Abstract
Predicting regions at risk from introductions of non-native species and the subsequent invasions is a fundamental aspect of horizon scanning activities that enable the development of more effective preventative actions and planning of management measures. The Asian cyprinid fish topmouth gudgeon Pseudorasbora parva has proved highly invasive across Europe since its introduction in the 1960s. In addition to direct negative impacts on native fish populations, P. parva has potential for further damage through transmission of an emergent infectious disease, known to cause mortality in other species. To quantify its invasion risk, in regions where it has yet to be introduced, we trained 900 ecological niche models and constructed an Ensemble Model predicting suitability, then integrated a proxy for introduction likelihood. This revealed high potential for P. parva to invade regions well beyond its current invasive range. These included areas in all modelled continents, with several hotspots of climatic suitability and risk of introduction. We believe that these methods are easily adapted for a variety of other invasive species and that such risk maps could be used by policy-makers and managers in hotspots to formulate increased surveillance and early-warning systems that aim to prevent introductions and subsequent invasions.
Similar content being viewed by others
Introduction
The introduction and establishment of species into novel ecosystems can result in rapid dispersal rates1,2, with the magnitude of subsequent impacts and associated management costs increasing sharply with the time since the initial introduction3. When eradication is no longer a feasible management option, long-term strategies aimed at control and containment are generally adopted4,5. Such approaches tend to be expensive and only prevent further damage rather than restoring the invaded systems to their former status6. Consequently, more effective management strategies focus more on preventing introductions rather than managing subsequent invasions7.
Through increased global connectivity and trade, opportunities for the introduction of non-native species have increased8, with resources at border control stretched and ineffective at preventing the entry of non-native species into new regions9. Whilst risk assessment frameworks can be a strong mechanism to help allocate finite resources between preventing, detecting and managing introductions of non-native species10,11, they sometimes fail to highlight specific areas within regions where risks of introduction and subsequent invasions are higher12 (although, a notable exception is the recent application of the FISK risk assessment tool to a single catchment in Hungary13). Risk maps summarize landscape suitability for non-native species within a region by incorporating multiple factors, such as climatic or habitat suitability and potential introduction pathways, and thus can highlight smaller spatial areas within the region, or continent, where the risk is greatest14.
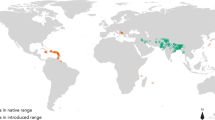
Freshwater ecosystems provide societies with valuable goods and services, yet are disproportionately vulnerable to biological invasions, compared to terrestrial systems, due to their high degree of isolation and endemism15,16. Introductions of non-native fishes represent a considerable threat to global freshwater fish conservation17 and can cause considerable economic damage16. For example, in North America the annual cost of invasive non-native fish species is estimated as at least US$ 5.4 billion18. In Europe, at least 650 fish species have been introduced into aquatic ecosystems, yet only a relatively small proportion of these are considered invasive and cause detrimental impacts19, such as the topmouth gudgeon Pseudorasbora parva (Temminck & Schlegel). This Asian cyprinid fish has been described as Europe’s most invasive freshwater fish (e.g.20), having achieved a pan-continental distribution in less than forty years, following its initial accidental introduction into Romania in the 1960s21. Its ability to invade new waters stems from traits that include high tolerance to degraded ecosystems, high reproductive effort and early sexual maturity alongside batch spawning and paternal nest guarding22. Moreover, it can rapidly colonize new waters23, with their larval stages particularly likely to disperse into the wider environment24. It is also a healthy carrier of an emergent infectious fungal disease Sphaerothecum destruens that has been implicated in declines in native European fishes25,26. Given its rapid rate of invasion in Europe and its disease risk, it is thus important to develop tools to predict additional locations at risk, outside of its current invasive range. Such tools would enable countries that contain potentially suitable habitats but have not yet been invaded to set up the necessary infrastructure to prevent an introduction and subsequent invasion.
Invasion risk is a combination of the likelihood of the species being introduced (e.g. the presence of an introduction pathway) and the suitability of the environment in the new region (e.g. climate suitability for the species). Consequently, the aim of this study was to predict the risk of P. parva invasion at the global scale in relation to its climate suitability and likely introduction pathways. Objectives were to (i) use climatic and other relevant abiotic data in both P. parva’s native and European invasive ranges to determine its realized niche; (ii) assess its potential global introduction pathways in relation to its known introduction pathways in Europe; and (iii) develop a niche model that characterizes its realized niche and combine this with data on introduction pathways to develop a global risk map of P. parva invasion. The implications of the risk map are then discussed in a global context.
Materials and Methods
Data on presence and absence of P. parva
Previous niche modelling studies suggest that presence location data from the invasive range can sometimes provide a significantly better approximation of the actual species niche breadth, thus producing more accurate suitability predictions beyond the native range27,28. Consequently, our presence dataset incorporated data from the European invasive range, compiled from European national monitoring schemes with consistent recording over the last 30+ years, as well as data from its native Asian range. When the European national monitoring data were combined with literature review and native and invasive presence records from the Global Biodiversity Information Facility (GBIF, www.gbif.org), this provided 2,882 confirmed P. parva observations from unique locations, across both native and invasive ranges, the latest of which was recorded in 2014. In order to minimise bias caused by multiple presence points in grid cells, i.e. spatial clustering of records, we applied a form of spatial filtering similar to that used by Boria et al.29, whereby we removed all but one record from grid cells, essentially leaving us with 2,048 “presence cells” in our dataset.
Absence data were generated on the principle of implied absence, as P. parva’s European distribution, after 40 years since introduction, suggested the species had already reached saturation point (cf.21) and we also assumed equilibrium in the native range. To avoid spatial autocorrelation with presence cells, random points were generated outside buffers of 50 km around presence cell centroids. These random points were limited to a maximum distance of 100 km from presence cell centroids to prevent the characterisation of areas far outside the species’ core areas, which can result in poor model performance30. These methods also maintained a degree of parity with regard to the ratio of presence to absence points across the two distributional aggregations (Asian and European). A total of 10,000 absence points were generated in 10 absence datasets, and weighted to give equal prevalence for the overall response variable dataset, which was coded in a binary (1/0, presence/absence) fashion. Due to the relatively high recording effort achieved in this monitoring, it was assumed these locations represented genuine absences, rather than just characterising the background environmental variation present within the invasive range.
Predictor variable data
We used a suite of abiotic variables as a basis for the description of P. parva’s niche. We used the 19 Bioclimatic datasets (Bio1 - Bio1931, see Table 1 for individual descriptions); Mean Potential Incoming Solar Radiation (INMSR); Topsoil pH (TpH); and Topographic Wetness Index (TWIS) as our initial suite of variables (see Table 1 for details of datasets). So as to promote parsimony and minimize over-fitting in models, we refined the list of variables used in the final modelling procedure, using Variance Inflation Factor to identify and remove collinearity in the final suite of variables used. Stepwise selection of variables was carried out using the r package ‘VIF’32 and left us with a refined suite of ten abiotic variables (see variables marked with asterisk in Table 1) to use in our models. All datasets were used at a spatial resolution of 10 km and a spatial reference of GCS WGS 1984. Conforming to accepted protocols in ecological niche modelling33, all of the remaining ten variables are ecologically relevant to P. parva.
Additional data
In order to produce a risk map accurately representing both the likelihood of introduction and climatic suitability, we incorporated a dataset representing the known major introduction pathways. Given that the spread of P. parva throughout Europe was strongly related to aquaculture-related introductions21, and because there is a strong correlation between non-native introductions and international trade34, the “Aquaculture Pressure” dataset from the “Global Threats to Human Water Security and River Biodiversity” database35 was used as a surrogate for introduction likelihood. This dataset was derived using national annual average aquaculture harvests from inland and diadromous fishes from 1997–2006 from the UN Food and Agriculture Organization’s FishStat Plus database (www.fao.org/fishery), which were then distributed proportionately to the grid-cell specific discharge for each nation. The original dataset was re-sampled using bilinear interpolation to match the resolution of all other datasets (10 km). All spatial datasets shared the same spatial reference (GCS WGS 1984).
Modelling methods
The first phase of modelling used nine modelling techniques: Artificial Neural Networks (ANN), Classification Tree Analysis (CTA), Flexible Discriminant Analysis (FDA36), Generalized Additive Models (GAM), Generalized Boosted Models (GBM), Generalized Linear Models (GLM), Multivariate Adaptive Regression Splines (MARS), Random Forests (RF) and Surface Range Envelope (SRE37). The use of GLM, GAM, CTA and ANN is described and discussed in Thuiller38. GBM incorporates interactive relationships between predictor variables, as well as being able to discern complex response curves39. MARS and ANN are good at deciphering complex relationships40 and RF models have been found to perform consistently well when evaluated against various other well established methods41. The models were implemented in the ‘R’ software42, using the ‘biomod2’ R package43. Modelling parameters were kept at default values, in the interest of repeatability. Model predictions were output as probability of presence (continuous values between 0 and 1).
Evaluation of model accuracy (predictive power) was carried out using the Area Under Curve (AUC) of the Receiver Operating Characteristic (ROC). A random 70% selection of the data was used to train each model before the remaining 30% of data were used to evaluate accuracy. To compensate for any possible spatial autocorrelation between training and evaluation data44, this cross-validation process was repeated ten times for each model, with the mean calculated to provide a value for a version of the model trained using all of the data.
The final ensemble forecast output was produced in ‘biomod2’ using a weighted average technique, based on the AUC values of each model. Models with higher AUC values were more influential in deciding final cell values. Evaluation of final ensemble model accuracy was conducted using the same cross-validation approach as that used for individual models. Variable importance for each individual model was assessed through the use of biomod2’s randomization function, where each variable’s values are randomized in turn. Pearson’s correlations of model predictions before and after randomization were used to infer relative importance of the variable being randomized44. Where the correlation is high, the relative importance of the variable to the prediction outcome is low. Final Ensemble Model (EMmw) variable importance was calculated by applying the weightings used for the ensemble forecasting to the variable importance scores for individual models, summing values by variable then dividing by the number of models used.
Creation of the P. parva risk map
The final risk map was created by multiplying the values of the EMmw layer, which represents suitability of conditions according to modelled realized niche, by those of the “Aquaculture pressure” layer, which represents likelihood of introduction. Both layers’ values range from 0–1, so the final Risk Map represents equal weightings of the two factors in assessment of the risk of successful invasion, with potential values ranging from 0 (no risk) to 1 (high risk).
Results
Predictive accuracy in models
Predictive accuracy across the nine individual modelling methods was consistently high and the mean of each modelling method always exceeded 0.847 in cross-validation runs, except for SRE (Fig. 1). SRE models were dropped from the final EM, as they were consistently lower scoring than the others. AUC scores indicated that all of the retained individual models were acceptable45 for predicting the climatic suitability of locations for P. parva. However, the weighted method of ensemble forecasting provided a significant increase in model accuracy (Welch’s t-test p ≤ 0.001, testing results of all retained individual models against ensemble model). Variability of predictive accuracy across the different absence datasets and the randomized subsets of data (i.e. partitioning for training and testing) was very similar across all modelling methods, indicating that the data for training were representative of the total dataset in each instance (i.e. for each absence dataset and for each randomization run, of which there were 100 in total for each modelling method; 900 in total).
Predictor variable importance
The three most important predictor variables, Bio14, Bio3 and TWIS (in that order) accounted for 66% of the sum of all importance values (Table 2). The density plot curve for Bio14, precipitation of driest month, (Fig. 2) suggested that P. parva prefer areas with values between 25 and 60 mm. The density plot for Bio3, isothermality, suggested that P. parva prefers areas with a mean monthly temperature range that is approximately 33% of the total annual range. Topographic wetness index was the most important of the non-‘bioclim’ variables used in the final modelling, ranking 3rd with a mean score of 11.64% in the randomization tests.
Density plots of top three most important variables in final mean-weighted Ensemble Model (EMmw), in order of importance; (a) Bio14 – Precipitation of driest month (mm); (b) Bio3 – Isothermality (mean diurnal temperature range divided by annual temperature range; (c) TWIS – Topographic Wetness Index.
Suitability predictions
Current known ranges, both invasive (Europe) and native (Asia), were generally well represented by the model output (Fig. 3a). However there were some areas within the native range where the suitability score was lower than expected, such as Taiwan. In addition to the known current invasive range, a number of other areas displayed medium to high suitability values. These areas included parts of central and southern Africa, Malaysia, Indonesia, New Guinea, New Zealand, Australia, North and South America. The highest values outside the current known range were in the USA (predominantly in the western and southern states), South America (focused around southern Brazil, eastern Paraguay and Argentina, and all of Uruguay), in Eastern Australia and in New Zealand.
(a,b) Suitability (a) and Risk (b) maps. Both maps use the same colour scale, from low value of 0 to high value of 1, indication niche suitability and risk of successful invasion, respectively. Final layout of map panels was generated using Esri ArcMap (Version 10.0, Build 2414. url: https://www.arcgis.com/).
Risk predictions
The process of incorporating introduction likelihood to the map refined the suitability map by producing a prediction of relative likelihood of a successful invasion (Fig. 3b). Areas with predicted high suitability and high levels of aquaculture pressure, a surrogate for introduction likelihood, were fairly widespread. The most significant areas of high and medium-high risk values included the USA, Brazil, Paraguay, Uruguay, Argentina, coastal South Africa, Malaysia, Indonesia and Australia. Although New Zealand possessed areas with high suitability values, the low values of aquaculture pressure meant that the risk levels were very low throughout the country. Similarly, the Island of New Guinea possessed medium-high suitability values, but aquaculture values in the country of Papua New Guinea meant that the eastern part of the island received generally low risk values, whilst the western part of the island, a region of Indonesia with higher levels of aquaculture, received a medium-high risk of successful invasion.
Discussion
The risk maps indicated that P. parva has a substantial scope to invade areas and regions beyond its current invasive range, and these cover all continents. There were several hotspots for climatic suitability and risk of introduction in each continent that could represent a potential future point of entrance. In North America for example, the key states at risk are Mississippi, Louisiana, Arkansas, N/S Carolina, Georgia, Alabama, and Tennessee. In South America, two geographically separated hotspots were identified, which included medium-high values in upper parts of the Amazon basin in Peru and Columbia and Brazil. Further south, high values were observed in the south of Brazil, Uruguay and north-east parts of Argentina. In Africa there were narrow fringes of medium-high risk in coastal areas of South Africa, Madagascar and in Liberia. However these were surrounded by more diffuse areas of medium risk, generally following the coast but also present in central countries such as the Democratic Republic of the Congo and Uganda, where the upper part of the Congo River is most at risk. Although already present in East Asia, our outputs indicated that areas in northern India would be suitable for P. parva, particularly the foot hills of the Himalayas, including the Indus River basin as far west as Srinagar in the Jammu and Kashmir state of India. Of particular note is the recent discovery of P. parva in the Brahmaputra River in India46, an area predicted to be climatically suitable and at medium-high risk by our analysis, thus providing confidence in the performance of the risk maps, particularly given that our training dataset contained only one presence cell in the entirety of India. Finally, in Oceania, predicted areas at medium-high to high risk were in Indonesia, Malaysia and parts of eastern Australia.
Evaluation of the model via cross-validation techniques indicated that the modelling method used to predict climatic suitability achieved a high degree of accuracy. However, there were areas where high suitability values were expected, due to known presence of P. parva, but were not predicted by the model. For instance there were locations in Belarus and southern Turkey that received low suitability scores, but contained P. parva records – although they are only present in relatively low abundances47,48. Equally, in the native range, Taiwan received generally low suitability scores, despite being known to host widespread P. parva populations21. This disparity between predicted and observed suitability in Taiwan may have been due to underrepresentation of distinct environmental conditions present there, owing to low numbers of presence points in this location. Such an issue can lead to underrepresentation of these conditions in the modelled niche. Because niche conservatism is a fundamental assumption of the ecological modelling techniques, the projected niche may not have included the full range of suitable conditions in Taiwan.
In time, introduced species will adapt and evolve within their new range, potentially shifting their climatic preferences and their ecological niche. Simon et al.49,50 revealed P. parva populations of Japan and Taiwan have been isolated from those found in continental China for some time (5–6 MYA and 1–1.5 MYA, respectively), allowing the adaptation of the Taiwanese populations to the more tropical climate there. Neither the Japanese, nor the Taiwanese populations have been introduced to the European invasive range, which could explain why the climatic niche of the invasive range may have not yet included the distinct part of the niche occupied only by the Taiwanese populations, which would compound the underrepresentation of this part of the niche in the data used to train our models. One implication of this is that the risk of invasion may be heavily dependent upon the source population for the introduction, particularly when the native ecological niche is distinct compared to that of other populations. In the case of the Taiwanese populations, this may mean that scope for successful invasion into areas with more tropical climates is greater, and for the species as a whole, this may mean that current risk predictions are underestimated for tropical regions.
Despite some of the invasion predictions in India already being realized, the risk level of individual countries will also be determined by extant policies on managing non-native species regarding aspects such as risk assessments and surveillance4. For example, the high level of awareness in Australia regarding non-native species introductions51 and the calls for more consistent use of appropriate risk assessment tools, as long ago as the 1990s (e.g.52), should have by now resulted in adoption of such practices for all major groups of taxa. However, particularly with regard to non-native fishes, this has not been the case; moreover the only application of a generic risk assessment scheme for non-native fishes, to date, has been that of Vilizzi and Copp53, in 2013. With this in mind, the addition of species-specific risk maps, such as the one produced in our study, present an important additional tool that can identify the specific regions of a country that are most at risk from an introduction and thus enable increased preventative measures such as import bans and additional screening at entry points to be implemented54,55. This increased prevention is preferable, as it should reduce the long-term management costs3. Currently, screening points for non-native species are often at ports or national/state borders7, locations which might not necessarily be identified as having a high risk of introduction and invasion but are entry points for species in transit and so can serve as useful points of inspection.
Failing to prevent future invasions of P. parva would greatly increase the associated threat to fish biodiversity. This species is a healthy carrier of a severe emerging fungal pathogen, S. destruens24, that is non species-specific (i.e. generalist) and has so far led to disease in all tested species including salmonids, cyprinids and percids25,56. Because of the life history traits that make P. parva invasive in so many locations (e.g. broad tolerances, high fecundity, etc) and because it is a healthy carrier of the disease (i.e. it doesn’t suffer mortality from infection), it constitutes a significant threat, as it can rapidly build dense populations which can act as persistent disease reservoirs for an outbreak. A recent study has highlighted the particular significance of the threat posed by such species57. Given the long distance of natural salmon migrations, the potential impact of an outbreak of S. destruens among North American salmonid populations (which are highly sensitive to the disease58,56), could have far reaching consequences. P. parva’s invasion could thus be the potential stepping stone to a much wider transmission of the disease. We now know that the rate of spread after initial introduction in Europe was about 0.5 countries per year21, and that invasion in a region comprising largely suitable areas is likely to result in an array of negative impacts.
Given the potential impacts of P. parva of disease transmission and the formation of highly abundant populations that can lead to increased inter-specific competition59, there are strong ecological drivers for the avoidance of introduction. Whilst simple climate-matching approaches can be useful, more representative models such as ours, accounting for critical factors such as introduction likelihood/ propagule pressure (cf.60,23) should be used in order to provide more accurate risk representations. Consequently, the outputs in this paper can be used to formulate stronger preventative measures and ensure the invasion of P. parva is limited to Europe and does not develop more globally.
Additional Information
How to cite this article: Fletcher, D. H. et al. Predicting global invasion risks: a management tool to prevent future introductions. Sci. Rep. 6, 26316; doi: 10.1038/srep26316 (2016).
References
Stohlgren, T. J. & Schnase, J. L. Risk Analysis for Biological Hazards: What We Need to Know about Invasive Species. Risk analysis : an official publication of the Society for Risk Analysis 26, 163–173 (2006).
Vitousek, P. M., Mooney, H. a, Lubchenco, J. & Melillo, J. M. Human domination of Earth’s ecosystems. Urban Ecology: An International Perspective on the Interaction Between Humans and Nature 3–13 (2008).
Norton, G. “I. Invasive species, databases, and decision making: critical issues.” Extension bulletin. Food and Fertilizer Technology Centre, Taiwan (2005).
Britton, J. R., Gozlan, R. E. & Copp, G. H. Managing non-native fish in the environment. Fish Fish. 12, 256–274 (2011).
Fernald, S. H., Caraco, N. F. & Cole, J. J. Changes in cyanobacterial dominance following the invasion of the zebra musselDreissena polymorpha: Long-term results from the Hudson River estuary. Estuaries Coasts 30, 163–170 (2007).
Myers, J. H., Simberloff, D., Kuris, A. M. & Carey, J. R. Eradication revisited: Dealing with exotic species. Trends Ecol. Evol. 15, 316–320 (2000).
Vander Zanden, M. J. & Olden, J. D. A management framework for preventing the secondary spread of aquatic invasive species. Can. J. Fish. Aquat. Sci. 65, 1512–1522 (2008).
Crowl, T. A., Crist, T. O., Parmenter, R. R., Belovsky, G. & Lugo, A. E. The spread of invasive species and infectious disease as drivers of ecosystem change. Front. Ecol. Environ. 6, 238–246 (2008).
Keith, M. & Jaarsveld, A.S. van Revisiting green data species lists. Biodivers. Conserv. 11, 1313–1316 (2002).
Leung, B. et al. An ounce of prevention or a pound of cure: bioeconomic risk analysis of invasive species. Proc. Biol. Sci. 269, 2407–2413 (2002).
Copp, G. H. et al. Calibration of FISK, an invasiveness screening tool for nonnative freshwater fishes. Risk Anal. 29, 457–467 (2009).
Jiménez-Valverde, A. et al. Use of niche models in invasive species risk assessments. Biol. Invasions 13, 2785–2797 (2011).
Ferincz, Árpád, et al. Risk assessment of non-native fishes in the catchment of the largest Central-European shallow lake (Lake Balaton, Hungary). Hydrobiologia 1–13 (2016).
Hulme, P. E. Trade, transport and trouble: Managing invasive species pathways in an era of globalization. J. Appl. Ecol. 46, 10–18 (2009).
Moorhouse, Tom P. & David W. Macdonald . Are invasives worse in freshwater than terrestrial ecosystems? WIREs Water 2, 1–8 (2015).
Gozlan, R. E., Britton, J. R., Cowx, I. & Copp, G. H. Current knowledge on non-native freshwater fish introductions. J. Fish Biol. 76, 751–786 (2010).
Villéger, S., Blanchet, S., Beauchard, O., Oberdorff, T. & Brosse, S. From current distinctiveness to future homogenization of the world’s freshwater fish faunas. Divers. Distrib. 21, 223–235 (2015).
Pimentel, D., Zuniga, R. & Morrison, D. Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol. Econ. 52, 273–288 (2005).
Savini, D. et al. The top 27 animal alien species introduced into Europe for aquaculture and related activities. J. Appl. Ichthyol. 26, 1–7 (2010).
Gozlan, R. E., Pinder, A. C. & Shelley, J. Occurrence of the Asiatic cyprinid Pseudorasbora parva in England. J. Fish Biol. 61, 298–300 (2002).
Gozlan, R. E. et al. Pan-continental invasion of Pseudorasbora parva: Towards a better understanding of freshwater fish invasions. Fish Fish. 11, 315–340 (2010).
Gozlan, R. E., Burnard, D., Britton, J. R. & Andreou, D. Evidence of female preference for hidden sex signals in distant fish species. Behav. Ecol. 25, 53–57 (2014).
Britton, J. R. & Gozlan, R. E. How many founders for a biological invasion? Predicting introduction outcomes from propagule pressure. Ecology 94, 2558–2566 (2013).
Pinder, A. C., Gozlan, R. E. & Britton, J. R. Dispersal of the invasive topmouth gudgeon, Pseudorasbora parva in the UK: a vector for an emergent infectious disease. Fish. Manag. Ecol. 12, 411–414 (2005).
Andreou, D., Arkush, K., Guegan, J. F. & Gozlan, R. E. Introduced Pathogens and Native Freshwater Biodiversity: A Case Study of Sphaerothecum destruens. PLoS One 7, 1–6 (2012).
Gozlan, R. E. et al. Current ecological understanding of fungal-like pathogens of fish: What lies beneath? Front. Microbiol. 5, 1–16 (2014).
Fitzpatrick, M. C., Weltzin, J. F., Sanders, N. J. & Dunn, R. R. The biogeography of prediction error: Why does the introduced range of the fire ant over-predict its native range? Glob. Ecol. Biogeogr. 16, 24–33 (2007).
Guisan, A., Petitpierre, B., Broennimann, O., Daehler, C. & Kueffer, C. Unifying niche shift studies: insights from biological invasions. Trends Ecol. Evol. 29, 260–269 (2014).
Boria, R. A., Olson, L. E., Goodman, S. M. & Anderson, R. P. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. Ecol. Modell. 275, 73–77 (2014).
Mainali, K. P. et al. Projecting future expansion of invasive species: comparing and improving methodologies for species distribution modeling. Glob. Chang. Biol. 21, 4464–4480 (2015).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Lin, D., Foster, D. P. & Ungar, L. H. VIF regression: a fast regression algorithm for large data. J. Am. Stat. Assoc. 493, 232–247 (2012).
Guisan, A. & Zimmermann, N. E. Predictive habitat distribution models in ecology. Ecol. Modell. 135, 147–186 (2000).
Bradley, B. A. et al. Global change, global trade, and the next wave of plant invasions. Front. Ecol. Environ. 10, 20–28 (2012).
Vörösmarty, C. J. et al. Global threats to human water security and river biodiversity. Nature 467, 555–561 (2010).
Hastie, T., Tibshirani, R. & Buja, A. Flexible Discriminant Analysis by Optimal Scoring. J. Am. Stat. Assoc. 89, 1255–1270 (1994).
Busby, J. R. Nature Conservation: Cost Effective Biological Surveys and Data Analysis. BIOCLIM - A Bioclimatic Analysis and Prediction System. In: Margules 64–68 (1991).
Thuiller, W., Araújo, M. B. & Lavorel, S. Generalized models vs. classification tree analysis: Predicting spatial distributions of plant species at different scales. J. Veg. Sci. 14, 669–680 (2003).
Wisz, M. S. et al. Effects of sample size on the performance of species distribution models. Divers. Distrib. 14, 763–773 (2008).
Moisen, G. G. & Frescino, T. S. Comparing five modelling techniques for predicting forest characteristics. Ecol. Modell. 157, 209–225 (2002).
Prasad, A. M., Iverson, L. R. & Liaw, A. Newer classification and regression tree techniques: Bagging and random forests for ecological prediction. Ecosystems 9, 181–199 (2006).
Ihaka, R. & Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 5, 299–314 (1996).
Thuiller W., George D. & Engler R. biomod2: Ensemble platform for species distribution modelling. R package version 3.1-64. http://CRAN.R-project.org/package=biomod2 (2014).
Thuiller, W., Lafourcade, B., Engler, R. & Araújo, M. B. BIOMOD - A platform for ensemble forecasting of species distributions. Ecography 32, 369–373 (2009).
Hosmer, D. W. & Lemeshow, S. Applied Logistic Regression. 2nd ed. Wiley Series in Probability and Statistics 23, (2000).
Makhrov A. A., Artamonova V. S. & Karabanov D. P. Finding of Topmouth Gudgeon Pseudorasbora parva (Temminck et Schlegel) (Actinopterygii: Cyprinidae) in the Brahmaputra River Basin (Tibetan Plateau, China). Russ. J. Biol. Invasions. 4, 174–179 (2013).
Ekmekçi, F. G. & Kirankaya, Ş. G. Distribution of an invasive fish species, Pseudorasbora parva (Temminck and Schlegel, 1846) in Turkey. Turk. J. Zool. 30, 329–334 (2006).
Semenchenko, V., Grabowska, J., Grabowski, M., Rizevsky, V. & Pluta, M. Non-native fish in Belarusian and Polish areas of the European central invasion corridor. Oceanol. Hydrobiol. Stud. 40, 57–67 (2011).
Simon, A. et al. Invasive cyprinid fish in Europe originate from the single introduction of an admixed source population followed by a complex pattern of spread. Plos One 6, e18560 (2011).
Simon, A., Gozlan, R. E., Britton, J. R., Van Oosterhout, C. & Hänfling, B. Human induced stepping-stone colonisation of an admixed founder population: the spread of topmouth gudgeon (Pseudorasbora parva) in Europe. Aquat. Sci. 77, 17–25 (2015).
Pyšek, P. & Richardson, D. M. Invasive Species, Environmental Change and Management, and Health. Annu. Rev. Environ. Resour. 35, 25–55 (2010).
Pheloung, P. C., Williams, P. A. & Halloy, S. R. A weed risk assessment model for use as a biosecurity tool evaluating plant introductions. J. Environ. Manage. 57, 239–251 (1999).
Vilizzi L. & Copp G. H. Application of FISK, an invasiveness screening tool for non-native freshwater fishes, in the Murray-Darling Basin (southeastern Australia). Risk Anal. 33, 1432–1440 (2013).
Robert Britton, J., Pegg, J. & Gozlan, R. E. Quantifying imperfect detection in an invasive pest fish and the implications for conservation management. Biol. Conserv. 144, 2177–2181 (2011).
Davies, G. D., Gozlan, R. E. & Robert Britton, J. Can accidental introductions of non-native species be prevented by fish stocking audits? Aquat. Conserv. Mar. Freshw. Ecosyst. 23, 366–373 (2013).
Arkush, K. D., Mendoza, L., Adkison, M. A. & Hedrick, R. P. Observations on the Life Stages of Sphaerothecurn destruens n. g., n. sp., a Mesomycetozoean Fish Pathogen Formally Referred to as the Rosette Agent. J. Eukaryot. Microbiol. 50, 430–438 (2003).
Al-Shorbaji, F., Roche, B., Gozlan, R. E., Britton, R. & Andreou, D. The consequences of reservoir host eradication on disease epidemiology in animal communities. Emerg. microbes & infect. (2016) (In Press).
Arkush, K. D., Frasca, S. & Hedrick, R. P. Pathology Associated with the Rosette Agent, A Systemic Protist Infecting Salmonid Fishes. J. Aquat. Anim. Health 10, 1–11 (1998).
Pinder, A. C., Davies, G. D., Brazier, M. & Britton, J. R. A Case Study of the Population Ecology of a Topmouth Gudgeon (Pseudorasbora parva) Population in the UK and the Implication for Native Fish Communities. Aquat. Conserv. Mar. Freshw. Ecosyst. 17, 749–759 (2007).
Colautti, R. I., Grigorovich, I. A. & MacIsaac, H. J. Propagule pressure: A null model for biological invasions. Biol. Invasions 8, 1023–1037 (2006).
Acknowledgements
This study was funded the Agence Nationale de la Recherche (GENESIS ANR-13- 0005-001, BIOADAPT) and the Probis Biodiversa programme.
Author information
Authors and Affiliations
Contributions
D.H.F., R.E.G., J.R.B. and P.K.G. designed the study; D.H.F. and P.K.G. analysed the data and completed the modelling; D.H.F., R.E.G., J.R.B., S.B. and P.K.G. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Fletcher, D., Gillingham, P., Britton, J. et al. Predicting global invasion risks: a management tool to prevent future introductions. Sci Rep 6, 26316 (2016). https://doi.org/10.1038/srep26316
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep26316
This article is cited by
-
Reconstructing the biological invasion of Tuta absoluta: evidence of niche shift and its consequences for invasion risk assessment
Journal of Pest Science (2024)
-
Modelled distribution of an invasive alien plant species differs at different spatiotemporal scales under changing climate: a case study of Parthenium hysterophorus L.
Tropical Ecology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.