Abstract
Salvia miltiorrhiza Bunge, which contains tanshinones and phenolic acids as major classes of bioactive components, is one of the most widely used herbs in traditional Chinese medicine. Production of tanshinones and phenolic acids is enhanced by methyl jasmonate (MeJA). Transcription factor MYC2 is the switch of jasmontes signaling in plants. Here, we focused on two novel JA-inducible genes in S. miltiorrhiza, designated as SmMYC2a and SmMYC2b, which were localized in the nucleus. SmMYC2a and SmMYC2b were also discovered to interact with SmJAZ1 and SmJAZ2, implying that the two MYC2s might function as direct targets of JAZ proteins. Ectopic RNA interference (RNAi)-mediated knockdown experiments suggested that SmMYC2a/b affected multiple genes in tanshinone and phenolic acid biosynthetic pathway. Besides, the accumulation of tanshinones and phenolic acids was impaired by the loss of function in SmMYC2a/b. Meanwhile, SmMYC2a could bind with an E-box motif within SmHCT6 and SmCYP98A14 promoters, while SmMYC2b bound with an E-box motif within SmCYP98A14 promoter, through which the regulation of phenolic acid biosynthetic pathway might achieve. Together, these results suggest that SmMYC2a and SmMYC2b are JAZ-interacting transcription factors that positively regulate the biosynthesis of tanshinones and Sal B with similar but irreplaceable effects.
Similar content being viewed by others
Introduction
Salvia miltiorrhiza Bunge, named Danshen in China, is a widely used traditional Chinese medicine with a remarkable clinical effect on cardiovascular diseases, cerebrovascular, hyperlipidemia and acute ischemic stroke1,2,3. Pharmacological investigations indicate that the active ingredients of S. miltiorrhiza are lipophilic tanshinones and hydrophilic phenolic acids3,4,5. Jasmonate acid (JA) and its oxylipin derivatives, collectively called jasmonates, are phytohormones that play an integral role in various defense responses, leading to the accumulation of secondary metabolites6. The biosynthesis of JA has been extensively studied and well-defined7. Under bioactive jasmonate stress, JA is conjugated to isoleucine. Jasmonoyl-L-isoleucine (JA-Ile) is formed and perceived by Coronatine Insensitive 1 (COI1) protein to form Skp1/Cullin1/F-box protein COI1 (SCFCOI1) complexes. Afterwards, Jasmonate ZIM-domain (JAZ) proteins are recruited, ubiquitinated and subsequently degraded, leading to the release and activation of MYC2 proteins8,9,10. However, under stress-free growth conditions, Jasmonate ZIM-domain (JAZ) proteins recruit its co-repressors, such as TOPLESS (TPL) or TPL-related proteins (TRPs)11,12,13, leading to the repression of JA responses.
Methyl jasmonate (MeJA) has been commonly used as an elicitor to explore the regulatory mechanisms underlying the biosynthesis of active compounds14. There have been reports indicating that the accumulation of tanshinone IIA and Salvianolic acid B (Sal B) in S. miltiorrhiza hairy root cultures is accelerated by applying exogenous MeJA or improving endogenous JA15,16,17. Multiple MeJA-responsive transcription factors (TFs) and genes involved in the regulation and biosynthesis of tanshinones and phenolic acids pathways were previously mined without advanced functional verifications yet18. The specific role that MeJA plays in the activation of TFs which regulate a wide array of pathway genes is still veiled in S. miltiorrhiza. Whether MYC2 regulates the biosynthesis of tanshinones and Sal B still remains poorly understood.
Here, we focused on two MYC2 genes, SmMYC2a and SmMYC2b, which belonged to bHLH gene family. SmMYC2a and SmMYC2b turned out to be positive JA-responsive genes, which not only affected multiple genes in tanshinones and Sal B biosynthetic pathway, but also dramatically influenced the output of tanshinones and Sal B. In electrophoretic mobility shift assay (EMS assay), SmMYC2a could bind with an E-box motif within promoters of hydroxy-cinnamoyl transferase 6 (SmHCT6) and cytochrome P450-dependent monooxygenase 98A14 (SmCYP98A14), while SmMYC2b bound with an E-box motif within SmCYP98A14 promoter. These results indicated that SmMYC2a and SmMYC2b were positive regulators that strongly regulated pathway genes in the biosynthesis of tanshinones and phenolic acids.
Results
Molecular Cloning, phylogenetic analysis and organ expression pattern of SmMYC2a and SmMYC2b
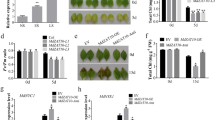
Based on the built MeJA-induced gene expression profiling database of S. miltiorrhiza18, two JA-responsive MYC2 members, SmMYC2a and SmMYC2b were successfully amplified. The genomic sequence of SmMYC2a contained an 1803 bp ORF without introns and SmMYC2b contained an ORF of 1809 bp without introns. Meanwhile, the two MYC2s contained conserved bHLH domain and JAZ interaction domain (JID) (Fig. 1A), which defined bHLH family. Alignment of SmMYC2a, SmMYC2b and Arabidopsis MYC2 was displayed at amino acid level, indicating that SmMYC2a and SmMYC2b shared a 53% and 47% identity with AtMYC2 respectively. For evolutionary relationship analysis, two SmMYC2s, together with 37 bHLH proteins from various plant species, were used to construct a Neighbour-joining (NJ) tree at the amino acid level. The dendrogram (Fig. 1B) showed that the selected MYC2s were clustered into two groups, group I and group II. SmMYC2a was clustered into a subgroup, which belonged to group I, with SmfMYC2, while SmMYC2b belonged to group II. Organ expression analysis was carried out to study the expression pattern of SmMYC2s. Both SmMYC2a and SmMYC2b were discovered to be constitutively expressed in roots, stems and leaves. Meanwhile, both genes were significantly highly expressed in leaves, followed by stems and roots (Fig. 1C).
Multiple alignment, phylogenetic analysis and organ expression analysis of SmMYC2a and SmMYC2b.
(A) Alignment of SmMYC2a, SmMYC2b and Arabidopsis MYC2 homologs. AtMYC2 (GenBank Accession: NM_102998.3) amino acid sequence was displayed. bHLH domain was boxed. Black highlighted residues indicated identical residues and gray boxes indicated similar residues. (B) Phylogenetic tree was constructed on MEGA using neighbor-joining method. Bootstrap values were obtained for 1000 replications. NJ tree included SmMYC2s and 37 bHLH proteins from other plant species (Supplementary Table S1). The bars represent evolutionary distance. (C) Organ expression pattern of SmMYC2a and SmMYC2b. Transcript abundance of each MYC2 is normalized to actin as control using 2−△△Ct method.
SmMYC2a and SmMYC2b were localized in the nucleus
To investigate the subcellular localization of SmMYC2a and SmMYC2b, the constructed pCAMBIA1301-GFP-SmMYC2a/b vector was transformed into prepared rice protoplasts. As shown in Fig. 2, the free green fluorescent protein (GFP) was detected in the nucleoplasm and cytoplasm. By contrast, the two TFs presented exclusively nuclear fluorescent signals in the observed protoplasts, indicating the localization in the nucleus. The nucleus localization characteristic confirmed the expectation of SmMYC2a and SmMYC2b as TFs.
SmMYC2a and SmMYC2b were localized in nucleus.
(A) A rice protoplast expressing MYC2a-GFP showed green fluorescent signals in the nucleus. (B) The same protoplast cell of (A) showing the chlorophyll autofluorescence signal in the plastids. (C) The merged signal of (A,B). (D) The same protoplast of (C) under white light. Bars = 10 μm. (E) A rice protoplast which expressed MYC2b-GFP showed the green fluorescent signal in the nucleus. (F) The same rice protoplast of (E) expressing red fluorescent signal in the plastids. (G) The merged signal of (E,F). (H) The same protoplast of (G) under white light. Bars = 10 μm. (I) Rice protoplast that expressed free GFP showed green fluorescent signals as control. (J) The same rice protoplast cell of (I) showing red fluorescent signals. (K) The merged signal of (I,K). (L) The same protoplast of (K) under white light. Bars = 10 μm.
SmMYC2a and SmMYC2b formed complexes with SmJAZ1 and SmJAZ2 repressors in yeast two-hybrid assay
Yeast two-hybrid (Y2H) assay using JAZ1 and JAZ2 as baits was implemented to investigate the interactive ability of SmMYC2 with SmJAZ repressors. The control strain, containing pAD-GAL4 (AD) and pBD-GAL4 (BD), could grow on double selection medium (SD/–Leu/–Trp) but not capable of growing on quadruple selection medium (SD/–Ade/–His/–Leu/–Trp) (Fig. 3), indicating the feasibility of the system. Strains where pGADT7-MYC2a/b and pGBKT7 vectors were transformed showed same growing states with above control strains, suggesting that SmMYC2a and SmMYC2b were not capable of auto-activating the reporting gene (Supplementary Fig. S1). However, transformed yeast cells which harbored pAD-GAL4-MYC2a/b (AD-MYC2a/b) plus pBD-GAL4-SmJAZ1/2 (BD-JAZ1/2) grew well on both double and quadruple selection medium, suggesting in vitro interactions between MYC2 and JAZs. Subsequently, α-galactosidase (α-gal) assay was applied to reconfirm their interactions. The appearance of blue colonies was observed, which was due to the activation of MEL1 reporter, indicating the binding of MYC2-JAZ. The Y2H result demonstrated that SmMYC2 regulators could interact with SmJAZ repressors.
The interactions of SmMYC2s and SmJAZs in vitro.
(A) AH109 yeast cells were grown on SD medium lacking leucine and tryptophan. (B) SD medium lacking adenine, histidine, leucine and tryptophan. (C) The 10-fold dilutions of yeast cells of (B). (D) 100-fold dilutions of yeast cells of (B). (E) Assayed for MEL1 activity.
SmMYC2a and SmMYC2b were inducible by MeJA in S. miltiorrhiza hairy roots
Both SmMYC2a and SmMYC2b were induced with exogenous MeJA, reaching a maximal level at 0.5 h after treatment. The transcript levels of SmMYC2a and SmMYC2b increased rapidly within 30 min (Fig. 4). Afterwards, the variation trends for transcript levels of SmMYC2a and SmMYC2b slightly differed. Transcript level of SmMYC2a declined slowly till 4 h and a minor increase occurred at 12 h post-treatment with MeJA. However, transcript level of SmMYC2b decreased sharply till 1 h and the second peak appeared at 2 h after MeJA treatment. The variation profiling of SmHCT6 and SmCYP98A14, two pathway genes of phenolic acid biosynthesis, was characterized in Fig. 4. The expression level of SmHCT6 and SmCYP98A14 increased 492.4 and 40.2 folds respectively at 0.5 h. Generally speaking, the variation trends were in parallel with changes in SmMYC2a and SmMYC2b transcript levels.
MeJA-induced transcript level changes of SmMYC2a and SmMYC2b.
Each data point was the average of three biological replicates. Bars indicated SD. Asterisks indicated statistically significant differences when compared with 0 h. “*” indicated p < 0.05 and “**” indicated p < 0.01. (A,B) showed qRT-PCR analysis of SmMYC2a/b expression at various times after MeJA treatment in S. miltiorrhiza hairy roots. (C,D) respectively showed qRT-PCR analysis of SmCYP98A14 and SmHCT6 expression at various times following MeJA treatment.
Expression of critical pathway genes was affected in MYC2-RNAi hairy roots
To further explore whether the biosynthesis of tanshinones and Sal B was regulated by SmMYC2a and SmMYC2b, RNAi-mediated expression knockdown experiments were carried out in S. miltiorrhiza. The RNAi silencing vectors p1300-pHANNIBAL-MYC2a/b recombinants (Supplementary Fig. S2A) were transformed into Agrobacterium tumefaciens C58C1 strain and subsequently introduced into S. miltiorrhiza leaf explants. Growing status of fresh hairy roots from leaf explant which carried pCAMBIA1300-pHANNBIAL vector was displayed in Supplementary Fig. S2B–D. PCR testing positive transgenic genes were harvested (Supplementary Fig. S2E,F). The expression of SmMYC2a and SmMYC2b decreased to 24.9% and 61.5% respectively of control in corresponding MYC2-RNAi hairy roots on average. In transgenic SmMYC2a-RNAi hairy roots, the expression of pathway genes was reduced, such as 3-hydroxy-3-methylglutaryl-CoA reductase (HMGR), mevalonate kinase (MK), 4-(cytidine 5’-diphospho)-2-C-methyl- D-erythritol kinase (CMK), 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MCS), geranyl diphosphate synthase (GPPS), geranylgeranyl diphosphate synthase (GGPPS), copalyl diphosphate synthase (CPS) and kaurene synthase-like protein (KSL) which were involved in the biosynthesis of tanshinones (Figs 5 and 6). Expression level of phenolic acid biosynthetic genes as phenylalanine ammonia-lyase (PAL), HCT6, 4-hydroxyphenylpyruvate reductase (HPPR) and CYP98A14 also decreased in SmMYC2a knockdown lines. In contrast to SmMYC2a knockdown effect, SmMYC2b not only affected several common genes (SmPAL, SmHPPR, SmHCT6, SmCYP98A14, SmMK, SmCMK, SmMCS, SmCPS and SmKSL), but also reduced the expression of SmDXR. The expression of SmMYC2a/b and pathway genes in control hairy roots and transgenic SmMYC2a/b suppressed hairy roots were characterized in Supplementary Fig. S3 in a form of histogram.
The production of phenolic acids and the expression of biosynthetic genes were repressed in MYC2-RNAi hairy roots compared with control.
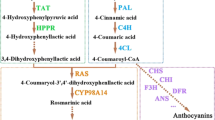
MYC2a, SmMYC2a suppressed hairy roots. MYC2b, SmMYC2b suppressed hairy roots. PAL, phenylalanine ammonialyase; C4H, cinnamic acid -4-hydroxylase; 4CL, hydroxycinnamate, coenzyme A ligase; TAT, tyrosine aminotransferase; HPPR, 4-hydroxyphenylpyruvate reductase; HPPD, 4-hydroxy phenyl pyruvate dioxygenase; HCT, hydroxy-cinnamoyl transferase; CYP98A14, cytochrome P450 monooxygenase 98A14. Red indicated enhanced expression or production; blue indicated repressed expression or production; white indicated the expression of control; darker color indicated more significant change. The relative expression values were converted into color array by Microsoft Excel software (version 2010). All the values were ratios of average gene expression level or compounds content in MYC2-RNAi hairy roots to control.
The production of tanshinones and the expression of biosynthetic genes were repressed in transgenic SmMYC2a/b suppressed hairy roots compared with control.
MYC2a, SmMYC2a suppressed hairy roots. MYC2b, SmMYC2b suppressed hairy roots. AACT, acetyl-CoA C-acetyltransferase; CDP-ME, 4-(cytidine 5’-diphospho)-2-C-methyl- D-erythritol; CDP-ME2P, 2-phospho-4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol; CMK, 4-(cytidine 5’-diphospho)-2-C-methyl- D-erythritol kinase; CPS, copalyl diphosphate synthase; DMAPP, dimethylallyl diphosphate; DXP, 1-deoxy-D-xylulose 5-phosphate; DXR, 1-deoxy-D-xylulose 5-phosphate reductoisomerase; DXS, 1-deoxy-D-xylulose 5-phosphate synthase; FPP, farnesyldiphosphate; FPPS, farnesyl diphosphate synthase; G3P, glyceraldehyde 3-phosphate; GA, gibberellin; GGPP, geranylgeranyl diphosphate; GGPPS, geranylgeranyldiphosphate synthase; GPP, geranyl diphosphate; GPPS, geranyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; HDS,4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HMBPP, 4-hydroxy-3-methylbut-2-enyl diphosphate; HMG-CoA, 3-Hydroxy-3-methylglutary l-CoA; HMGR, hydroxymethylglutaryl-CoA reductase; HMGS, hydroxymethylglutaryl-CoA synthase; IDI, isopentenyl diphosphate isomerase; IDS, isoprenyl diphosphate synthase; IPP, isopentenyl diphosphate; KS, kaurene synthase; KSL, kaurene synthase-like; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; MDC, mevalonatepyrophosphate decarboxylase; MDS, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; ME-cPP, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate; MeJA, methyljasmonate; MEP, 2-C-methyl-D-erythritol 4-phosphate; MK, mevalonate kinase; MVA, mevalonate; MVAP, mevalonate-5-phosphate; MVAPP, mevalonate-5-diphosphate; PMK, 5-phosphomevalonate kinase; CYP76AH1, cytochrome P450 monooxygenase 76AH1. Red indicated enhanced expression or production; blue indicated repressed expression or production; white indicated the expression of control; darker color indicated more significant change. All the values were ratios of average gene expression level or compounds content in MYC2-RNAi hairy roots to control.
Accumulation of tanshinones and salvianolic acids was affected by RNAi silencing of SmMYC2a or SmMYC2b
To assess the effects of reduced SmMYC2a and SmMYC2b expression on tanshinones and phenolic acids biosynthetic gene expression, 10 compounds involved in tanshinones and Sal B synthetic pathways were determined and the corresponding chromatogram map was shown in Supplementary Fig. S4. Three SmMYC2a and four SmMYC2b knockdown lines with significant reduction of MYC2a or MYC2b were examined for their effects on production of tanshinone and phenolic acids. The ratios of the gene expression level or compounds content in MYC2-RNAi hairy roots to control were displayed in Figs 5 and 6. A significant decrease in the biosynthesis of phenolic acids (Sal A and Sal B) and intermediate products (L-phenylalanine, homogentisic acid, rosmarinic acid) was observed in SmMYC2a or SmMYC2b knockdown lines, showing a consistent repression effect. Moreover, both SmMYC2a and SmMYC2b knockdown lines presented an extremely low yield of tanshinone I, dihydrotanshinone I, tanshinone IIA, cryptotanshinone. The content of tanshinone IIA reduced to 6.4% and Sal B dropped to 17.2% compared with control lines on average in SmMYC2a knockdown hairy roots. The silencing of SmMYC2b caused similar changes in tanshinones accumulation with the production of tanshinone IIA reduced to 45.2% and Sal B dropped to 5.0% compared with control lines on average. Determination of compounds in transgenic SmMYC2a/b knockdown hairy roots was characterized in Supplementary Fig. S5 in a form of histogram. These results indicated that SmMYC2a and SmMYC2b were positive regulators that affected the biosynthesis of tanshinones and Sal B.
SmMYC2a and SmMYC2b could bind with the E-Box within the SmCYP98A14 or SmHCT6 promoter in vitro
Previous studies indicated that G-box (5′-CACGTG-3′) fragment was the preferred core binding site of MYC2, followed by 5′-CACATG-3′ and 5′-CACGTT-3′ motifs19. Thus, the screening of candidate MYC2 activated genes took the preferential order into consideration. SmCYP98A14, namely SmCYP98A78 was reported as a JA-responsive gene20. Pathway genes as SmCPS1 and SmCYP76AH1 were previously investigated and proven to be JA inducible21,22. SmHCT6 was also illustrated as a JA-inducible gene. The above four genes were discovered to contain E-box motifs in their promoters. Thus, they were selected as candidates of MYC2 activated genes.
Purified fusion proteins consisting of SmMYC2a/b protein and cleavable His-tags were prepared (Supplementary Fig. S6). Promoters of pathway genes (SmCYP98A14, SmHCT6, SmCPS1 and SmCYP76AH1) which contained E-box element were potential binding sites of MYC2. The promoters of the candidate genes were amplified by genomic walking and biotin-modified for EMS assay. EMS assays were performed using purified target proteins and biotin-modified promoter fragments containing the E-box sequences to test binding behavior. As presented in Fig. 7, C-terminally His-tagged SmMYC2a protein (named as MYC2a-His protein) bound to the HCT6 oligonucleotide probe with intact E-box motif (lane 2), but was not capable of binding to promoter fragments in which the whole E-box was replaced by T residues (lane 1). Both MYC2a-His and MYC2b-His (C-terminally His-tagged SmMYC2b) proteins bound to G-box motif-containing SmCYP98A14 promoter.
Discussion
MeJA is widely used as an elicitor to investigate the biosynthetic pathway of active compounds and underlying regulatory mechanisms14. Previous research indicated that MeJA treatment promoted the accumulation of secondary metabolites, tanshinones and phenolic acids15,16,17,23. MYC2 is a master regulator of JA signaling pathway which plays a vital role in the activation of pathway genes in various model plants19,24,25. Whether MYC2 regulates the biosynthesis of tanshinones and phenolic acids still remains unknown yet. How MYC2 fine-tunes the pathway hasn’t been explored before in S. miltiorrhiza. In this study, two bHLH TFs, SmMYC2a and SmMYC2b were obtained from S. miltiorrhiza. Phylogenetic analysis result revealed that SmMYC2a and SmMYC2b showed high homologs to MYC2 proteins of many other species. Previous studies indicated that MYC2 was a master regulator with extensive regulating effect in JA signaling pathway. Among the well studied MYC2s, AtMYC2 was reported as a regulator of glucosinolate (GS) biosynthesis, insect performance and feeding behavior26. NtMYC2a and NtMYC2b regulated multiple jasmonate-inducible steps in nicotine biosynthesis27.
Previous investigations demonstrated that JAZ repressors targeted bHLH TFs to control JA-dependent anthocyanin accumulation and trichome initiation28. As the SmMYC2a and SmMYC2b contained conserved JID, we reasoned that a similar complex might be formed to control the accumulation of tanshinones and phenolic acids. Y2H assay was implemented to investigate the interactive ability of SmMYC2 with SmJAZ repressors. The binding of the two proteins could enable the growth of host AH109 yeast strain on quadruple selection medium and activate the expression of MEL1 reporter gene, making α-gal turn blue. Both SmMYC2s exhibited interactions with SmJAZ1 and SmJAZ2, implying that the two MYC2 TFs might function as direct targets of JAZ proteins. The binding effect of MYC2-JAZ was in accordance with previous findings in Arabidopsis26, which might result in the repression of MYC2 proteins that could bind to the promoters of critical pathway genes and activate their transcription. Both SmMYC2a and SmMYC2b were discovered to be JA-inducible, reaching a maximal level at 0.5 h after MeJA treatment. The early response pattern was in accordance with that in N. benthamiana and the maximum response occurred at 0.5 h29. Afterwards, the variation trends of transcript levels of SmMYC2a and SmMYC2b slightly differed. Transcript level of SmMYC2a declined slowly till 3 h, which was similar to changes of NaMYC2. However, transcript level of SmMYC2b decreased sharply till 1 h after MeJA treatment. The different reduction velocity of SmMYC2a and SmMYC2b transcripts indicated that compared to SmMYC2b, SmMYC2a could maintain a high expression longer and might serve as a regulator with more extensive modulatory effects. Our results also demonstrated that SmHCT6 and SmCYP98A14 were remarkable JA-responsive genes and the response occurred relatively early. The JA-mediated regulation profile of positive pathway genes was in parallel with that in N. benthamiana29.
After silencing SmMYC2a or SmMYC2b, qRT-PCR analysis was performed to seek for evidence of the relationships between the two TFs and critical pathway genes. Not only the phenolic acids biosynthetic genes were affected, but also the tanshinone pathway was repressed in SmMYC2a knockdown lines. Most of the pathway genes influenced by SmMYC2a had been previously studied. SmCMK helped to enhance the accumulation of tanshinones30. SmHMGR and SmDXR were critical pathway genes whose overexpression could significantly enhance the yield of tanshinone in hairy root lines, while co-expressing produced evidently higher levels of total tanshinone, which provide a useful strategy to improve tanshinone content31. SmGGPPS played a more important role in stimulating tanshinone accumulation than the upstream enzyme SmHMGR32. The decreased expression of SmCPS caused a decrease in tanshinone levels which confirmed SmCPS as a key enzyme for tanshinone biosynthesis in S. miltiorrhiza33. Rosmarinic acid synthase (RAS), namely HCT also played a vital role in the biosynthesis of Sal B. In SmRAS antisense transgenic S. miltiorrhiza hairy root lines, the accumulation of RA and Sal B obviously decreased to 59% and 42% of control on average and in CYP98A14-antisense transgenic lines, the content of RA and Sal B decreased to 44% and 81% of control on average34. In MYC2-RNAi hairy roots, several genes were found to be dramatically reduced, including SmPAL, SmHPPR, SmHCT6, SmCYP98A14, SmMK, SmCMK, SmMCS, SmCPS and SmKSL. Additionally, the expression of SmDXR decreased only in SmMYC2b knockdown hairy roots, while SmHMGR, SmGPPS and SmGGPPS were found to be repressed in SmMYC2a knockdown hairy roots, indicating SmMYC2a and SmMYC2b were positive regulators that strongly regulated pathway genes in the biosynthesis of tanshinones and phenolic acids. To assess the effects of reducing SmMYC2a/b expression on tanshinone and phenolic acid biosynthesis, all the PCR-tested positive hairy roots were subjected to compounds analysis. A significant decrease was observed in the biosynthesis of intermediate product (L-phenylalanine, homogentisic acid, rosmarinic acid) and end products (Sal A, Sal B) in SmMYC2a or SmMYC2b knockdown lines, showing a consistent repression effect of phenolic acid biosynthesis pathway. The biosynthesis of tanshinones (tanshinone I, dihydrotanshinone I, tanshinone IIA, cryptotanshinone) was remarkably depressed in SmMYC2a knockdown transgenic lines and knockdown of SmMYC2b caused similar changes in the production of tanshinones. The positive regulating effect of MYC2 was in parallel with previous studies on Arabidopsis and Artemisia annua19,25,35.
To further explore the genes that MYC2 could transcriptionally activate, EMS assays were performed using purified target proteins and biotin-modified promoter fragments containing the E-box. Purified SmMYC2a fusion protein was discovered to bind to the E-box containing SmHCT6 and SmCYP98A14 oligonucleotide probes. SmMYC2b fusion protein could bind to E-box motif in SmCYP98A14 promoter. The binding effect revealed that SmMYC2a might regulate the biosynthesis pathway of phenolic acids by activating the transcription of SmHCT6 and SmCYP98A14 while the regulation of SmMYC2b might be via the binding to SmCYP98A14.
Our current understanding of biosynthetic regulation of tanshonone and phenolic acid pathway by MYC2 was illustrated in Fig. 8. In the presence of bioactive jasmonate stress, JA was conjugated to isoleucine to form JA-Ile and subsequently bound SCFCOI1 complex and JAZ proteins, leading to the degradation of JAZ proteins and the release of MYC2 TFs. SmMYC2a might regulate the biosynthesis pathway of phenolic acids by activating the transcription of SmHCT6 and SmCYP98A14 while the regulation of SmMYC2b might be via the binding to SmCYP98A14. JA-MYC2-genes-active compounds network was roughly constructed. However, it is still far away from fully understanding for regulating role of MYC2 in secondary metabolites. More attention should be paid on the characterization of MYC2 TFs in JA pathway to provide additional evidence for the extensive regulating effect and help to better understand the complex JA regulatory network.
In conclusion, our results indicate that SmMYC2a and SmMYC2b are positive regulators which exert similar but also irreplaceable functions in the regulation of JA-responsive genes, leading to the biosynthetic regulation of tanshinones and phenolic acids. The functional characterization of SmMYC2a and SmMYC2b which regulate multiple jasmonate-inducible steps in the biosynthesis of active compounds in S. miltiorrhiza will definitely facilitate better understanding of regulating mechanism of JAs on active compounds. As SmMYC2a and SmMYC2b are positive regulators which affect a wide array of pathway genes, high yield of target compounds in S. miltiorrhiza may be achieved by metabolic engineering strategy using SmMYC2a and SmMYC2b as candidates.
Materials and Methods
Plant growth and hairy root cultures
S. miltiorrhiza plants were grown in the botanical garden of the Second Military Medical University and identified by Professor Hanming Zhang. Fresh roots, stems and leaves were separated from 2-month-old S. miltiorrhiza seedling, washed with distilled water, dried with filter paper and stored immediately at −70 °C for organ expression analysis. To obtain sterilized seedlings, seeds of S. miltiorrhiza were thoroughly sterilized utilizing 75% ethanol for 5 min and subsequent 1% HgCl2 solution for 10 min. After rinsed thoroughly with sterile distilled water for 5 times, the seeds were sown on prepared Murashige and Skoog (MS) medium (Sigma-Aldrich, St Louis, MO, USA) plus 3% sucrose and 0.5% agar with the pH value adjusted to 5.6.
Hairy roots of S. miltiorrhiza were derived from plantlets infected with Agrobacterium rhizogenes bacterium of C58C1 strain and maintained subculturing after sterilization. MeJA treatment assay was performed on S. miltiorrhiza hairy roots which were shaking cultured and reached exponential growth phase. Dissolved in ethanol, MeJA was added to a final concentration of 50 μM into S. miltiorrhiza hairy roots medium. Hairy roots from MeJA-treated group and solvent control group were harvested at selected times (0, 0.5, 1, 2, 4, 6, 12 and 24 h) and stored at −70 °C for gene expression analysis27.
Subcellular localization experiment was performed on rice protoplast which was derived from rice seedlings. Rice seeds were sterilized in 75% ethanol for 1 min and then disposed with 33% sodium hypochlorite solution for 5 min. After thoroughly sterilized using 0.1% HgCl2, the seeds were rinsed thoroughly with sterile distilled water for 5 times and placed on MS medium plus 0.5% sucrose and 0.5% agar with the pH value adjusted to 5.8. After culturing under light at 25 °C for 3 days, the height of shoots reached 1 cm. Then, the rice seedlings were placed in a dark environment and used for protoplast separation after 14-day-culture in dark.
Cloning and phylogenic analysis of SmMYC2a and SmMYC2b
Based on transcription profiling database previously established18, we amplified SmMYC2a with primers 5′-ATGATTGATTACCGCACGCC-3′ and 5′-CTATCTAATCTCAGCAACTTTAG-3′ and SmMYC2b with 5′-ATGGGGGTTGTTGGTTGG-3′ and 5′-TTACCCGAGAGATAACTGATG-3′ using total RNA from S. miltiorrhiza seedling as amplification template. The two novel SmMYC2 genes were used to create multiple sequence alignments with AtMYC2 (GenBank accession: NM_102998.3) on ClustalX 2 at amino acid level36. To establish the evolutionary relationships between SmMYC2s and their best homologs, amino acid sequences of SmMYC2a and SmMYC2b were used as query sequences to search for homologs in NCBI (http://www.ncbi.nlm.nih.gov/) using BLAST. The two SmMYC2s, together with 37 closest MYC2s from other species (Supplementary Table S1), were used to construct a Neighbour-joining (NJ) tree using MEGA 5.05 (http://www.megasoftware.net) with 1000 bootstrap replicates using full-length amino acid sequences.
Vector construction and plant transformation
DNA fragment of GUS in pCAMBIA1301 vector was changed into GFP to get the modified pCAMBIA1301-GFP vector which could serve as a subcellular localization vector. cDNA of SmMYC2 without termination codon was inverted into pCAMBIA1301-GFP vector to get pCAMBIA1301-GFP-SmMYC2 which expressed corresponding recombinant protein.To construct RNAi silencing vectors of SmMYC2, a 400-bp DNA fragment circumventing conserved bHLH domain was amplified. The modified pCAMBIA1300-pHANNBIAL vector which contained a 35S promoter, a PDK intron and an OCS terminal was used as the RNAi vector. The amplified 400-bp SmMYC2 fragment was forwardly and reversely inverted to the two ends of the PDK intron to get pCAMBIA1300-pHANNBIAL-MYC2 vector. The constructed vectors were separately introduced into the disarmed Agrobacterium tumefaciens C58C1 strain which carried the rooting plasmid pRiA437. The pCAMBIA1300-pHANNBIAL vector was also transformed as control using the same method. Modified PHB-FLAG vector was constructed by introducing the FLAG tag and Spe I restriction site into the MCS region with primers Bam HI and Sac I. The complete coding region of SmMYC2a/b was amplified and inserted into the modified binary vector PHB-Flag. The final construct was transferred into Agrobacterium tumefaciens (C58C1 strain) and introduced into leaf explant from S. miltiorrhiza. The plants which carried PHB-Flag vector were used as control. The prokaryotic expression vectors which expressed SmMYC2a-His and SmMYC2b-His proteins were constructed by introducing the whole length-amplified SmMYC2a or SmMYC2b into pET32a vector using Bam HI and Hind III, Eco RV and Sal I restriction sites. Yeast two-hybrid assay was applied to investigate the interaction of SmJAZ1/2 and SmMYC2a/b in vitro. The whole coding sequences of SmJAZ1/2 and SmMYC2a/b were introduced into pGADT7 vectors between Nde I and Eco RI, Nde I and Xma I restriction sites while SmJAZ1/2 were inserted into pGBKT7 vectors between Nde I and Eco RI, Nde I and Xma I restriction sites, respectively. All the primers used in the construction of vectors were shown in Supplementary Table S2.
Subcellular localization of SmMYC2a and SmMYC2b
Transient expression assay was performed to explore subcellular localization. The pCAMBIA1301-GFP-MYC2 vector which expressed GFP-tagged fusion protein was transformed into freshly prepared rice protoplast using PEG transfection method38. Subcellular localizations of GFP, MYC2a-GFP and MYC2b-GFP proteins were determined by microscopy assay with a fluorescence microscopy. GFP fluorescent signals were visualized with an excitation wave length of 488 nm and an emission wavelength of 505–530 nm. The red autofluorescence resulting from chlorophylls was captured by an emission wavelength longer than 650 nm. The processing of captured images was conducted using Photoshop software.
Yeast two-hybrid assay
Y2H assay was conducted to detect the interaction of JAZ with bHLH TF in vitro. Recombinant vector of pGBKT7-JAZ1/2 that contained GAL4 DNA-binding domain served as a bait protein and pGADT7-MYC2a/b vector that contained GAL4 activation domain acted as a prey protein. The two types of recombinant vectors were co-transformed into yeast strain AH109 using the PEG/LiAC method. Empty vectors of pGADT7 and pGBKT7 were also co-transformed as a negative control. After selection on synthetic SD dropout medium lacking leucine and tryptophan (SD-Leu-Trp), single transformant colonies were then screened for growth on SD selection medium lacking adenine, histidine, leucine and tryptophan (SD-Ade-His-Leu-Trp). α-gal was dissolved in DMF to a concentration of 4 mg/ml and then smeared uniformly on quadruple SD medium. Transformant colonies that turned blue indicated the interactions of SmMYC2a/b and SmJAZ1/2. To test potential auto-activation of the preys, vectors of pGADT7-MYC2a/b and pGBKT7 were also co-transformed as controls.
Gene expression analysis
Total RNA was isolated using a total RNA Isolation Kit (TIANGEN BIOTECH (BEIJING) CO., LTD, Beijing, China) and DNA contamination was eliminated by DNase treatment. After measuring quality and concentration of RNA obtained, 1μg of total RNA was reverse-transcribed to obtain cDNA using TransScript First-Strand cDNA Synthesis SuperMix Kit (TransGen Biotech, Beijing, China) according to manufacturer’s instructions. Quantitative RT-PCR (qRT-PCR) was performed following the instructions of SYBR®PreMix Ex Taq (Takara Bio Inc., Dalian, China) on Thermal Cycler Dice Real Time System TP800 (Takara Bio Inc., Dalian, China), with primers listed in Supplementary Table S3. Efficiency-corrected comparative Ct method was applied and relative expression was calculated by normalizing interested genes according to the abundance of the housekeeping gene (18S ribosomal subunit, namely SmActin). All qRT-PCR experiments were performed with three independent replicates. Expression levels of SmMYC2a, SmMYC2b and main pathway genes were determined by qRT-PCR.
Compounds extraction and analysis
The hygromycin selected hairy roots were cultured in liquid medium for 45 days. Afterwards, positive transgenic strains were confirmed by PCR testing with primers listed in Supplementary Table S4. All the independent control clones and typical transgenic hairy roots were subjected to gene and compound analysis. The hairy roots were dried at room temperature for 2 days to a constant dry weight and milled to a homogeneous size of which No. 100 mesh could be sieved through. Active compounds (tanshinones and Sal B) were extracted according to the reported method16,39. 0.02 g powder sample was extracted in 70% methanol under sonication for 30 min and then centrifuged at 10,000 rpm for 5 min. The extraction volume was 5 mL and the weight loss was made up with 70% methanol after extraction. The extracted solution was filtered through a 0.22 μm organic membrane before determination.
A method of performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) has been developed for the determination analysis of 10 compounds in S. miltiorrhiza hairy root cultures. The separation of lipophilic tanshinones and hydrophilic phenolic acids was performed by a common Waters xSELECT CSHTM C18 column (2.1 mM × 50 mM, 2.5 μM, Agilent Corporation, MA USA) in two ion monitoring modes. When analyzing 6 hydrophilic phenolic acid compounds, we selected the gradient elution approach by using a mobile phase acetonitrile (A) - H2O (B) (containing 2 mmol/L ammonium acetate and 0.1% formic acid) 0 min (7% A) →1 min (7% A) →2.5 min (93% A) →7 min (93% A) and the flow rate was 0.3 mL/min in the progress. For lipophilic tanshinone analysis, an isocratic mobile phase consisting of acetonitrile (A) - H2O (B) (containing 2 mmol/L ammonium acetate and 0.1% formic acid) (60:40) was applied with a flow rate of 0.3 mL/min. The column temperature was 35 °C and the sample injection volume was 10 μL. The components were detected by an Agilent G6410A triple quadrupole LC/MS system equipped with a Mass hunter interface. The 10 hydrophilic phenolic acid compounds were detected using ESI in negative-ion mode and the lipophilic tanshinones in positive-ion mode, quantified by multiple-reaction monitoring (MRM) mode using following transition mass of m/z, 164→147 for L-phenylalanine, 167→123 for homogentisic acid, 163→119 for 4-coumaric acid, 359.1→161.1 for rosmarinic acid (RA), 493→294.9 for Sal A, 717.1→519.1 for Sal B, 297.2→254.0 for cryptotanshinone, 277.1→249.1 for tanshinone I, 295.1→276.8 for tanshinone IIA, 279.1→233.1 for Dihydrotanshinone I. All the standards were purchased from Sigma-Aldrich (St. Louis, MO). Data analysis was carried out using the Agilent Mass Hunter Workstation software.
Electronic mobility shift assay
The PCR-amplified cDNA fragment of SmMYC2 was inserted into pET32a vector and the constructed plasmid pET32a-MYC2 was introduced into E. coli strain BL21 (DE3). Transformants were cultured at 37 °C until OD 600 reached about 0.5. Afterwards, to induce the expression of target protein, isopropyl thiob-D-galactoside (IPTG) was added to a final concentration of 1 mM. Further incubation was conducted at 18 °C for 2 days. After ultrasonic cell disruption, target proteins were purified based on immobilized metal affinity chromatography technique according to manufacturer’s instruction (Bio-Rad Laboratories, Hercules, CA). E-box elements, especially G-box were universally known as well-characterized binding sites of bHLH TFs which mediated JA responsiveness in Arabidopsis19,40. As genomic regions flanking E-Box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape41, DNA fragments of 30 bp length which contained the preferred binding site and flanking genomic regions were designed to detect protein-nucleic acid interactions in EMSA.
Wild-type HCT6: 5′-AAGCCTAAAGCTCACATGGCTGCTAAACCT-3′. m-HCT6 (G-box mutation in HCT6 promoter): 5′-AAGCCTAAAGCTTTTTTTGCTGCTAAACCT-3′. Wild-type CYP98A14: 5′-ATTAGTGATTAACACGTGCACATTTATTAA-3′. m-CYP98A14 (G-box mutation in CYP98A14 promoter): 5′-ATTAGTGATTAATTTTTTCACATTTATTAA-3′.
To detect protein-nucleic acid interactions in vitro, EMS assay was performed. 2 μg of purified recombinant protein was incubated with 2 μg of 3′ biotin-labeled probes at room temperature for 10 min. After binding reaction, free nucleic acid was separated from the formed complexes by polyacrylamide gel electrophoresis. Subsequently, the gel was transferred to nylon membrane, crosslink transferred and chemiluminescence detected according to manufacturer’s protocol (TransGen Biotech, Beijing, China).
Statistical analysis
Statistical analysis was conducted using SPSS 16.0 software (e.g. analysis of variance (ANOVA) for between-treatment assessment and Student’s t test for pair comparisons to determine significant differences between means).
Additional Information
How to cite this article: Zhou, Y. et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 6, 22852; doi: 10.1038/srep22852 (2016).
Change history
24 April 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Zhou, L., Zuo, Z. & Chow, M. S. S. Danshen: an overview of its chemistry, pharmacology, pharmacokinetics and clinical use. J. Clin. Pharmacol. 45, 1345–1359 (2005).
Zeng, M. et al. Systematic review of recent advances in pharmacokinetics of four classical Chinese medicines used for the treatment of cerebrovascular disease. Fitoterapia 88, 50–75 (2013).
Su, C. Y., Ming, Q. L., Khalid, R., Han, T. & Qing, L. P. Salvia miltiorrhiza: Traditional medicinal uses, chemistry and pharmacology. Chin. J.Nat. Med. 13, 163–182 (2015).
Chen, H., Chena, F., Chiu, F. C. & Lo, C. M. The effect of yeast elicitor on the growth and secondary metabolism of hairy root cultures of Salvia miltiorrhiza. Enzyme Microb. Tech. 28, 100–105 (2001).
Gao, C. & Zou, S. Progress on pharmacological researches of Salvia miltiorrhiza and its chief components in coronary heart disease and pancreatitis. Chin. J. Integr. Med. 30, 1222 (2010).
Browse, J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant. Biol. 60, 183–205 (2009).
Wasternack, C. & Hause, B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann. Bot. 111, 1021–1058 (2013).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Thines, B. et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature 448, 661–665 (2007).
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 (2010).
Kazan, K. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends Plant Sci. 11, 109–112 (2006).
Kagale, S., Links, M. G. & Rozwadowski, K. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant physio. 152, 1109–1134 (2010).
Shyu, C. et al. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. Plant Cell 24, 536–550 (2012).
Gundlach, H., Müller, M. J., Kutchan, T. M. & Zenk, M. H. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Pro. Natl. Acad. Sci. 89, 2389–2393 (1992).
Xiao, Y. et al. Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol. Plant. 137, 1–9 (2009).
Gu, X. C. et al. Overexpression of allene oxide cyclase promoted tanshinone/phenolic acid production in Salvia miltiorrhiza. Plant Cell Rep. 31, 2247–2259 (2012).
Kai, G. et al. Metabolic engineering and biotechnological approaches for production of bioactive diterpene tanshinones in Salvia miltiorrhiza. Biotechnol. Adv. 10.1016/j.biotechadv.2014.10.001 (2014).
Luo, H. et al. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol. Plant. 152, 241–255 (2014).
Dombrecht, B. et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245 (2007).
Wang, B. et al. Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 241, 711–725 (2015).
Guo, J. et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. 110, 12108–12113 (2013).
Ma, Y. et al. Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 63, 2809–2823 (2012).
Hao, X. et al. Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Biochem. 62, 24–31 (2015).
Kazan, K. & Manners, J. M. MYC2: the master in action. Mol. plant 6, 686–703 (2012).
Hong, G. J., Xue, X. Y., Mao, Y. B., Wang, L. J. & Chen, X. Y. Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648 (2012).
Schweizer, F. et al. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3 and MYC4 regulate glucosinolate biosynthesis, insect performance and feeding behavior. Plant Cell 25, 3117–3132 (2013).
Zhang, H. B., Bokowiec, M. T., Rushton, P. J., Han, S. C. & Timko, M. P. Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol. Plant 5, 73–84 (2011).
Qi, T. et al. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814 (2011).
Zhai, Q. et al. Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 9, e1003422 (2013).
Wang, X., Cui, G., Huang, L., Gao, W. & Yuan, Y. A full length cDNA of 4-(cytidine 5’-diphospho)-2-C-methyl-D-erythritol kinase cloning and analysis of introduced gene expression in Salvia miltiorrhiza. Acta. Pharm. Sin. 43, 1251–1257 (2008).
Shi, M. et al. Increased accumulation of the cardio-cerebrovascular disease treatment drug tanshinone in Salvia miltiorrhiza hairy roots by the enzymes 3-hydroxy-3-methylglutaryl CoA reductase and 1-deoxy-d-xylulose 5-phosphate reductoisomerase. Funct. Integr. Genomics 14, 603–615 (2014).
Kai, G. et al. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 13, 319–327 (2011).
Cheng, Q. et al. RNA interference-mediated repression of SmCPS (copalyldiphosphate synthase) expression in hairy roots of Salvia miltiorrhiza causes a decrease of tanshinones and sheds light on the functional role of SmCPS. Biotechnol. Lett. 36, 363–369 (2014).
Di, P. et al. 13C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 8, 1537–1548 (2013).
Shen, Q. et al. The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 10.1111/nph.13874 (2016).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Mozo, T. & Hooykaas, P. J. Electroporation of megaplasmids into Agrobacterium. Plant Mol. Biol. 16, 917–918 (1991).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Zhu, Z. et al. Rapid separation and identification of phenolic and diterpenoid constituents from Radix Salvia miltiorrhiza by high-performance liquid chromatography diode-array detection, electrospray ionization time-of-flight mass spectrometry and electrospray ionization quadrupole ion trap mass spectrometry. Rapid Commun. Mass Spectrom. 21, 1855–1865 (2007).
Boter, M., Ruíz-Rivero, O., Abdeen, A. & Prat, S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591 (2004).
Gordân, R. et al. Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep. 3, 1093–1104 (2013).
Acknowledgements
This work was financially supported by the Natural Science Foundation of China (Grant nos 81325024, 81303160, 81403046 and 31300159), Shanghai Science and Technology Development Funds (14QB1402700) and program 15391900500 from Science and Technology Commission of Shanghai Municipality.
Author information
Authors and Affiliations
Contributions
W.C. and L.Z. conceived and organized the study; Y.Z., W.S., H.T. and L.C. performed the experiments; W.S., J.C., Y.X., Q.L., H.T., S.G. analyzed the data; Q.L., Q.J. and S.C. contributed reagents/materials/analysis tools; Y.Z. and W.S. wrote the manuscript; Y.Z. and H.T. prepared the figures. All authors discussed and commented on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhou, Y., Sun, W., Chen, J. et al. SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci Rep 6, 22852 (2016). https://doi.org/10.1038/srep22852
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep22852
This article is cited by
-
Integrative effects of phytohormones in the phenolic acids production in Salvia verticillata L. under multi-walled carbon nanotubes and methyl jasmonate elicitation
BMC Plant Biology (2024)
-
A high-efficient protoplast transient system for screening gene editing elements in Salvia miltiorrhiza
Plant Cell Reports (2024)
-
Contemporary understanding of transcription factor regulation of terpenoid biosynthesis in plants
Planta (2024)
-
Advances in studies on transcription factors in regulation of secondary metabolites in Chinese medicinal plants
Plant Cell, Tissue and Organ Culture (PCTOC) (2022)
-
A tracking work on how Sm4CL2 re-directed the biosynthesis of salvianolic acids and tanshinones in Salvia miltiorrhiza hairy roots
Plant Cell Reports (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.