Abstract
Tumor necrosis factor-α (TNF-α) is a proinflammation cytokine secreted by various cells. Understanding its secretive pathway is important to understand the biological functions of TNF-α and diseases associated with TNF-α. TNF-α is one of the first proteins known be modified by lysine fatty acylation (e.g. myristoylation). We previously demonstrated that SIRT6, a member of the mammalian sirtuin family of enzymes, can remove the fatty acyl modification on TNF-α and promote its secretion. However, the mechanistic details about how lysine fatty acylation regulates TNF-α secretion have been unknown. Here we present experimental data supporting that lysine fatty acylation promotes lysosomal targeting of TNF-α. The result is an important first step toward understanding the biological functions of lysine fatty acylation.
Similar content being viewed by others
Introduction
TNF-α is a proinflammation cytokine produced by various types of cells, including macrophages, lymphocytes, dendritic cells, and neurons1,2,3. It binds to TNF-α receptors and triggers signaling pathways that affect apoptosis and cell survival4,5. Dysregulation of TNF-α is associated with many inflammatory diseases, including rheumatoid arthritis, psoriatic arthritis, and inflammatory bowel disease2,6,7.
There are two protein forms of TNF-α, membrane TNF-α (mTNF-α, 26 kD) and secreted TNF-α (sTNF-α, 17 kD)8. mTNF-α is a type II transmembrane protein, with a single transmembrane domain linking the intracellular N-terminal domain and extracellular C-terminal domain. It is synthesized at the endoplasmic reticulum (ER) and transported to plasma membrane through Golgi and recycling endosomes9,10,11,12. At the plasma membrane, it is quickly digested by the metalloprotease TNF-α converting enzyme (TACE) to release the C-terminal sTNF-α13,14. The native mTNF-α and sTNF-α are trimers15,16,17,18,19 and both of them have been demonstrated to have biological activities and bind to TNF-α receptors4,8.
Because TNF-α has important immune functions, there has been significant research interest in the regulation of TNF-α. TNF-α is known to be regulated by many protein posttranslational modifications. In addition to proteolytic cleavage by TACE, proteases SPPL2a and SPPL2b were found to cleave the intramembrane part of mTNF-α and promote the release of the intracellular domain to induce the expression of pro-inflammatory cytokine interleukin-1220,21. TNF-α is also modified by O- and N-glycosylation, cysteine fatty acylation, and lysine fatty acylation. It was reported that human TNF-α has O-glycosylation at Ser8022 and mouse TNF-α has N-glycosylation at Asn8623. Recently, glycosylation at Ser80 of rat TNF-α was shown to regulate the sorting of TNF-α into mast cell granules24. Cysteine fatty acylation was found on human TNF-α at Cys3025. Fatty acylation of Cys30 is thought to facilitate the association of TNF-α with plasma membrane and its integration into lipid raft26. Although lysine fatty acylation on mTNF-α at Lys19 and Lys20 was discovered in early 1990s27, the effect of this modification on TNF-α was only recently elucidated. Lysine fatty acylation on TNF-α decreases its secretion and this modification is regulated by SIRT6, a nicotinamide adenine dinucleotide (NAD+)-dependent deacylase28. This was the first demonstration that lysine fatty acylation can regulate protein secretion. However, how lysine fatty acylation affects TNF-α secretion has been unknown. Here we demonstrate that lysine fatty acylation helps to target TNF-α to the lysosomes.
Results
Lysine fatty-acylated TNF-α localizes to large membrane vesicles
Previously we demonstrated that lysine fatty acylation downregulated TNF-α secretion (the production of sTNF-α)28. The secretion of sTNF-α in Sirt6 knockout (KO) or knockdown (KD) cells was lower than that in control cells. When Sirt6 KD cells were treated with palmitic acid to increase the level of lysine fatty acylation on TNF-α, the secretion efficiency of TNF-α in Sirt6 KD cells decreased even more dramatically28.
In order to find out how lysine fatty acylation affects TNF-α secretion, we overexpressed wildtype (WT) TNF-α with a C-terminal green fluorescent protein (GFP), termed TNF-α_WT_GFP, in WT and Sirt6 KO MEF cells and examined the cellular localization of TNF-α. Confocal imaging results (Fig. 1) showed that TNF-α localized to distinct vesicles near the nucleus and throughout the cytosol in WT MEF cells. The size of TNF-α containing vesicles remained small when cells were treated with palmitic acid. While in Sirt6 KO MEF cells, TNF-α existed in large vesicles, which became even larger when the cells were treated with palmitic acid.
Lysine fatty acylation retains TNF-α in lysosomes
We next set out to find out the identities of the TNF-α containing vesicles in both WT and Sirt6 KO MEF cells. We examined the co-localization of TNF-α with protein markers of various organelles involved in protein secretion29, including Rab11 (recycling endosome marker), EEA1 (early endosome marker), Rab7 (late endosome marker), and LAMP1 (lysosome marker) (Fig. 2). In Sirt6 WT cells, TNF-α co-localized with Rab11, EEA1 and Rab7 but not LAMP1 (Fig. 2, quantification of co-localization is shown in Fig. 3), in both perinuclear regions (arrows) and nucleus-distal regions (squares). These results indicated that TNF-α resided in recycling endosomes, early endosomes, and late endosomes but not in lysosomes in WT MEF cells. This is consistent with early report that TNF-α is secreted via the recycling endosomes9. In contrast, in Sirt6 KO cells, TNF-α co-localized with the lysosomal marker LAMP1 very well, suggesting that TNF-α localized more in lysosomes in Sirt6 KO cells (Fig. 2, right, and Fig. 3). In the Sirt6 KO cells, we still found co-localization of TNF-α with Rab11, EEA1, and Rab7. However, co-localization with the recycling endosome marker, Rab11, was significantly decreased in Sirt6 KO cells compared to that in Sirt6 WT cells (Figs 2 and 3). This suggested that the TNF-α localization in the recycling endosomes was decreased by Sirt6 KO, most likely due to the increased localization of TNF-α in the lysosomes.
Co-localization of TNF-α_WT_GFP with different intracellular organelle markers in WT and Sirt6 KO MEF cells treated with 50 μM palmitic acid. Rab11, Rab7, EEA1 and LAMP1 are the protein markers of recycling endosomes, late endosomes, early endosomes and lysosomes, respectively. Arrows point to perinuclear regions while squares highlight nucleus-distal regions containing TNF-α_WT_GFP.
Overlap coefficients of TNF-α_WT_GFP and TNF-α_KR_GFP with Rab11 (recycling endosomes), Rab7 (late endosomes), EEA1 (early endosomes), and LAMP1 (lysosome). Co-localization was quantified using ImageJ. Each bar is the mean ± SD (error bars) for three images. Two asterisks indicate significance with p < 0.01.
The different localizations of TNF-α in WT and Sirt6 KO cells could be caused by the different levels of lysine fatty acylation on TNF-α or by differences in the secretory pathway. In order to find out whether the different localizations of TNF-α in WT and Sirt6 KO MEF cells were caused by differences in TNF-α lysine fatty acylation, we further examined the localization of the TNF-α mutant, TNF-α_KR, in which both Lys19 and Lys20 were mutated to Arg (Fig. 4). TNF-α_KR is defective in lysine fatty acylation and thus should not be affected by SIRT6-catalyzed defatty-acylation. In both WT and Sirt6 KO cells, the localization of TNF-α_KR was similar to that of TNF-α_WT (Fig. 4, quantification of co-localization is shown in Fig. 3). In particular, TNF-α_KR did not co-localize with LAMP1 in either WT or Sirt6 KO cells (Figs 3 and 4). This confirmed that localization of TNF-α_WT in the lysosomes of Sirt6 KO MEF cells was directly caused by lysine fatty acylation on TNF-α_WT.
To further confirm the results obtained with confocal imaging, we also employed lysosome fractionation to determine the lysosomal localization of TNF-α WT and KR in WT and Sirt6 KO MEF cells. Consistent with the imaging data, more TNF-α WT was found in the lysosomal fraction of Sirt6 KO cells than WT cells (Fig. 5). In contrast, the levels of TNF-α KR mutant were similar in the lysosomal fractions of WT and Sirt6 KO MEF cells (Fig. 5).
A portion of unacylated TNF-α is also targeted to lysosome for degradation
Although no TNF-α was observed in the lysosomes in Sirt6 WT cells (Fig. 2), we wondered whether unacylated TNF-α molecules in Sirt6 WT cells were also targeted to lysosomes but they were either degraded quickly or exit the lysosome quickly. To investigate this, we treated cells with E6430, a lysosomal protease inhibitor, and then examined the localization of TNF-α_WT_GFP and TNF-α_KR_GFP via confocal imaging (Fig. 6). Interestingly, when treated with E64, we observed significant amount of co-localization of TNF-α_WT and TNF-α_KR with LAMP1 (Figs 3 and 6) in both WT and Sirt6 KO MEF cells.
These results suggest that unacylated TNF-α can also be targeted to the lysosome, but they are degraded very quickly. If TNF-α molecules are targeted to the lysosome and degraded, we would predict that E64 would not affect the amount of TNF-α secreted. However, it is also possible that the TNF-α molecules are targeted to lysosome as part of the normal secretory pathway (for example, cleavage of the transmembrane domain by lysosome proteases can actually create the sTNF-α). In that case, we would predict that E64 would decrease the amount of sTNF-α secreted. To differentiate the two possibilities, we measured the sTNF-α secretion in WT and Sirt6 KO MEF cells with or without E64 treatment (Fig. 7). The secretion of sTNF-α was not significantly affected by E64 treatment in either WT or Sirt6 KO cells (Fig. 7A). The only consequence of E64 treatment was that there was significantly increased TNF-α_WT inside of WT MEF cells (49% increase, Fig. 7B), which was consistent with the increased localization of TNF-α_WT_GFP in the lysosomes of WT MEF cells under E64 treatment (Fig. 6). Thus, the secretion results suggest that in WT cells, a certain percentages of TNF-α molecules are delivered to lysosomes, but they are quickly degraded and cannot be seen in the lysosomes without E64 treatment. In the Sirt6 KO cells, more TNF-α molecules are targeted to the lysosomes, which allows the detection of TNF-α in the lysosomes even without E64 treatment.
The effect of E64 on TNF-α secretion. (A) Relative (with E64 treatment versus without E64 treatment) levels of sTNF-α from WT and Sirt6 KO MEF cells. (B) Relative (with E64 treatment versus without E64 treatment) levels of intracellular mTNF-α in WT and Sirt6 KO MEF cells. Each bar is the mean ± SD (error bars) for three independent experiments. One asterisk indicates significance with p < 0.05.
Endogenous TNF-α in THP-1 cells is also targeted to lysosome by lysine fatty acylation
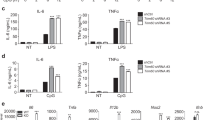
All the above experiments were carried out with over-expressed TNF-α. To make sure endogenous TNF-α are also regulated similarly by SIRT6 and lysine fatty acylation, we examined the localization of endogenous TNF-α in THP-1 cells with control or Sirt6 knockdown (Fig. 8B). Similar to the localization in MEF cells, endogenous TNF-α existed in large vesicles in THP-1 cells with Sirt6 knockdown (Fig. 8A). In THP-1 cells with control shRNA, endogenous TNF-α co-localized with EEA1, Rab11, Rab7, but very little co-localized with LAMP1 (Fig. 8A,C). When SIRT6 was knocked down, TNF-α showed significantly increased co-localization with lysosome marker LAMP1 (Fig. 8A,C). Different from the results obtained in MEF cells, TNF-α also showed increased EEA1 co-localization in SIRT6 knockdown THP-1 cells than that in control THP-1 cells (Fig. 8A,C). Overall, these results demonstrate that lysine fatty acylation also helps to target endogenous TNF-α to lysosomes in THP-1 cells.
Co-localization of endogenous TNF-α with different intracellular organelle markers in THP-1 cells with control shRNA or Sirt6 shRNA. (A) Confocal imaging of endogenous TNF-α, Rab11 (recycling endosome marker), Rab7 (late endosome marker), EEA1 (early endosome marker), and LAMP1 (lysosome marker) in THP-1 cells. (B) Western blot of endogenous SIRT6 in THP-1 cells with control shRNA or Sirt6 shRNA. (C) Overlap coefficients of TNF-α with Rab11, Rab7, EEA1, and LAMP1. Co-localization was quantified using coloc 2 plugin in ImageJ. Each bar is the mean ± SD (error bars) for three images. Two asterisks indicate significance with p < 0.01.
Discussion
Understanding the mechanism that regulates cytokine secretion is essential to understand and treat cytokine-associated diseases. There are canonical and non-canonical secretion pathways for various cytokines10,11,12. TNF-α is generally believed to be secreted via the canonical pathway, from the ER/Golgi to plasma membrane through recycling endosomes10,11,12. We previously reported that secretion of sTNF-α is regulated by reversible lysine fatty acylation28. Lysine fatty acylation was discovered over 20 years ago, and there are only a few mammalian proteins known to have this modification, including interleukin 1α and TNF-α27,31. Regulating TNF-α secretion is the only known physiological function of lysine fatty acylation. However, how lysine fatty acylation regulates the secretion of TNF-α has been unknown. Our current study now provides more in-depth insights into how lysine fatty acylation may affect protein secretion.
The data presented here demonstrate that lysine fatty acylation helps to target TNF-α to the lysosomes for degradation and at the same time decrease its localization in the recycling endosome (Fig. 9), which explains why Sirt6 KO cells have decreased TNF-α secretion. The lysosomal localization of TNF-α in Sirt6 KO cells is directly determined by TNF-α lysine fatty acylation, as the TNF-α_KR mutant that cannot be lysine fatty-acylated is not localized in the lysosomes in Sirt6 KO cells. Some unfatty-acylated TNF-α molecules in Sirt6 WT cells are also targeted to the lysosomes, but they are quickly degraded and cannot be seen in the lysosomes unless a lysosomal protease inhibitor E64 is used.
The observation that TNF-α can only be observed in the lysosomes of Sirt6 KO cells may be caused by a combination of increased lysosomal targeting and resistance to lysosomal degradation. Given that lysine fatty acylation decreases TNF-α secretion, we are certain that lysosome targeting is increased by lysine fatty acylation (if lysine fatty acylation only increases resistance to lysosomal protease cleavage, the overall secretion would not be affected by lysine fatty acylation). However, we cannot rule out that lysine fatty acylation also increases resistance to lysosomal degradation. Whether TNF-α degradation by lysosomal proteases is affected by lysine fatty acylation awaits further studies.
Interestingly, it was previously reported that hypoxia reduced TNF-α secretion and enhanced co-localization of TNF-α with LAMP132. Our results here may explain how TNF-α lysosomal localization increased under hypoxia condition32. It is possible that the level of lysine fatty acylation on TNF-α increases in hypoxia because cellular NAD+ level decreases in hypoxia33, which reduces SIRT6 enzymatic activity and increases lysine fatty acylation and thus lysosomal localization of TNF-α28.
It is not clear at this point whether lysine fatty acylation is a general mechanism for lysosomal targeting or not as only one defatty-acylation substrate for SIRT6 has been reported thus far. This question will be addressed when more defatty-acylation substrates of SIRT6 or other sirtuins are discovered in the future.
Methods
Reagents
Palmitic acid, E64, phorbol 12-myristate 13-acetate (PMA) and lipopolysaccharides from Escherichia coli 0111:B4 (LPS) were purchased from Sigma-Aldrich (St. Louis, MO). Anti-Flag antibody conjugated with horseradish peroxidase (A8592) was purchased from Sigma-Aldrich. Rat monoclonal anti-mouse LAMP1 antibody (1D4B) was purchased from BD Biosciences (San Jose, CA). Goat anti-rat IgG conjugated with Alexa Fluor 647 was purchased from Life Technologies (Grand Island, NY). Rabbit monoclonal anti-Rab11 antibody (D4F5), rabbit monoclonal anti-Rab7 antibody (D95F2), rabbit monoclonal anti-EEA1 antibody (C45B10), rabbit monoclonal anti-SIRT6 antibody (D8D12), goat anti-rabbit IgG conjugated with Alexa Fluor 647, and ProLong Gold antifade reagent with DAPI were purchased from Cell Signaling Technology (Danvers, MA). Anti-human TNF-α antibody (MAB610) was purchased from R&D systems (Minneapolis, MN). Human TNF-α ELISA kit was purchased from eBioscience. ECL plus western blotting detection reagent and Lysosome enrichment kit were purchased from Thermo Fisher Scientific Inc. SIRT6 shRNA lentiviral plasmid (pLKO.1-puro vector) was purchased from Sigma, the sequence of shRNA was: TRCN0000378253 (ccggcagtacgtccgagacacagtcctcgaggactgtgctcggacgtactgtttttg). Plasmid of pEGFP-N1 vector was a kind gift from Professor Barbara A. Baird.
Cloning of TNF-α_WT and TNF-α_KR into pCMV4A and pEGFP-N1 vector
For construction of the expression vector for wild-type human TNF-α protein (TNF-α_WT) or the double lysine mutant (TNF-α_KR, K19R, K20R) with N-terminal Flag or C-terminal EGFP tag, full-length TNF-α_WT or TNF-α_KR cDNA28 were generated by PCR and inserted into pCMV4A vector at EcoRV and XhoI sites or pEGFP-N1 vector at Xho I and BamH I sites.
Cells and Cell Transfection
THP-1 cells were maintained in Roswell Park Memorial Institute Medium (RPMI) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies). WT and Sirt6 KO MEF cells28 were maintained in Dulbecco’s Modified Eagle Medium (DMEM) containing glucose and L-glutamine (Life Technologies, Grand Island, NY) supplemented with 10% heat-inactivated fetal bovine serum. Human Embryonic Kidney (HEK) 293T cells were maintained in DMEM supplemented with 10% heat-inactivated fetal bovine serum. The TNF-α_WT and TNF-α_KR in pCMV4A or pEGFP-N1 vector were transfected into WT and Sirt6 KO MEF cells (50% confluency) using FuGene 6 transfection reagent (Promega, Madison, WI) according to the manufacture’s protocol.
Generation of stable SIRT6 knockdown THP-1 cells
SIRT6 shRNA plasmid, pCMV-dR8.2, and pMD2.G were co-transfected into HEK 293T cells. After 24 hr, the medium was collected to infect the THP-1 cells. After infection for 6 hr, the medium was changed to fresh medium and the cells were cultured for 72 hr. Then 1.5 μg/mL of puromycin was added into the cells for at least three generations to select stable SIRT6 knockdown cells.
Confocal imaging of TNF-α_WT_GFP or TNF-α_KR_GFP in WT and Sirt6 KO MEF cells
After WT and Sirt6 KO MEF cells were transfected with TNF-α_WT_GFP or TNF-α_KR_GFP for 12 hr, cells were incubated with fresh medium containing 50 μM palmitic acid or 50 μM E64 or DMSO (as control) for 6 hr. Then cells were washed with 1 mL 1× PBS buffer and fixed with 1 mL 4% paraformaldehyde (PFA) for 15 min at room temperature. After washing cells 3 times with 1 mL 1× PBS buffer, cells were permeabilized with 1 mL Saponin-BSA solution (0.1% Saponin with 3% BSA in 1× PBS buffer with 0.02% NaN3) for 1 hr at room temperature and then incubated with primary antibody (against Rab7, Rab11, EEA1 or LAMP1) solution (100 μL in Saponin-BSA solution) at 4 °C for 12 hr. After washing 4 times with 1 mL Saponin-BSA solution, cells were incubated with the secondary antibody (goat anti-rabbit or anti-rat IgG conjugated with Alexa Fluor 647) solution (100 μL in Saponin-BSA solution) for 2 hr at room temperature. After washing 4 times with 1 mL Saponin-BSA solution, cells were incubated with 250 μL ProLong Gold antifade reagent with DAPI for confocal imaging experiments. For quantification of colocalization, Mander’s overlap coefficients in images were determined using ImageJ coloalization plugin Intensity Correlation Analysis34 with manual setting of thresholds.
Confocal imaging of endogenous TNF-α_in THP-1 cells
THP-1 cells (with control shRNA or SIRT6 shRNA) were treated with PMA (200 ng/mL) for 24 hr. Then cells were cultured in fresh RMPI medium with palmitic acid (50 μM) and LPS (1 μg/mL) for 4 hr. After washing with 1 mL 1× PBS buffer, the cells were fixed with 1 mL 4% PFA for 15 min at room temperature. The cells were then washed twice with 1× PBS buffer and permeabilized with 1 mL Saponin-BSA solution (0.1% Saponin with 3% BSA in 1× PBS buffer with 0.02% NaN3) for 1 hr at room temperature. The cells were then incubated with the primary antibodies (against TNF-α, Rab7, Rab11, EEA1 or LAMP1) in Saponin-BSA solution at 4 °C for 12 hr. After washing 3 times with 1 mL Saponin-BSA solution, the cells were incubated with the secondary antibodies (goat anti-rabbit or anti-rat IgG conjugated with Alexa Fluor 647) solution in Saponin-BSA solution for 1 hr at room temperature. After washing 3 times with 1 mL Saponin-BSA solution, confocal imaging experiments were performed. For quantification of co-localization, Mander’s overlap coefficients in images were determined using ImageJ coloc 2 plugin with manual setting of thresholds.
Lysosome fractionation of WT and Sirt6 KO MEF cells
After WT and Sirt6 KO MEF cells were transfected with TNF-α WT or TNF-α KR for 24 hr, the cells were collected. Lysosome enrichment kit was used to fractionate the lysosome fraction. Flag western blot was then performed to determine the TNF-α expression in lysosome fraction and total lysates. The chemiluminescence signal in membrane was recorded after developing in ECL plus western blotting detection reagents using Typhoon 9400 Variable Mode Imager (GE Healthcare Life Sciences).
Secretion of TNF-α_WT and TNF-α_KR from WT and Sirt6 KO MEF Cells treated with or without E64
After transient transfection of TNF-α_WT or TNF-α_KR28 for 6 hr, WT and Sirt6 KO MEF cells were further incubated with fresh medium containing 10 μM E64 or DMSO (as control) for 12 hr. Then the culture medium and cells were collected separately for detection of TNF-α using human TNF-α ELISA kit. Three independent experiments were carried out.
Statistical Analysis
Data were expressed as mean ± SD (standard deviation, shown as error bars). Differences were examined by Student’s t test between two groups. P value below 0.05 was attributed to significance and represented by one asterisk. P value below 0.01 was attributed to most significance and represented by double asterisks.
Additional Information
How to cite this article: Jiang, H. et al. Lysine fatty acylation promotes lysosomal targeting of TNF-α. Sci. Rep. 6, 24371; doi: 10.1038/srep24371 (2016).
References
Locksley, R. M., Killeen, N. & Lenardo, M. J. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104, 487–501 (2001).
Croft, M. The role of TNF superfamily members in T-cell function and diseases. Nature reviews. Immunology 9, 271–285, 10.1038/nri2526 (2009).
Caminero, A., Comabella, M. & Montalban, X. Tumor necrosis factor alpha (TNF-alpha), anti-TNF-alpha and demyelination revisited: an ongoing story. Journal of neuroimmunology 234, 1–6, 10.1016/j.jneuroim.2011.03.004 (2011).
Wajant, H., Pfizenmaier, K. & Scheurich, P. Tumor necrosis factor signaling. Cell death and differentiation 10, 45–65, 10.1038/sj.cdd.4401189 (2003).
Gaur, U. & Aggarwal, B. B. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochemical pharmacology 66, 1403–1408 (2003).
Aggarwal, B. B., Gupta, S. C. & Kim, J. H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 119, 651–665, 10.1182/blood-2011-04-325225 (2012).
Palladino, M. A., Bahjat, F. R., Theodorakis, E. A. & Moldawer, L. L. Anti-TNF-alpha therapies: the next generation. Nature reviews. Drug discovery 2, 736–746, 10.1038/nrd1175 (2003).
Horiuchi, T., Mitoma, H., Harashima, S., Tsukamoto, H. & Shimoda, T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology 49, 1215–1228, 10.1093/rheumatology/keq031 (2010).
Murray, R. Z., Kay, J. G., Sangermani, D. G. & Stow, J. L. A Role for the Phagosome in Cytokine Secretion. Science 310, 1492–1495, 10.1126/science.1120225 (2005).
Stanley, A. C. & Lacy, P. Pathways for cytokine secretion. Physiology 25, 218–229, 10.1152/physiol.00017.2010 (2010).
Duitman, E. H., Orinska, Z. & Bulfone-Paus, S. Mechanisms of cytokine secretion: a portfolio of distinct pathways allows flexibility in cytokine activity. European journal of cell biology 90, 476–483, 10.1016/j.ejcb.2011.01.010 (2011).
Stow, J. L., Low, P. C., Offenhauser, C. & Sangermani, D. Cytokine secretion in macrophages and other cells: pathways and mediators. Immunobiology 214, 601–612, 10.1016/j.imbio.2008.11.005 (2009).
Black, R. A. et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature 385, 729–733, 10.1038/385729a0 (1997).
Moss, M. L. et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature 385, 733–736, 10.1038/385733a0 (1997).
Eck, M. J. & Sprang, S. R. The structure of tumor necrosis factor-alpha at 2.6 A resolution. Implications for receptor binding. The Journal of biological chemistry 264, 17595–17605 (1989).
Jones, E. Y., Stuart, D. I. & Walker, N. P. Structure of tumour necrosis factor. Nature 338, 225–228, 10.1038/338225a0 (1989).
Smith, R. A. & Baglioni, C. The active form of tumor necrosis factor is a trimer. The Journal of biological chemistry 262, 6951–6954 (1987).
Tang, P., Hung, M. C. & Klostergaard, J. Human pro-tumor necrosis factor is a homotrimer. Biochemistry 35, 8216–8225, 10.1021/bi952182t (1996).
Wingfield, P., Pain, R. H. & Craig, S. Tumour necrosis factor is a compact trimer. FEBS letters 211, 179–184 (1987).
Friedmann, E. et al. SPPL2a and SPPL2b promote intramembrane proteolysis of TNFalpha in activated dendritic cells to trigger IL-12 production. Nature cell biology 8, 843–848, 10.1038/ncb1440 (2006).
Fluhrer, R. et al. A gamma-secretase-like intramembrane cleavage of TNFalpha by the GxGD aspartyl protease SPPL2b. Nature cell biology 8, 894–896, 10.1038/ncb1450 (2006).
Takakura-Yamamoto, R., Yamamoto, S., Fukuda, S. & Kurimoto, M. O-glycosylated species of natural human tumor-necrosis factor-alpha. European journal of biochemistry 235, 431–437 (1996).
Sherry, B., Jue, D. M., Zentella, A. & Cerami, A. Characterization of high molecular weight glycosylated forms of murine tumor necrosis factor. Biochemical and biophysical research communications 173, 1072–1078 (1990).
Olszewski, M. B., Trzaska, D., Knol, E. F., Adamczewska, V. & Dastych, J. Efficient sorting of TNF-alpha to rodent mast cell granules is dependent on N-linked glycosylation. European journal of immunology 36, 997–1008, 10.1002/eji.200535323 (2006).
Utsumi, T. et al. Transmembrane TNF (pro-TNF) is palmitoylated. FEBS letters 500, 1–6 (2001).
Poggi, M. et al. Palmitoylation of TNF alpha is involved in the regulation of TNF receptor 1 signalling. Biochimica et biophysica acta 1833, 602–612, 10.1016/j.bbamcr.2012.11.009 (2013).
Stevenson, F. T., Bursten, S. L., Locksley, R. M. & Lovett, D. H. Myristyl acylation of the tumor necrosis factor alpha precursor on specific lysine residues. The Journal of experimental medicine 176, 1053–1062 (1992).
Jiang, H. et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496, 110–113, 10.1038/nature12038 (2013).
Andrews, N. W. Regulated secretion of conventional lysosomes. Trends in cell biology 10, 316–321 (2000).
Hashida, S., Towatari, T., Kominami, E. & Katunuma, N. Inhibitions by E-64 derivatives of rat liver cathepsin B and cathepsin L in vitro and in vivo . Journal of biochemistry 88, 1805–1811 (1980).
Stevenson, F. T., Bursten, S. L., Fanton, C., Locksley, R. M. & Lovett, D. H. The 31-kDa precursor of interleukin 1 alpha is myristoylated on specific lysines within the 16-kDa N-terminal propiece. Proceedings of the National Academy of Sciences of the United States of America 90, 7245–7249 (1993).
Lahat, N. et al. Hypoxia enhances lysosomal TNF-alpha degradation in mouse peritoneal macrophages. American journal of physiology. Cell physiology 295, C2–12, 10.1152/ajpcell.00572.2007 (2008).
Lim, J. H. et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Molecular cell 38, 864–878, 10.1016/j.molcel.2010.05.023 (2010).
Li, Q. et al. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24, 4070–4081 (2004).
Acknowledgements
We thank Professor Barbara A. Baird for providing the pEGFP-N1 vector, Dr. Frederick Maxfield and Dr. Fenghua Hu for helpful discussions. Imaging data was acquired in the Cornell BRC-Imaging Facility using the shared, NYSTEM (CO29155) and NIH (S10OD018516)-funded Zeiss LSM880 confocal/multiphoton microscope. This work was supported in part by grants from NIH to H.L. (GM086703 and DK107868).
Author information
Authors and Affiliations
Contributions
H.J., X.Z. and H.L. designed experiments, H.J. carried out most experiments with MEF cells and X.Z. carried out the experiments with THP-1 cells and lysosome fractionation experiments, H.J., X.Z. and H.L. analyzed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jiang, H., Zhang, X. & Lin, H. Lysine fatty acylation promotes lysosomal targeting of TNF-α. Sci Rep 6, 24371 (2016). https://doi.org/10.1038/srep24371
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep24371
This article is cited by
-
Golgi stress induces SIRT2 to counteract Shigella infection via defatty-acylation
Nature Communications (2022)
-
Nicotine promotes neuron survival and partially protects from Parkinson’s disease by suppressing SIRT6
Acta Neuropathologica Communications (2018)
-
Organelle membrane derived patches: reshaping classical methods for new targets
Scientific Reports (2017)
-
The role of caloric load and mitochondrial homeostasis in the regulation of the NLRP3 inflammasome
Cellular and Molecular Life Sciences (2017)
-
Spatio-Temporal Control of Cellular and Organismal Physiology by Sirtuins
Journal of the Indian Institute of Science (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.