.5) farms when compared to the commercial farms 2.8% (95% CI: 0.3–6.0). Based on these ranges, there is a significant difference in estimated prevalence between the small and commercial scales. Logistic regression indicates that medium- and small-scale farming are significant risk factors for Shiga toxin producing E. coli (STEC) with commercial as a reference (Table 1). Out of the 73 E. coli isolates from human patients with diarrhoea for which good quality sequence information was generated, no Shiga toxin genes were detected.
As an additional investigation, the enrichment cultures for all the animals (n = 371) were streaked onto sorbitol MacConkey agar plates and any non-sorbitol fermenting colonies tested for O157 agglutination. Only one animal yielded a positive strain (ZB-2213N0194) and this was then added to the study.
Phylogenetics
In order to understand the genetic backgrounds of the STEC strains isolated in this study, including their potential threat to human health, their relationship to other human disease-associated EHEC were tested by phylogenetic methods. The WGS of 297 of the Zambian isolates (224 bovine and 73 human) were determined. This included 41 STEC, 37 of the 41 defined as stx+ by PCR from main study, three STEC strains from a pilot study and the single positively selected E. coli O157 strain. These were compared with 262 E. coli sequences from human, cattle, avian and canine hosts; one hundred and twenty nine strains in this second collection were human clinical STEC isolates (see Supplementary Table 1).
Alignment to a reference genome (E. coli O157:H7 str. Sakai, RefSeq assembly accession: GCF_000008865) resulted in 715,632 core positions with 68,327 single nucleotide polymorphism (SNPs) across all 559 sequences. A maximum likelihood phylogeny revealed the population structure of the E. coli strains (Fig. 1). While there was no clear clustering of the strains based on geographical location or host, there was, as anticipated, good correlation with established E. coli phylogroups, with only minor discordance. All possible phylogroups and cryptic clades were identified, however the majority of the E. coli strains (97%) were distributed across 5 phylogroups (Fig. 1).
The tree depicts the phylogenetic relationship of E. coli isolates from Zambia (bovine - red and human - blue) with other E. coli isolates (grey). The ML tree is based on core SNPs as described in Materials and Methods. The tree is un-rooted and grey circles on branches represent bootstrap values higher than 80. Vertical columns demonstrate: (1) Diversity of the sequence types (ST) based on MLST analysis where each colour represents a different ST; (2) Diversity of O-serogroups for which each colour represents a different group; (3) Phylogroups: A-yellow, B1-red, B2-blue, C-green, D-turquoise, E-pink, F-grey, cryptic clades-light green. The phylogroups are consistent with core SNP clustering with some minor discordance. White spaces on all columns indicate sequences that were untypable.
The Zambian bovine strains (n = 224) predominately associated with the B1 ‘commensal’ cluster (67%) with the remainder present as: A (9%); B2 (4%); C (8%); D (9%); other (3%). By contrast, the Zambian human strains (n = 73) had equivalent representation across the 5 main phylogroups: A (22%); B1 (19%); B2 (16%); C (22%); D (16%); other (5%). The Zambian cattle STEC strains were also predominately in the B1 phylogroup (27/41).
Serotyping
The majority of EHEC strains that are a threat to human health are associated with 7 specific serogroups. A bioinformatic approach was used to serotype the sequenced strains. H typing was possible for 550/559 strains and O-typing for 483/559 (summarized in Supplementary Tables 3 and 4). Failure to detect specific genes in some of the strains was most likely due to assembly issues with short read sequences.
With the exception of the positively selected E. coli O157 strain, none of the Zambian bovine STEC strains (0/40) were assigned to any of these seven serogroups. In fact, only 3 strains across the whole set of cattle isolates could be assigned within these serogroups (ZB-244; serogroup O45 and ZB-2213N0112; serogroup O111 and ZB-2213N0194; serogroup O157). Overall, the Zambian strains (cattle and human) exhibited an extensive array of serogroups (Fig. 1 and Supplementary Table 4) and H:O combinations were unique for each strain with little clustering or association with established human clinical isolates (Fig. 2).
The same ML core SNP tree as in Fig. 1 plotted in a circular manner to depict relationships between Shiga toxin encoding strains. The strain designations are Zambian bovine (red), Zambian human (blue), other E. coli (grey). For the Zambian strains, the coloured bars indicate the presence of Stx genes: stx2 (purple) and stx1 (green). Black blocks around the tree indicate non-Zambian E. coli encoding stx (1 or 2). Orange blocks highlight the presence of intimin (eae) and sepL indicating the possession of a type 3 secretion system. It is apparent that with the exception of one positively selected EHEC O157 (ZB-2213N0194), that the Zambian cattle STEC do not encode this system.
Toxicity analysis of stx+ strains
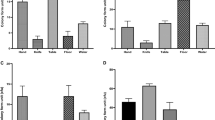
To determine if the genotypically positive stx strains were able to express Stx, eighteen of the bovine STEC were examined for Vero cell cytotoxicity with and without mitomycin C (MMC) induction. 89% (16/18) of the MMC-induced STEC strains had a cytotoxic effect on Vero cells (Fig. 3). These samples were verified as Stx positive using a commercial ELISA (Fig. 3), with only one strain (4) exhibiting toxicity on Vero cells without any detection of Stx by ELISA.
The top panel graph indicates the cytotoxic effect of selected STEC strain supernatants on Vero cells. Increased cell survival results in higher values. 0.2% Triton X-100 was used as a positive control; RPMI + LB and RPMI alone were used as negative controls. Values below the dashed line indicate a cytotoxic effect on the cells. 89% (16/18) of the STEC supernatants tested demonstrated a cytotoxic effect. Supernatants were prepared as described in Materials and Methods. Stx subtypes are shown in the lower panel along with ELISA results for detection of Stx. Isolates with both stx1a and stx2a are associated with higher toxicity. Sample number ZB-4-stx contains stx2a and exhibited cytotoxicity on Vero cells but was negative by ELISA.
Shiga toxin subtyping
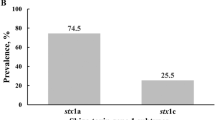
Forty one STEC positive strains were included in the WGS analysis (Fig. 2) and from this stx alleles could be further subtyped using a published BLAST-based methodology15 (Supplementary Table 5). It was evident that the most cytotoxic strains (Fig. 3) were those encoding Stx2a often in combination with Stx1a, in line with studies of cytotoxicity and pathology induced by enterohaemorrhagic strains with different Stx variants16,17.
Stx association with type 3 secretion and enteroaggregative virulence factors
Typical enterohaemorrhagic E. coli strains are defined by the co-association of stx genes with a type 3 secretion system (T3SS)7. In the present study, the presence of a T3SS was determined by detection of both eae and sepL. Based on BLAST analysis, 3.6% (8/224) and 2.7% (2/73) of the Zambian bovine and human isolates respectively may encode a T3SS (Fig. 2). Excluding the positively selected O157 strain, neither intimin (eae) nor sepL were detected in the bovine STEC. Other non-STEC but intimin positive strains were present within the Zambian strains analysed and some were in close proximity to clinical human STEC strains (Fig. 2). The cattle strains were also checked for the presence of the enteroaggregative E. coli (EAEC) adherence factors AggR and AA probe by PCR but all were negative indicating that these dairy cattle are not a common reservoir of enteroaggregative E. coli as associated with the atypical EHEC O104 outbreak in Northern Germany in 2011.
Discussion
Shiga toxins (Stx) can pose a serious threat to human health, and human infections are usually restricted to a subset of serogroups that can express Stx and a type 3 secretion system (T3SS) or other adherence mechanisms that can facilitate colonisation of the human gastrointestinal tract. In this study, E. coli isolates were obtained from cattle associated with dairy production in a region of central Zambia. A total of 371 isolates, each from an individual animal, covering 104 farms were tested for the presence of stx by PCR. Of these, 41 (11%) were positive. This gives an estimated prevalence (taking into account sampling and the study design effect) of 6% (95% CI: 2.5–10.2). To our knowledge, this is one of the first surveys to systematically analyse the proportion of random E. coli from a farm animal source that are positive for stx as other studies usually use positive selection methods from animals. Our survey does indicate that STEC are common in these dairy cattle. It was also evident that the small and medium sized production units had a higher prevalence of STEC than the commercial units sampled. This is of interest as it does indicate that management practices potentially influence the selection of STEC. On-going work will investigate these differences including the influence of breed which can differ between the farming scales13 and/or diet which can change EHEC O157 prevalence18.
Our study also examined 73 E. coli strains from human patients with diarrhoea presenting at the University Teaching Hospital (UTH) in Lusaka over the same time period. While the human sample numbers were relatively low, stx was not detected, indicating that STEC are unlikely to be common in the local human population. The overall phylogenetic analyses of the strains into phylogroups was as anticipated, with the majority of the Zambian bovine strains being present in the B1 group associated more with commensal strains, although this cluster contains non-O157 EHEC serotypes causing infections in humans that likely originate from cattle. A greater proportion of the human Zambian isolates clustered within the phylogroups associated with human disease, reflecting that the strains were collected from patients with diarrhoea and in some cases the strain may be the etiological agent.
Based on bioinformatic analyses of WGS, there was marked diversity of serotypes in the cattle and human sample populations (Fig. 1 and Supplementary Tables 3 and 4). None of the randomly selected bovine STEC were allocated to a serogroup commonly associated with human EHEC infections. Furthermore, none of these bovine STEC encoded a T3SS based on detection of intimin (eae) and sepL alleles (Fig. 2), as both genes are present on the locus of enterocyte effacement that encodes the system. An additional study to positively select for E. coli O157 isolates from the faecal pat enrichments only identified one positive sample from a farm. Taken together, this study indicates that while STEC are common in the Zambian dairy cattle these strains would not be classified as EHEC and are unlikely to be associated with serious human disease.
While it is encouraging that EHEC strains were extremely rare, many of the supernatants from the STEC strains were cytotoxic and appropriate backgrounds for EHEC emergence are present. As such, we should remain vigilant in case Stx-encoding prophages from this reservoir do emerge in other strain backgrounds that have a higher capacity to cause disease in the human host. Continuing work on factors driving the maintenance of STEC strains in the bovine host will hopefully clarify approaches to reducing the threat from this emerging group of pathogens.
Methods
Bovine and human isolates from Zambia
Bovine E. coli isolates (n = 371) were collected as part of a previously published study investigating antimicrobial resistance13. In addition, a further 81 E. coli isolates from cattle were collected as part of a pilot study in 2013 in the same region13. Faecal sampling and animal handling of the farm animals was carried out in accordance with the approved guidelines issued by The Roslin Institute Animal Welfare and Ethical Review Body which approved this study13. In the main study, 376 dairy cattle from 104 farms representing about 20% of the dairy herds in the study area were randomly sampled and an E. coli was isolated from 371 animals (E. coli was not isolated from 5 animals) based on growth characteristics on both MacConkey agar and Bile-X-Glucuronide (TBX) plates (Oxoid, UK). Subsequent phylotyping indicated that 97% (361/371) could be allocated to established E. coli phylogroups19. In terms of subsequent studies the isolates were chosen as follows: From the main study all isolates (n = 371) were tested by the stx PCR to allow the prevalence to be estimated. 188 were sequenced but 186 were used in the phylogenetic analyses due to quality issues with 2 sets of reads. The sub-selected strains for sequencing were as follows (those with poor reads removed): (1) All strains showing phenotypic antibiotic resistance in the original study (n = 61)13; Strains positive for stx by PCR (37/41); the rest (n = 88) were randomly chosen from the remainder. From the pilot study 40 from 81 strains were sequenced including three that were stx+ as determined by PCR; 37/40 were included in the phylogenetic study as three had read quality issues. As a separate study, enrichments from all animals (n = 371) were plated onto sorbitol MacConkey and any non-sorbitol fermenting colonies (3 per plate) tested for O157 agglutination. Only one animal yielded a positive isolate (ZB-2213N0194) and this positively selected isolated was then sequenced and added into the phylogenetic analysis. In total there were 224 bovine E. coli good quality whole genome sequences that were analysed in this study.
E. coli isolates (n = 79) from patients presenting with diarrhoea were collected at Lusaka hospital between 4th December 2014 and 7th January 2015 as part of another project managed by Prof. J.B. Muma and generously supplied for sequencing. Informed consent was obtained from all subjects. Six of the isolate sequences were not analysed due to read quality leaving n = 73 for phylogenetic and virulence determinant analysis. Further strain and sequence details are provided in the Supplementary Table 1.
DNA extraction
DNA extraction was carried out using either a Wizard Genomic DNA Extraction Kit® or a Qiagen® DNA extraction kit from 1 ml of bacterial culture as defined in the manufacturers’ protocols.
PCR detection of virulence determinants
All the bovine strains were screened by a published multiplex PCR for Shiga toxin genes and intimin14. The strains were also screened with a multiplex PCR for aggR and AA probe genes as markers for enteroaggregative E. coli20. The PCR products were visualised and captured using multi imaging software (Fluorchem HD2) following electrophoresis in 1.5% w/v agarose gel (Agarose, Melford, UK) and staining with Gelred®.
Verocytotoxicity assays
Established method21,22, with these minor variations: Single colonies were selected from LB agar plates and suspended in 10 ml of LB broth for 24 h (overnight). 50 μl of overnight culture was added to 5 ml (1:100) of fresh LB broth and incubated for 60 min. Then 20 μl of 5 μg/ml mitomycin C (MMC) was added followed by overnight incubation.
Supernatant samples were screened for the presence or absence of Stx using a commercial ELISA kit (RIDASCREEN® Verotoxin ELISA (C2201), R-Biopharm AG, Darmstad, Germany) according to the manufacturer’s instructions.
Statistical analysis
The adjustment of prevalence estimates per farming scale and the risk factor analysis were carried out using logistic regression in ‘survey package’23 in R software environment version 3.1.1 (http://cran.r-project.org/), p < 0.05 values were taken as statistically significant. The statistical analyses and more information on the definition of the different level farming systems were as described previously13.
E. coli whole genome sequence analyses
To better understand how the Zambian E. coli strains dataset compared with other E. coli, the Zambian strain genomic sequences were analysed with a larger strain collection that consisted of 559 E. coli genomes, including clinical and commensal isolates from 4 different broad categories of animal and human hosts (Supplementary Table 1). New short read sequence files have been uploaded to European Nucleotide Archive under the study accession number: PRJEB11782, PRJEB11950, PRJEB11956. Some genome sequences from the Zambian strain sets were removed due to poor read and/or assembly quality, resulting in 297 Zambian genome sequences (224 bovine and 73 human) available for analysis.
Sequencing analysis
All reads were generated by Illumina 1.9 paired-end read sequencing with read lengths from 36 to 251 bp. FASTQC24 was used for quality assessment and where necessary trimming was done with cutadapt25. Short reads were aligned to a reference E. coli O157:H7 str. Sakai (RefSeq assembly accession: GCF_000008865) by combining BWA26, SAMtools and SnpEff27 in a custom-made python script. The consensus sequence for each alignment of 5,590,092 bp was produced using the majority rule.
Consensus sequences for each alignment were concatenated into one multifasta file that were then parsed to find core positions. Multifasta files of concatenated core nucleotides for each strain were used for recombination analysis with GUBBINS28. The recombinatorial regions were removed from the final alignment. The final alignment was then used to construct a Maximum Likelihood (ML) tree with RAxML29 under a GAMMA model of heterogeneity with 100 bootstrap replicates (BS). The trees were visualised with ITOL30.
An established phylotyping scheme31 was used as a starting point to develop a programme that assigned each strain to one of the 4 possible phylogroups (A, B1, B2, D) based on the presence or absence of one of 3 genes chuA, yjaA, arpA and one genetic fragment TspE4.C2. To further distinguish between groups and assign strains to an additional 4 phylogroups (C, E, F or cryptic clades), it was necessary to check for the presence of a fifth gene trpA and for the presence of specific alleles for the above genes. arpA alleles were used to distinguish between phylogroups D and E based on specific primer sequences described in31.
To establish gene presence or absence a database that includes all sequences from the collection were built with BLAST+15. Query gene’s sequences of intimin, sepL, chuA, yjaA, arpA, trpA, genetic fragment TspE4.C2 were downloaded from the NCBI website. Gene identifiers are presented in the (Supplementary Table 2). Query Shiga toxin sequences identified in32 also were downloaded from the NCBI website. Gene’s presence were established based on a E-value = 0 and similarity match at >90% coverage of the query sequence. For Shiga toxins blast results were filtered based on bit score above 1000, if multiple contigs were involved only the highest result was kept.
Serogroups were identified based on presence of one or several alleles from the following genes: for O-typing - wzx, wzy wzm and wzt; for H-typing the flagellin genes fliC, flkA, fllA, flmA and flnA. Databases were provided by Dr Flemming Scheutz and colleges32. Multi locus sequence type were identified using SRST2 software33.
Additional Information
Accession codes: Short read sequence files have been uploaded to European Nucleotide Archive under the study accession number: PRJEB11782, PRJEB11950, PRJEB11956.
How to cite this article: Mainda, G. et al. Phylogenomic approaches to determine the zoonotic potential of Shiga toxin-producing Escherichia coli (STEC) isolated from Zambian dairy cattle. Sci. Rep. 6, 26589; doi: 10.1038/srep26589 (2016).
References
Islam, M. Z. et al. Regional variation in the prevalence of E. coli O157 in cattle: A meta-analysis and meta-regression. PloS One 9 10.1371/journal.pone.0093299 (2014).
Chase-Topping, M. E. et al. Pathogenic potential to humans of bovine Escherichia coli O26, Scotland. Emerging Infectious Diseases 18, 439–448 (2012).
Pruimboom-Brees, I. M. et al. Cattle lack vascular receptors for Escherichia coli O157: H7 Shiga toxins. Proceedings of the National Academy of Sciences 97, 10325–10329 (2000).
Karch, H. et al. In Zoonoses-Infections Affecting Humans and Animals 235–248 (Springer, 2015). URL: http://link.springer.com/chapter/10.1007/978-94-017-9457-2_9#page-1. (Date of access: 15/11/2015).
Pearce, M. et al. Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111, and O145 shed by cattle in Scotland. Applied and Environmental Microbiology 72, 653–659 (2006).
Friesema, I. et al. Emergence of Escherichia coli encoding Shiga toxin 2f in human Shiga toxin-producing E. coli (STEC) infections in the Netherlands, January 2008 to December 2011. Euro Surveill 19, 26–32 (2014).
Kaper, J. B., Nataro, J. P. & Mobley, H. L. Pathogenic Escherichia coli . Nature Reviews Microbiology 2, 123–140 (2004).
Fruth, A., Prager, R., Tietze, E., Rabsch, W. & Flieger, A. Molecular epidemiological view on Shiga toxin-producing Escherichia coli causing human disease in Germany: Diversity, prevalence, and outbreaks. International Journal of Medical Microbiology (2015). URL: http://www.sciencedirect.com/science/article/pii/S1438422115000831. (Date of access: 15/11/ 2015).
Clermont, O., Gordon, D. & Denamur, E. A guide to the various phylogenetic classification schemes for Escherichia coli and the correspondence among schemes. Microbiology, mic. 0.000063 (2015). URL: http://mic.microbiologyresearch.org/content/journal/micro/10.1099/mic.0.000063. (Date of access: 15/11/2015).
Hasman, H. et al. Rapid whole genome sequencing for the detection and characterization of microorganisms directly from clinical samples. Journal of Clinical Microbiology, JCM. 02452–02413 (2013).
Mulemba, H. The Livestock Sector in Zambia and Rising Food Prices Country Briefing–Zambia. International Institute for Sustainable Development, Manitoba, Canada (2009). http://www.iisd.org/sites/default/files/pdf/ag_scenarios_south_africa_zambia.pdf. (Date of access: 15/11/ 2015).
Mumba, C. Economic Analysis of the Viability of Small holder Dairy Farming in Zambia, The University of Zambia, (2012). URL: http://dspace.unza.zm:8080/xmlui/handle/123456789/1804. (Date of access: 15/11/2015).
Mainda, G. et al. Prevalence and patterns of antimicrobial resistance among Escherichia coli isolated from Zambian dairy cattle across different production systems. Scientific Reports 5 (2015).
Paton, A. W. & Paton, J. C. Detection and Characterization of Shiga ToxigenicEscherichia coli by Using Multiplex PCR Assays forstx 1, stx 2, eaeA, Enterohemorrhagic E. coli hlyA, rfb O111, andrfb O157. Journal of Clinical Microbiology 36, 598–602 (1998).
Camacho, C. et al. BLAST+: architecture and applications. BMC bioinformatics 10, 421 (2009).
Zweifel, C., Cernela, N. & Stephan, R. Detection of the Emerging Shiga Toxin-Producing Escherichia coli O26: H11/H-ST29 in Human Patients and Healthy Cattle in Switzerland. Applied and Environmental Microbiology, AEM. 01728–01713 (2013). http://aem.asm.org/content/early/2013/06/24/AEM.01728-13.short. (Date of access: 15/11/2015).
Amézquita-López, B. et al. Detection of Shiga Toxin Variants, Virulence Genes and the Relationship to Cytotoxicity of Shiga Toxin-Producing Escherichia coli (STEC) from Domestic Farm Animals. in Meeting Abstract. URL: http://sistemanodalsinaloa.gob.mx/archivoscomprobatorios/_14_resumeneventoscientificos/694.pdf. (Date of access: 17/12/2015).
Fox, J., Depenbusch, B., Drouillard, J. & Nagaraja, T. Dry-rolled or steam-flaked grain-based diets and fecal shedding of O157 in feedlot cattle. Journal of Animal Science 85, 1207–1212 (2007).
Clermont, O., Bonacorsi, S. & Bingen, E. Rapid and simple determination of theEscherichia coli phylogenetic group. Applied and Environmental Microbiology 66, 4555–4558 (2000).
Cerna, J. F., Nataro, J. P. & Estrada-Garcia, T. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. Journal of Clinical Microbiology 41, 2138–2140 (2003).
Padhye, V., Zhao, T. & Doyle, M. Production and characterisation of monoclonal antibodies to Verotoxins 1 and 2 from Escherichia coli of serotype O157: H7. Journal of Medical Microbiology 30, 219–226 (1989).
Krüger, A., Lucchesi, P. M. & Parma, A. E. Verotoxins in bovine and meat verotoxin-producing Escherichia coli isolates: type, number of variants, and relationship to cytotoxicity. Applied and Environmental Microbiology 77, 73–79 (2011).
Lumley, T. Analysis of complex survey samples. Journal of Statistical Software 9, 1–19 (2004).
Andrews, S. (2011). URL: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (Date of access: 20/11/2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal 17, pp. 10–12 (2011).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Research, gku1196 (2014). URL: http://nar.oxfordjournals.org/content/early/2014/11/20/nar.gku1196.short. (Date of access: 15/11/2015).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007).
Clermont, O., Christenson, J. K., Denamur, E. & Gordon, D. M. The Clermont Escherichia coli phylo‐typing method revisited: improvement of specificity and detection of new phylo‐groups. Environmental Microbiology Reports 5, 58–65 (2013).
Scheutz, F. et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. Journal of Clinical Microbiology 50, 2951–2963 (2012).
Inouye, M. et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6, 90 (2014).
Acknowledgements
We are grateful to the Government of the Republic of Zambia (GRZ) and the University of Zambia for facilitating the field and laboratory research while in Zambia. We are indepted to the help of Geoffrey Kwenda and Sydney Malama to enable collection of the human E. coli isolates. We acknowledge access to pathogenic avian strains supplied by Zoetis. G.M. is grateful to the Commonwealth Scholarship Commission (CSC) for providing funding (ZMSC-2012-640). N.L. acknowledges support from a University of Edinburgh College studentship. D.L.G., S.P.M., M.P.S. and B.M.B. receive core strategic funding to The Roslin Institute from the Biotechnology & Biological Sciences Research Council (BB/J004227/1). The sequencing by PHE was funded by the National Institute for Health Research scientific research development fund (108601).
Author information
Authors and Affiliations
Contributions
G.M., D.L.G., M.P.S., J.B.M. and B.M.B. designed the project and developed the survey. G.M., D.L.G., L.S. and B.M.B. carried out the sampling work. S.P.M., J.G., J.B.M., L.S., D.L.G. and G.M. assisted with the microbiology design and laboratory work. K.G., N.J.W., S.K.S., R.L.R., G.C., S.A.A. and S.W. supplied unpublished strains and sequences for analysis. G.M., P.B., D.V.H., M.E.C.T. and B.M.B. carried out statistical analyses. N.L., G.M., T.J.D. and D.L.G. carried out bioinformatics analysis and drafted the manuscript, and all the authors read and helped edit the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Mainda, G., Lupolova, N., Sikakwa, L. et al. Phylogenomic approaches to determine the zoonotic potential of Shiga toxin-producing Escherichia coli (STEC) isolated from Zambian dairy cattle. Sci Rep 6, 26589 (2016). https://doi.org/10.1038/srep26589
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26589
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.