Abstract
Intrauterine infection of hepatitis B virus (HBV), which accounts for the majority of mother-to-child transmission, is one of the main reasons for the failure of combined immunoprophylaxis against the transmission. Recent studies have identified that genetic background might influence the susceptibility to intrauterine infection of HBV. We conducted this study to investigate the associations between 10 genetic variants in 9 genes (SLC10A1, HLA-DP, HLA-C, CXCR5, CXCL13, TLR3, TLR4, TLR9 and UBE2L3) of mothers and their neonates and HBV intrauterine infection. A significantly decreased risk of HBV intrauterine transmission were found among mothers who carried the rs355687 CT genotypes in CXCL13 gene compared to those with CC genotypes (OR = 0.25, 95% CI, 0.08–0.82, P = 0.022); and a marginally significantly decreased risk was also observed under the dominant model (OR = 0.34, 95% CI, 0.11–1.01, P = 0.052). Besides, neonatal rs3130542 in HLA-C gene was found to be marginally significantly associated with decreased risk of HBV intrauterine infection under the additive model (OR = 0.55, 95% CI, 0.29–1.04, P = 0.064). However, we found no evidence of associations between the remaining 8 SNPs and risk of HBV intrauterine infection among mothers and their neonates. In conclusion, this study suggested that genetic variant in CXCL13 gene was associated with susceptibility to intrauterine infection of HBV.
Similar content being viewed by others
Introduction
Hepatitis B virus (HBV) infection is a global public health problem with 350 million people being chronically infected worldwide. People with hepatitis B are at increased risk of developing hepatic decompensation, cirrhosis and hepatocellular carcinoma1. China is an endemic region, with a 7.2% prevalence rate of chronic HBV infection in individuals aged 1–592. Mother-to-child transmission (MTCT), including intrauterine, labor, postnatal, is considered as one of the main modes of HBV infection. The rate of intrauterine transmission, which accounts for the majority of mother-to-child transmission, among HBsAg positive pregnant women ranges from 5% to 40% in different areas of China3,4. Infants with intrauterine HBV infection have a considerably high risk of becoming chronic carriers. Currently, combined immunoprophylaxis with the utilization of hepatitis B vaccine and hepatitis B immunoglobulin (HBIG) can block most cases of labor and postnatal transmission of HBV. Whereas, HBV intrauterine infection still occurs at high incidence due to lack of effective preventive measures and its exact underlying mechanisms have not been completely elucidated. Therefore, exploring the risk factors of susceptibility to intrauterine HBV infection will help effectively control the spread of HBV infection among neonates.
Previous studies regarding the factors associated with HBV intrauterine infection were mostly focused on maternal characteristics (i.e. mode of delivery, maternal HBeAg status and viral load)5,6. In addition to maternal characteristics, genetic background may also play an important role in HBV intrauterine infection. However, few studies7,8,9 had explored the genetic susceptibility to intrauterine infection of HBV and still, little is known about the associations between genetic factors and HBV intrauterine infection.
Sodium taurocholate cotransporting polypeptide (NTCP), identified as a functional receptor for human hepatitis B virus10, is encoded by the SLC10A1 gene. Variation of the p.S267F (rs2296651) in SLC10A1 gene results in loss of HBV receptor function in vitro and was found to be significantly associated with resistance to chronic hepatitis B11. Meanwhile, Zhenzhen Su et al.12 demonstrated that AA genotype of the other locus (rs7154439) in SLC10A1 gene was also associated with HBV clearance. Recently, genome-wide association studies (GWAS) in Japanese and Korean populations determined that single nucleotide polymorphisms (SNPs) in the HLA-DP gene were associated with persistent HBV infection13. Subsequently, one locus (rs3128917) in the HLA-DP was observed to be associated with chronic HBV infection in the Chinese population14. The other GWAS conducted in Han Chinese population had identified rs3130542 in HLA-C and rs4821116 in UBE2L3 to be associated with chronic HBV infection15. Follicular T helper (TfH) cells are newly recognized lineage of CD4+ T cells16, which have been reported to be partially responsible for the dysfunction of immune responses during HBV infection17. CXCR5 and CXCL13 are two of TfH-associated genes and genetic variants in the CXCR5 gene (rs3922) and CXCL13 gene (rs355687) were reported to be associated with non-responsiveness to the hepatitis B vaccine18. Toll-like receptors (TLRs), which act as bridging molecules between innate and adaptive immunity, are able to recognize different viruses and/or bacteria by pathogen-associated molecular patterns (PAMPs) and activate a pathogen specific immune response19,20. TLR3, TLR4 and TLR9 are important members of TLR gene family, with TLR3 recognizing virus-derived dsRNA as well as poly I: C, a synthetic dsRNA analogue, TLR4 recognizing lipopolysaccharide and TLR9 recognizing unmethylated CpG motifs present in bacteria and viruses. Genetic variants in these three genes may influence its biological functions. Rs3775291 in the TLR3 gene was shown to be associated with dsRNA binding capacity21, rs1927914 in the TLR4 gene was found to influence the binding sites of transcriptional factor, promoter activity and gene expression22 and rs352140 in the TLR9 gene was observed to affect the expression of TLR9 gene23. Hence, we hypothesized that the above 10 SNPs in the 9 genes may modify the risk of intrauterine HBV infection.
Thus we performed this study which aimed to investigate the associations between the 10 SNPs and risk of HBV intrauterine infection. Furthermore, we evaluated the associations of these loci from both maternal and neonatal aspects with HBV intrauterine infection, respectively.
Results
Characteristics of mothers and neonates
706 mother-neonate pairs were included in this study and 44 neonates were tested positive for HBsAg and/or HBV DNA. The HBV intrauterine infection rate was 6.23% (44/706). Characteristics of mothers and neonates of cases and controls are summarized in Table 1. Mothers in case group were younger than those in control group (P = 0.002). While, no statistically significant difference was observed between mothers of cases and controls in the distribution of profession (P = 0.804). Additionally, there were no statistically significant differences in birth weight, birth length, gestational weeks and gender between neonates of cases and controls.
Associations between maternal characteristics and HBV intrauterine transmission
As shown in Table 2, maternal HbeAg positivity (P < 0.001), maternal HBV DNA positivity (P < 0.001) and caesarean section (P = 0.004) were significantly associated with HBV intrauterine transmission. No statistically significant associations were observed between other maternal characteristics and HBV intrauterine transmission.
Associations between maternal and neonatal SNPs and HBV intrauterine infection
10 SNPs were successfully genotyped and basic information about these SNPs is shown in Supplementary Table S1. The call rate of all the maternal and neonatal SNPs were >95% and the genotypes for all SNPs of mothers and neonates in control group conformed to Hard-Weinberg equilibrium (HWE, FDR-P > 0.05). The maternal and neonatal minor allele frequencies (MAFs) in the control group were similar to those in the HapMap database of Han Chinese in Beijing except for the SNP (rs3128917) in HLA-DP which were similar to those in HapMap database of Southern Han Chinese. Distribution of genotypes of the 10 SNPs of mothers and neonates was presented in Supplementary Tables S2 and S3. Moreover, rs2296651 and rs7154439 were not in strong LD with each other of mothers (r2 = 0.008) or neonates (r2 = 0.008) in the control group. As shown in Table 3, after adjusting for maternal age, maternal HBeAg, maternal HBV DNA and mode of delivery, mothers who carried the rs355687 CT genotypes in CXCL13 gene had a lower risk of HBV intrauterine transmission compared to those who carried CC genotypes (OR = 0.25, 95% CI, 0.08–0.82, P = 0.022). Besides, a marginally significantly decreased risk of HBV intrauterine transmission was also found under the dominant model (OR = 0.34, 95% CI, 0.11–1.01, P = 0.052). Then, we performed the association analysis for each neonatal SNP under general genetic model, dominant, recessive and additive models. Unfortunately, none of the P value was statistically significant after adjustment for maternal age, maternal HBeAg, maternal HBV DNA and mode of delivery. However, the rs3130542 in HLA-C gene of neonates was found to be marginally significantly associated with decreased risk of susceptibility to intrauterine HBV infection under the additive model (OR = 0.55, 95% CI, 0.29–1.04, P = 0.064).
Gene-virus interactions with respect to the risk of HBV intrauterine infection
Table 4 shows the results of multiplicative and additive interaction analyses between maternal rs355687, neonatal 3130542 and maternal HBeAg, maternal HBV DNA. Unfortunately, no significant multiplicative or additive interactions were detected.
Interactions between genes and mode of delivery with respect to the risk of HBV intrauterine infection
The results of multiplicative interaction analyses between maternal rs355687, neonatal 3130542 and mode of delivery are shown in Table 5. No significant multiplicative interactions between genes and mode of delivery were detected.
Discussion
In this study, we investigated the associations between the genetic variants in 9 candidate genes and risk of HBV intrauterine infection. Our results showed that maternal rs355687 in CXCL13 gene was significantly associated with decreased risk of susceptibility to intrauterine infection of HBV.
Maternal characteristics play an important role in the intrauterine infection of HBV. In our study, we observed that mothers with younger age had a significantly higher risk of HBV intrauterine transmission, which is similar with the results obtained by Schillie et al.24. The possible explanation is that younger mothers were more often HBeAg positive (Supplementary Table S4) and had longer labors than older mothers25. Majority of previous studies investigating the influencing factors of HBV intrauterine infection showed that maternal HBeAg positivity and maternal HBV DNA positivity were two main risk factors5,26,27, which is consistent with our results. Furthermore, we also found that caesarean delivery was a protective factor of HBV intrauterine transmission, which is in agreement with earlier studies5,9, indicating that caesarean delivery might help to reduce HBV infection rate of neonates intrauterinely.
Apart from these above factors associated with HBV intrauterine infection, recent studies have suggested that host genetic factors may also have an influence on the susceptibility to HBV intrauterine infection7,9,28. In the current study, maternal rs355687 in CXCL13 gene was found to be associated with decreased risk of HBV intrauterine transmission. CXCL13, a molecule of TfH cells, was found to play an integral role in facilitating an effective immune response against HBV by Publicover et al.29. In addition, their study also showed that CXCL13 expression increases in an age-dependent manner30. The rs355687 is located in the intron of CXCL13 gene, variation of which may affect the transcription of CXCL13 gene. Zhaojun Duan et al. found that the rs355687 in CXCL13 gene was associated with non-responsiveness to the hepatitis B vaccine and the CT genotype in rs355687 appeared more frequently in the group defined as HBV responders (P = 0.038, OR = 0.30, 95% CI, 0.09–0.97)18. Based on the above findings, we speculated that CT genotype in maternal rs355687 might up-regulate the expression of CXCL13 gene, increase CXCL13 level and clear HBV in the blood of mothers and thus interrupt HBV transmission from mothers to newborns. Furthermore, no significant association between neonatal rs355687 in CXCL13 gene and HBV intrauterine infection was observed in our study, which was possibly due to the age-dependent manner of CXCL13 production. Notably, we found that the rs3130542 in HLA-C gene of neonates was marginally significantly associated with decreased risk of susceptibility to HBV intrauterine infection under the additive model (OR = 0.55, 95% CI, 0.29–1.04, P = 0.064). HLA-C encodes a type I HLA molecule. Increased HLA-C expression in hepatocytes stimulates attack by cytotoxic T cells (CTLs), a crucial component of HBV clearance31,32. Zhibin Hu et al. conducted a genome-wide association studies (GWAS) in Chinese population and identified the A allele of rs3130542 in HLA-C gene was associated with chronic HBV infection (OR = 1.33, P = 9.49 × 10−14)15. And Montgomery SB et al. showed that the A allele of rs3130542 was associated with lower expression of HLA-C33. Hence, we inferred that neonates carrying the G allele of rs3130542 in HLA-C gene might have a higher expression of HLA-C, which resulted in a decreased risk of HBV intrauterine infection. Subsequently, we ran interaction analyses of the relationship between maternal rs355687, neonatal 3130542 and maternal HBeAg, maternal HBV DNA, mode of delivery and found no significant interactions, which indicated that virological markers (HBeAg, HBV DNA), mode of delivery and host genetic factors might confer susceptibility of HBV intrauterine infection individually. Besides, we did not observe statistically significant associations between other 8 maternal and neonatal SNPs (rs2296651, rs7154439, rs3128917, rs3922, rs3775291, rs1927914, rs352140 and rs4821116) and risk of HBV intrauterine infection. One plausible reason was that the sample size of neonates with HBV intrauterine infection was small in the current study, which resulted in failure to detect the statistically significant associations between them. Another possible explanation was that these SNPs had no effect on the susceptibility to intrauterine infection of HBV essentially.
Some potential limitations in this study should be considered. Firstly, the low incidence of HBV intrauterine infection results in the relatively small sample size of neonates with HBV intrauterine infection and further studies with larger sample size are needed. Secondly, only one or two SNPs in a gene, reported or identified by previous studies, were selected, so we could not reveal the role of a gene in HBV intrauterine infection comprehensively. Further studies covering all probably functional SNPs in candidate genes are needed. Thirdly, the significant associations found were only at the statistical level and their biological functions were not evaluated. Further biological experiments are therefore warranted to clarify its mechanism underlying the etiology of HBV intrauterine infection and are among the future studies we plan to pursue.
In summary, our study suggested that maternal rs355687 variant in CXCL13 gene was associated with decreased risk of HBV intrauterine infection. To the best of our knowledge, this is the first study on exploring the roles of host genetic variants in HBV intrauterine infection from both maternal and neonatal aspects. The characterization of functional significance of this particular CXCL13 molecule in HBV intrauterine infection may provide insight leading to the development of novel prophylactic and therapeutic approaches.
Materials and Methods
Study subjects
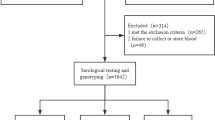
719 HBsAg positive pregnant women were consecutively enrolled in this study from May 2012 to October 2014 at Wuhan Women and Children Medical and Healthcare Center, Wuhan, China. In total, these women delivered 731 neonates, including 12 twins. Women who co-infected with HCV and/or HIV or who gave multiple births were excluded, which yielded 706 mother-neonate pairs eventually.
Maternal information (maternal demographics, characteristics before and during pregnancy), maternal complications, mode of delivery and neonatal characteristics (birth weight, birth length and gender et al.) were obtained from medical records. 2-mL procoagulant and 2-mL anticoagulant peripheral venous blood samples of the mothers were collected before delivery, while 2-mL procoagulant and 2-mL anticoagulant femoral venous blood samples of their newborns were collected before the administration of passive-active immunoprophylaxis. Procoagulant blood samples were used for the detection of HBV DNA levels, while anticoagulant blood samples were stored at −80 °C for laboratory testing. After blood collection, the newborns received hepatitis B immune globulin (HBIG) intramuscularly immediately and they were also administered with hepatitis B vaccines at month 0, 1 and 6. This study was approved by the ethics committee of Tongji Medical College of Huazhong University of Science and Technology and the methods were carried out in accordance with the approved guidelines. Written informed consent was obtained from all mothers.
Serological HBV markers and HBV DNA
Information about HBV serological markers of mothers which were detected at the prenatal care visit were acquired from their medical records. HBV DNA levels were tested by RT-PCR (Da’an Gene Co. Ltd, Sun Yat-Sen University, Guangdong, China), which was performed by the staffs of clinical laboratory in the hospital. HBsAg of neonates were measured using ELISA (Kehua biotechnology, Shanghai, China). All procedures were performed according to the manufacturers’ instructions. HBV DNA loads >1 × 103 copies/ml were defined as positive. HBV intrauterine infection was defined as HBsAg and/or HBV DNA positive in neonatal peripheral venous blood within 24 hours of birth and before administration of active or passive immune prophylaxis34. Newborns with intrauterine HBV infection were classified as case group, those without were classified as control group.
DNA extraction and genotyping
Genomic DNA was extracted from the blood samples using a TIANamp Blood DNA Kit DP318 (TIANGEN BIOTECH (BEIJING) CO., LTD, China) under the instruction of manufacturer. DNA concentration and optical density were determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). SNPs of each sample were genotyped by the TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA, USA) with a 7900 HT Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. For quality control, genotyping was performed without knowing case control status. In addition, samples with a call rate <80% were removed to make sure the call rate of each SNP was over 95%.
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) for genotypes was assessed by a goodness-of-fit χ2 test among controls. The difference in distribution of demographic characteristics, genotype frequencies between cases and controls were examined by Pearson’s χ2 test or t-test, when appropriate. The association between each SNP of mothers and neonates and risk of intrauterine HBV infection was evaluated by the odds ratio (OR) and its 95% confidence interval (CI) using unconditional multivariable logistic regression model with adjustment for maternal age, maternal HBeAg, maternal HBV DNA level and mode of delivery. In order to avoid the assumption of genetic models, general genetic model, dominant, recessive and additive models were all calculated. Multiplicative and additive interactions were tested to evaluate interactions between genes and virological markers (HBeAg and HBV DNA). Multiplicative interaction was tested to evaluate interactions between genes and mode of delivery. Multiplicative interactions were evaluated by a likelihood ratio test in the logistic regression model. Synergy index (SI), attributable proportion due to interaction (AP) and relative excess risk due to interaction (RERI) and its 95% CIs were calculated in additive interaction analyses. Lack of interaction is indicated by 95% CI of SI containing 1 and 95% CIs of AP and RERI containing 0. All statistical analyses were conducted by SPSS v18.0 (SPSS, Chicago, Illinois, USA). Linkage disequilibrium (LD) was performed using the Haploview v4.2 software, by determining D’ and γ2 values. The additive interaction analysis was performed using the excel sheet made by Andersson et al.35. All P values were two-tailed with a statistically significant level set at 0.05. False discovery test was performed to take into account the multiple comparisons.
Additional Information
How to cite this article: Wan, Z. et al. Genetic variant in CXCL13 gene is associated with susceptibility to intrauterine infection of hepatitis B virus. Sci. Rep. 6, 26465; doi: 10.1038/srep26465 (2016).
References
Dienstag, J. L. Hepatitis B virus infection. N Engl J Med 359, 1486–1500 (2008).
Liang, X. et al. Epidemiological serosurvey of hepatitis B in China–declining HBV prevalence due to hepatitis B vaccination. Vaccine 27, 6550–6557 (2009).
Zhang, S. L., Yue, Y. F., Bai, G. Q., Shi, L. & Jiang, H. Mechanism of intrauterine infection of hepatitis B virus. World J Gastroenterol 10, 437–438 (2004).
Shao, Z. J. et al. Mother-to-infant transmission of hepatitis B virus: a Chinese experience. J Med Virol 83, 791–795 (2011).
Guo, Z. et al. Risk factors of HBV intrauterine transmission among HBsAg-positive pregnant women. J Viral Hepat 20, 317–321 (2013).
Wen, W. H. et al. Mother-to-infant transmission of hepatitis B virus infection: significance of maternal viral load and strategies for intervention. J Hepatol 59, 24–30 (2013).
Yu, H., Zhu, Q. R., Gu, S. Q. & Fei, L. E. Relationship between IFN-gamma gene polymorphism and susceptibility to intrauterine HBV infection. World J Gastroenterol 12, 2928–2931 (2006).
Xu, Y. Y. et al. Association between the frequency of class II HLA antigens and the susceptibility to intrauterine infection of hepatitis B virus. Int J Biol Sci 4, 111–115 (2008).
Gao, Y. et al. Evaluation of neonatal Toll-like receptors 3 (c.1377C/T) and 9 (G2848A) gene polymorphisms in HBV intrauterine transmission susceptibility. Epidemiol Infect 143, 1868–1875 (2015).
Yan, H. et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife 1, e49 (2012).
Peng, L. et al. The p.Ser267Phe variant in SLC10A1 is associated with resistance to chronic hepatitis B. Hepatology 61, 1251–1260 (2015).
Su, Z. et al. Association of the gene polymorphisms in sodium taurocholate cotransporting polypeptide with the outcomes of hepatitis B infection in Chinese Han population. Infect Genet Evol 27, 77–82 (2014).
Kamatani, Y. et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet 41, 591–595 (2009).
Wong, D. K. et al. Role of HLA-DP polymorphisms on chronicity and disease activity of hepatitis B infection in Southern Chinese. Plos One 8, e66920 (2013).
Hu, Z. et al. New loci associated with chronic hepatitis B virus infection in Han Chinese. Nat Genet 45, 1499–1503 (2013).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu Rev Immunol 29, 621–663 (2011).
Xing, T., Xu, H. & Yu, W. Role of T follicular helper cells and their associated molecules in the pathogenesis of chronic hepatitis B virus infection. Exp Ther Med 5, 885–889 (2013).
Duan, Z. et al. Genetic polymorphisms of CXCR5 and CXCL13 are associated with non-responsiveness to the hepatitis B vaccine. Vaccine 32, 5316–5322 (2014).
Boehme, K. W. & Compton, T. Innate sensing of viruses by toll-like receptors. J Virol 78, 7867–7873 (2004).
Takeda, K. & Akira, S. Toll-like receptors in innate immunity. Int Immunol 17, 1–14 (2005).
Zhou, P., Fan, L., Yu, K. D., Zhao, M. W. & Li, X. X. Toll-like receptor 3 C1234T may protect against geographic atrophy through decreased dsRNA binding capacity. Faseb J 25, 3489–3495 (2011).
Hsieh, Y. Y., Wan, L., Chang, C. C., Tsai, C. H. & Tsai, F. J. STAT2*C related genotypes and allele but not TLR4 and CD40 gene polymorphisms are associated with higher susceptibility for asthma. Int J Biol Sci 5, 74–81 (2009).
Xu, C. J. et al. Association study of a single nucleotide polymorphism in the exon 2 region of toll-like receptor 9 (TLR9) gene with susceptibility to systemic lupus erythematosus among Chinese. Mol Biol Rep 36, 2245–2248 (2009).
Schillie, S. et al. Outcomes of infants born to women infected with hepatitis B. Pediatrics 135, e1141–e1147 (2015).
Marion, S. A., Tomm, P. M., Pi, D. W. & Mathias, R. G. Long-term follow-up of hepatitis B vaccine in infants of carrier mothers. Am J Epidemiol 140, 734–746 (1994).
Lv, N. et al. Analysis on the outcomes of hepatitis B virus perinatal vertical transmission: nested case-control study. Eur J Gastroenterol Hepatol 26, 1286–1291 (2014).
Bai, H. et al. Relationship of hepatitis B virus infection of placental barrier and hepatitis B virus intra-uterine transmission mechanism. World J Gastroenterol 13, 3625–3630 (2007).
Xu, Y. Y. et al. Association between the frequency of class II HLA antigens and the susceptibility to intrauterine infection of hepatitis B virus. Int J Biol Sci 4, 111–115 (2008).
Publicover, J. et al. Age-dependent hepatic lymphoid organization directs successful immunity to hepatitis B. J Clin Invest 123, 3728–3739 (2013).
Leavy, O. Antiviral immunity: A ‘mature’ way of controlling HBV. Nat Rev Immunol 13, 616 (2013).
Ballardini, G. et al. HLA-A,B,C, HLA-D/DR and HLA-D/DQ expression on unfixed liver biopsy sections from patients with chronic liver disease. Clin Exp Immunol 70, 35–46 (1987).
Zhang, Y. et al. Hepatitis B virus core antigen epitopes presented by HLA-A2 single-chain trimers induce functional epitope-specific CD8+ T-cell responses in HLA-A2.1/Kb transgenic mice. Immunology 121, 105–112 (2007).
Montgomery, S. B. et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 (2010).
Li, X. M. et al. Effect of hepatitis B immunoglobulin on interruption of HBV intrauterine infection. World J Gastroenterol 10, 3215–3217 (2004).
Andersson, T., Alfredsson, L., Kallberg, H., Zdravkovic, S. & Ahlbom, A. Calculating measures of biological interaction. Eur J Epidemiol 20, 575–579 (2005).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC-81172678). We are particularly grateful to the mothers and their neonates. We also acknowledge the staff in Wuhan Women and Children Medical and Healthcare Center, Wuhan, China for their assistance.
Author information
Authors and Affiliations
Contributions
Z.H.W. and X.F.L. performed the experiments, collected the data, conducted the statistical analysis and drafted the initial manuscript. T.Y.L. collected the data and performed the experiments. A.F.Z., M.Y., D.H., L.F., S.X.P., L.L.F. and S.T. assisted with data and sample collection, reviewed and revised the manuscript. B.Z. helped collect the data and samples. Y.K.D. conceived and designed the study, directed the statistical analysis. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wan, Z., Lin, X., Li, T. et al. Genetic variant in CXCL13 gene is associated with susceptibility to intrauterine infection of hepatitis B virus. Sci Rep 6, 26465 (2016). https://doi.org/10.1038/srep26465
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26465
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.