Abstract
Atherosclerosis (AS) is chronic inflammation in response to lipid accumulation. MicroRNA-155 (miR-155) is being increasingly studied to evaluate its potential as diagnostic biomarkers and therapeutic targets in many diseases. However, delineating the role of miR-155 in AS remains difficult. Here, we detected constitutive expression of several microRNAs (miRNAs) possibly associated with cardiovascular disease in foam cells and clinical specimens from patients with AS. Among them, we found that the level of miR-155 in foam cells was the most significantly elevated in a dose- and time-dependent manner. In addition, the expression of miR-155 was elevated in the plasma and plaque of patients with AS. We also reported for the first time that miR-155 targets calcium-regulated heat stable protein 1 (CARHSP1), which regulates the stability of tumor necrosis factor alpha (TNF-α) mRNA. Furthermore, we investigated the mechanism by which the miR-155 level is elevated. miR-155 upregulation is due to transcriptional regulation by nuclear factor (NF)-κB, which is activated by the inflammatory factor TNF-α. In summary, increased miR-155 relieves chronic inflammation by a negative feedback loop and plays a protective role during atherosclerosis-associated foam cell formation by signaling through the miR-155–CARHSP1–TNF-α pathway.
Similar content being viewed by others
Introduction
AS is one of the most common cardiovascular diseases, leading to severe cardiovascular complications1. This disease is chronic inflammation in response to lipid accumulation and macrophages are the main effector cells that stimulate the vascular inflammatory reaction throughout the pathological process2,3,4. The role of cholesterol loading in macrophage inflammation raises the broader issue of mechanisms and consequences of macrophage-mediated inflammation in AS5. In the early stages of AS, monocytes respond to inflammatory factors and oxidative stress and subsequently differentiate into macrophages6,7. In this process, macrophages are recruited to take up oxLDL to form lipid-overloaded foam cells8, which play a crucial role in the formation of atherosclerotic plaque and the progression of AS.
miRNAs are endogenous, noncoding, single-stranded RNAs, ~22 nucleotides in length which silence gene expression by binding to the 3′-UTR of target mRNAs, thereby suppressing their translation or promoting their degradation9,10,11. miRNA sequences are highly conserved between different species12. Recent studies suggest that these molecules are closely involved in the pathogenesis and progression of cardiovascular disease and are good diagnostic or prognostic biomarkers and therapeutic targets13,14. In this research, we focused on miR-155, which is encoded by the MIR155 host gene (MIR155HG) and transcribed from the B-cell integration cluster (BIC), located on chromosome 2115. Various reports have indicated that miR-155 is a typical multi-functional miRNA that is closely linked to several important inflammation diseases, including AS16,17. This molecule has an anti-inflammatory effect and plays a part in the prevention of AS development and progression by targeting MAP3K10 in macrophages18. On the other hand, miR-155 may promote AS, as it has been observed to derepress BCL6-mediated inhibition of CCL2 transcription in the bone marrow cells of ApoE–/– mice19. These investigations have therefore demonstrated that miR-155 has opposite effects in different cell types, pathological stages and animal models of AS.
CARHSP1, also known as CRHSP-24, is a cytoplasmic protein located in processing bodies or exosome granules and has been identified as a cold shock domain (CSD) protein family member. These proteins are composed of an evolutionarily conserved CSD, which functions as a transcriptional or translational regulator20,21. CARHSP1 contains a CSD, making it possible for it to bind to polypyrimidine regions and regulate the stability of single-stranded RNA or DNA22,23. CARHSP1 was first identified as a physiological substrate for the Ca2+/calmodulin-regulated protein phosphatase calcineurin (PP2B). Recently, it was found to be an mRNA stability enhancer for tumor necrosis factor alpha (TNF-α), which is the central mediator of inflammation in macrophages and several studies have demonstrated that CARHSP1 binds to the AU-rich element (ARE) of the TNF-α 3′-UTR through the CSD24,25,26. The ARE binding site is critical for the regulation of mRNA translation, degradation and stability and therefore these studies indicate that CARHSP1 is critical for TNF-α mRNA stabilization and has a significant role in the regulation of inflammation.
In this study, we found that miR-155 is overexpressed in the serum and atherosclerotic lesions of AS patients and that expression of miR-155 in oxLDL-treated THP-1 inflammatory macrophages was increased in a dose- and time-dependent manner. We also demonstrated that miR-155 inhibits CARHSP1, which affects the stability of TNF-α mRNA, thus involving this cytokine in the process of AS. Furthermore, we confirmed that the transcription factor NF-κB, which is activated by TNF-α, binds to the promoter of miR-155 and stimulates the transcription of miR-155. In summary, we determined that an increased miR-155 level relieves chronic inflammation and plays a protective role by signaling through the miR-155–CARHSP1–TNF-α pathway.
Results
miR-155 is overexpressed in the plasma and atherosclerotic lesions of patients with AS and is induced by oxLDL in human macrophages
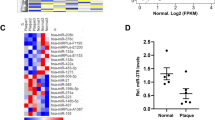
To identify novel miRNAs that are involved in atherogenesis, we established a foam cell model by inducing THP-1 cells with oxLDL and detected seven miRNAs possibly associated with cardiovascular disease by qRT-PCR. Among these miRNAs, miR-155 was the most significantly elevated in oxLDL-treated THP-1 cells (Fig. 1A). To confirm that the expression of miR-155 was increased in patients with AS, we measured the level of miR-155 in the plasma of patients with AS and in the normal control group and the results showed that the miR-155 level was indeed upregulated in patients with AS (Fig. 1B). The relative expression level of miR-155 in 17 pairs of atherosclerotic lesions and normal veins from the same patients were also detected by qRT-PCR and the results showed that the miR-155 level was significantly increased in the atherosclerotic lesions compared with the normal veins (Fig. 1C). To determine whether overexpression of miR-155 is induced by oxLDL, THP-1 cells were first treated with 100 nM PMA, then stimulated with oxLDL at the indicated concentrations (10, 50 and 100 μg/ml) for 24 h or with 50 μg/ml oxLDL for specific times (0, 6, 12, 24 and 48 h), while control cells were treated with PBS27. The results showed that the expression of miR-155 is increased by oxLDL stimulation in a dose- and time-dependent manner (Fig. 1D,E).
The expression of miR-155 is increased in human macrophages in clinical specimens and is induced by oxLDL.
(A) The expression of miRNAs in oxLDL (50 μg/ml)-treated macrophages was determined by qRT-PCR. Phosphate-buffered saline was used as control. (B) Levels of miR-155 in plasma from patients with AS (n = 70) and normal controls (n = 55). Values were normalized to U6. In this dot plot, the horizontal line indicates the mean. (C) qRT-PCR assay of miR-155 in atherosclerotic lesions (n = 17) and normal veins (n = 17) from the same patients with AS (P = 0.0095). (D) qRT-PCR analysis of miR-155 expression in THP-1 cells, which were first stimulated with propylene glycol monomethyl ether acetate (PMA, 100 nM) to induce them to differentiate into macrophages, then treated with oxLDL at the indicated doses (0, 10, 50, or 100 μg/ml). (E) qRT-PCR analysis of miR-155 expression in THP-1 cells, which were first stimulated with PMA (100 nM) to induce them to differentiate into macrophages, then treated with oxLDL (50 μg/ml) for the indicated times (0, 6, 12, 24, or 48 h). *P < 0.05, **P < 0.01, ***P < 0.001. Results are presented as mean ± SD of 3 independent experiments as determined by Bonferroni’s multiple comparison test after one-way ANOVA (D,E) or Student’s t test (A–C).
miR-155 attenuates lipid uptake and suppresses the inflammatory response by repressing the expression of TNF-α in foam cell formation
To investigate the role of miR-155 in the inflammatory response of AS, the macrophages that were stimulated by 100 nM PMA-treated THP-1 cells were transfected with 100 nM chemically synthesized miR-155 mimic or miR-155 inhibitor for 0, 6, 12, 24, or 48 hours, followed by treatment with 50 μg/ml oxLDL for 24 hours. The effect of miR-155 overexpression and knockdown on foam cell formation was analyzed by Oil Red O staining and inflammatory factor secretion was examined by ELISA. The transfection efficiency of miR-155 mimic or inhibitor was examined by qRT-PCR (Fig. 2A,C). When cells were transfected with miR-155 mimic for 0, 6, or 12 hours, the secretion of TNF-α was invariant and similar to the control group. After transfection with miR-155 mimic for 24 or 48 hours, the secretion of TNF-α was significantly suppressed (Fig. 2B). By contrast, when cells were transfected with miR-155 inhibitor for 24 or 48 hours, the secretion of TNF-α increased (Fig. 2D). Furthermore, to determine the effect of miR-155 on foam cell formation, Oil Red O staining showed that, after transfection with miR-155 mimic for 12, 24, or 48 hours, lipid uptake was remarkably attenuated (Fig. 2E). By contrast, transfection with miR-155 inhibitor led to the inverse outcome of enhanced lipid uptake (Fig. 2F).
The role of miR-155 in countering inflammation and lipid uptake by repressing the expression of TNF-α.
(A,C) The expression of miR-155 was examined by qRT-PCR for different times (0, 6, 12, 24, 48 h) following transfection of a miR-155 mimic, inhibitor and control (100 nM) into macrophages that had been previously differentiated from THP-1 cells. (B,D) After transfection of the miR-155 mimic, inhibitor and control (100 nM) into macrophages, these cells were then treated with oxLDL (50 μg/ml) for 24 h and the secreted protein concentration of TNF-α was determined by ELISA. (E,F) Lipid uptake was measured by Oil Red O staining after transfection of the miR-155 mimic, inhibitor and control (100 nM) into macrophages, then treated with oxLDL (50 μg/ml) for 24 h (magnification 400 × ). ***P < 0.001. Results are presented as mean ± SD of 3 independent experiments as determined by Student’s t test (A–D).
CARHSP1 is required for TNF-α mRNA stabilization
CARHSP1 was previously identified as a TNF-α 3′-UTR-interacting protein by luciferase assays and mRNA stability assays after CARHSP1 overexpression or CARHSP1 knockdown26,28. To verify that CARHSP1 is required for TNF-α mRNA stabilization in foam cells, we transfected siRNA control or CARHSP1 siRNA into THP-1 cells and the cells were then treated with oxLDL to form foam cells. qRT-PCR and western blotting were then used to measure the effect of CARHSP1 knockdown on TNF-α mRNA and protein. The transfection efficiency of CARHSP1 siRNA was examined by qRT-PCR (Fig. 3A) and western blotting (Fig. 3D) and the results indicate that CARHSP1 knockdown down-regulates TNF-α protein production and reduces the level of TNF-α mRNA (Fig. 3B–D). Additionally, we cloned the CARHSP1 ORF into the pCMV6-AC-GFP vector to overexpress CARHSP1 and transfected it into THP-1 cells and the transfection efficiency of the CARHSP1 overexpression plasmid was examined by qRT-PCR (Fig. 3E). The results showed that CARHSP1 overexpression increases the level of TNF-α mRNA and up-regulates TNF-α protein production (Fig. 3F–H). We also validated the role of CARHSP1 in TNF-α regulation using an mRNA stability assay. The TNF-αmRNA half-life was 94 min in cells with the siRNA control and 57 min in the CARHSP1 knockdown cells (Fig. 3I). These results suggest that CARHSP1 is required for TNF-α mRNA stabilization.
CARHSP1 is required for TNF-α mRNA stabilization.
(A,E) qRT-PCR analysis of CARHSP1 expression to examine the efficiency of overexpression and knockdown. Overexpression group was transfected with pCMV6-AC plasmid (Vector, 1 μg), pCMV6-AC-CARHSP1 plasmid (CARHSP1, 1 μg). Knockdown group was transfected with CARHSP1 siRNA (siCARHSP1, 50 pmol/ml), siRNA control (siCtrl, 50 pmol/ml) for 24 h. The control group was a mock transfection. (B,F) qRT-PCR analysis of TNF-α mRNA expression to determine the effect of CARHSP1 overexpression or knockdown on the TNF-α mRNA level. (C,G) The secreted protein concentration of TNF-α was determined by ELISA to determine the effect of CARHSP1 overexpression or knockdown on the TNF-α secreted protein level. (D,H) The intracellular protein concentration of TNF-α was determined by western blot to determine the effect of CARHSP1 overexpression or knockdown on TNF-α intracellular protein level, with β-actin as a control. (I) TNF-αmRNA half-life (t1/2) in cells treated with actinomycin D (ActD) post transfection with CARHSP1 siRNA or the control. **P < 0.01, ***P < 0.001. Results are presented as mean ± SD of 3 independent experiments as determined by Bonferroni’s multiple comparison test after one-way ANOVA (A–C,E–G) and RNA half-life (t1/2) was calculated by linear regression analysis (I). Full-length blots are presented in Supplementary Figures S1 and S2.
CARHSP1 is a functional target of miR-155
To elucidate the mechanism by which miR-155 affects macrophage-derived foam cell formation and the inflammatory response, we performed several computational analyses available in online miRNA target databases, including MirDB, Targetscan and PicTar, to identify potential direct mRNA targets of miR-155. Among many predicted target genes of miR-155 in miRBase, CARHSP1 (also known as CRHSP-24), which acts as an mRNA stability enhancer for TNF-α, is a newfound target for miR-155 and has a binding site for miR-155. The binding sites to the miR-155 seed sequence are highly conserved in human, mouse, rat, rabbit and horse (Figure S3). To validate the predicted miRNA binding site, we performed a luciferase reporter assay in HEK293T cells by transfecting luciferase reporters with either the wild-type CARHSP1 3′-UTR or a mutant lacking the miR-155 binding site (Fig. 4A). We found that transfection with the miR-155 mimic and a CARHSP1 3′-UTR reporter vector resulted in a significant reduction of luciferase activity (Fig. 4B). By contrast, transfection with the miR-155 inhibitor caused a significant increase in luciferase activity (Fig. 4C).
Validation of CARHSP1 as a functional target of miR-155.
(A) The predicted binding site of miR-155 in the 3′-UTR of CARHSP1 is indicated. A CARHSP1 3′-UTR mutant with a mutation in the miR-155 binding site is also shown. (B,C) After co-transfection with the pRL-TK plasmid carrying a wild type or mutant 3′-UTR sequence and a miR-155 mimic (10 pmol) or a miR-155 inhibitor (10 pmol) into HEK-293T cells for 48 h, the luciferase activity was measured. (D,E) The protein and mRNA level of CARHSP1 was measured by western blot and qRT-PCR after transfection with the miR-155 mimic (100 nM), the miR-155 inhibitor (100 nM), the mimic control, or the inhibitor control into THP-1 cells at 48 h. (F,G) Lipid uptake by foam cells was detected by Oil Red O staining after transfection of the pCMV6-AC-CARHSP1 plasmid (1 μg), CARHSP1 siRNA (siCARHSP1, 50 pmol/ml), plasmid control, or siRNA control (siCtrl) for 24 h in macrophages, which were treated with oxLDL for 24 h (magnification, 400 × ). (H,I) Western blot analysis and Oil Red O staining were employed to examine the protein level of CARHSP1 and TNF-α after transfection with miR-155 mimic control, miR-155 mimic, pCMV6-AC-GFP (defined as Vector), or pCMV6-AC-CARHSP1 or co-transfection with miR-155 mimic and pCMV6-AC-CARHSP1, with β-actin as western blot control. ***P < 0.001. Results are presented as mean ± SD of 3 independent experiments as determined by performing a Student’s t test (B,C,E). Full-length blots are presented in Supplementary Figures S4 and S5.
To identify the potential relationship between miR-155 and CARHSP1, qRT-PCR was employed to measure the mRNA levels of CARHSP1 in oxLDL-treated macrophages with or without miR-155 involvement. The cytoplasmic protein was collected from the cells, which were transfected with miR-control, miR-155 mimic, or miR-155 inhibitor for 24 h and subsequently stimulated with oxLDL for 24 h to determine whether miR-155 affects translation of CARHSP1. Western blot results revealed that overexpressed miR-155 also inhibits CARHSP1 protein expression (Fig. 4D). Furthermore, the qRT-PCR results indicate that the mRNA levels of CARHSP1 are downregulated by overexpressed miR-155 (Fig. 4E).
To further examine whether CARHSP1 is a functional target of miR-155 during foam cell formation, THP-1 cells were transfected with miR-155 mimic, CARHSP1 siRNA, or a CARHSP1 overexpression vector and it was found that overexpression of CARHSP1 increased the protein level of TNF-α and lipid uptake in oxLDL-induced foam cells (Figs 3H and 4F), while knockdown of CARHSP1 significantly decreased the protein level of TNF-α and strongly prevented lipid uptake, as shown by Oil Red O staining (Figs 3D and 4G). Moreover, overexpression of CARHSP1 rescued the protein level of CARHSP1 that had been decreased by overexpressed miR-155 (Fig. 4H) and reversed the protective effects caused by overexpression of miR-155 on lipid uptake in oxLDL-induced foam cells (Fig. 4I). All these results indicate that CARHSP1 is a functional target of miR-155, which directly targets its 3′UTR and that CARHSP1 serves as a mediator of miR-155 by regulating the production of TNF-α and thereby affecting foam cell formation.
Overexpression of miR-155 is induced by NF-κB, which is activated by TNF-α
Previous studies have demonstrated that TNF-α activates NF-kB29,30. To investigate how miR-155 is induced during foam cell formation, THP-1 cells were stimulated by TNF-α for 6 hours. We measured the mRNA and protein levels of NF-κB to confirm that endogenous NF-kB is induced by TNF-α at the indicated concentrations and times (Fig. 5A,B). Next, a ChIP assay was performed to determine whether NF-κB directly binds to the pri-miR-155 genomic locus and results showed that this was the case (Fig. 5C,D). Moreover, using the TRANSFAC database tool, we found that there were three potential binding sites (referred to as sites 1, 2 and 3) for NF-κB in the promoter region of pri-miR-155, which were located in the region (−2000 to −1 bp) upstream of pri-miR-155/BIC (Fig. 5E). Next, wild type or each of three different mutant reporter plasmids were transfected into TNF-α-treated THP-1 cells and the luciferase results showed that site 1 was the strongest binding site (Fig. 5F). All these results suggest that NF-κB, which is activated by TNF-α in THP-1 cells, is a transcriptional enhancer of miR-155.
NF-κB p65 is activated by TNF-α and subsequently induces transcription of miR-155.
(A,B) The mRNA and protein levels of NF-κB p65 were measured by qRT-PCR and western blot after treatment with TNF-α (20 ng/ml) for 6 h, with phosphate-buffered saline used as control. (C,D) A ChIP assay was performed with nuclear extracts of macrophages, which were differentiated from THP-1 cells treated with TNF-α (20 ng/ml) for 6 h using a specific p65 antibody. The ChIP assay input served as a positive control and normal IgG was used as a negative control. (E) Three potential binding sites (referred to as sites 1, 2 and 3) for NF-κB in the promoter region of pri-miR-155 were located in the region (–2000 to –1 bp) upstream of pri-miR-155/BIC. (F) Luciferase activity measurements in macrophages (differentiated from THP-1 cells) treated with TNF-α (20 ng/ml) for 6 h and transfected with the pRL-TK-pri-miR-155-luciferase wild type reporter or three pRL-TK-pri-miR-155-luciferase mutant reporters for 24 h. ***P < 0.001. Results are presented as mean ± SD of 3 independent experiments as determined by performing Bonferroni’s multiple comparison test after one-way ANOVA (D,F) or Student’s t test (A). Full-length blots/gels are presented in Supplementary Figures S6 and S7.
Discussion
Chronic inflammation driven by cholesterol accumulation in macrophages is instrumental in atherosclerotic lesion formation. It is also known that macrophage exposure to oxLDL leads to formation of lipid-laden foam cells31. In this study, we found that oxLDL-induced miR-155 plays a crucial role in the development of AS by regulating foam cell formation. We showed that miR-155 functions as an anti-inflammatory rather than pro-inflammatory factor in foam cells at the advanced stages of the atherosclerotic condition. The expression of miR-155 is critical for the production of TNF-α by directly targeting CARHSP1, which was previously shown to be required for the stabilization of TNF-α mRNA26. The absence of miR-155 in macrophages induced an inflammation reaction and foam cell formation by upregulating CARHSP1. Moreover, we found that it was NF-κB which was activated by TNF-α that bound to the promoter of miR-155 and positively regulated the expression of miR-155. Therefore, all the results suggest that increased miR-155 levels relieve chronic inflammation and foam cell formation by signaling through the miR-155–CARHSP1–TNF-α pathway.
Many researchers have demonstrated that miRNAs play a vital role in the pathogenesis of AS. Recently, miR-126-5p was discovered to maintain a proliferative reserve in endothelial cells (ECs) through repression of the Notch1 inhibitor delta-like 1 homolog (Dlk1), thereby preventing atherosclerotic lesion formation32. Similarly, another group reported that miR-223 was transported to ECs in high-density lipoprotein (HDL) and decreased expression of ICAM-1 in ECs, indicating that the anti-inflammatory function of HDL is conferred through miRNA33. By contrast, Menghini et al. reported that miR-217 was expressed in human atherosclerotic lesions and is negatively correlated with silent information regulator 1 (Sirt1), resulting in endothelial senescence34. Interestingly, certain miRNAs, including miR-155, have contradictory effects on the pathogenesis of AS. Most reports have supported that miR-155 acts as an anti-inflammatory, atheroprotective microRNA18,27,35,36,37,38,39. Yet several studies reported a pro-atherogenic role for this microRNA19,40,41. We believe that the function of miR-155 is highly dependent on the context and cell types because of its dynamic expression pattern and its suppression of multiple target genes. Zhang et al. use carotid artery ligation model to assess the effect of Tongxinluo (TXL), a traditional Chinese medicine, exerts its inhibitory action on neointima formation by suppressing miR-15541. Tian et al. use macrophages from mice and Raw264.7 mouse cells to detect lipid uptake42. We use a classic human foam cell model in vitro to study the function of miR-155 in AS. Most likely, there is a specie-specific role of miR-155 in AS.
CARHSP1 was originally identified in pancreatic acinar cells by 32P metabolic labeling43 and a group has speculated that CARHSP1 participates in the oxidative stress response via a dynamic and temporal association between stress granules and processing bodies44. Although a recent study has demonstrated that CARHSP1 is a TNF-α mRNA stability enhancer required for effective TNF-α production and the importance of both stabilization and destabilization pathways in regulating TNF-α mRNA half-life26, the biological effects of CARHSP1 have remained elusive. Here we found for the first time that miR-155 targets the 3′-UTR of CARHSP1 and inhibits CARHSP1 at the mRNA and protein level, thus indirectly down-regulating the expression of TNF-α. We also found that down-regulation of TNF-α attenuates the inflammation reaction and lipid uptake by macrophages. We therefore speculated that CARHSP1 is a functional target of miR-155, which directly targets its 3′-UTR and that CARHSP1 serves as a mediator of miR-155 by regulating the production of TNF-α and thereby affecting foam cell formation.
After investigating the biological effects of miR-155 and the relationship with its target, we were also interested in the question of how miR-155 is upregulated during the pathological process of AS. Recent studies have shown that several transcription factors, including AP-1, STAT3 and MYB, up-regulate the expression of miR-155 in the immune system45,46,47. Since TNF-α is overexpressed at the early stages of foam cell formation, we hypothesized that NF-κB is activated by up-regulated TNF-α and bioinformatic tools revealed that there were several binding sites in the region of the miR-155 promoter29,30. Our ChIP assay and luciferase results were previously supported by some researchers and showed that NF-κB, which is activated by TNF-α in THP-1 cells, is a transcriptional enhancer of miR-15548,49. We suggest that, at the initial stages of foam cell formation, inflammatory factors such as TNF-α are overexpressed and NF-κB is then activated by up-regulated TNF-α, thus resulting in a sharp increase in the miR-155 level. Based on our data, it appears that at the early stage of AS, the infiltrating monocytes are stimulated by inflammatory factors like TNF-α and differentiate into macrophages; due to the hyperlipidemia and oxidative modification, macrophages uptake oxLDL and differentiate into foam cells that deposit in endothelium, resulting atherosclerotic plaques. During the process, TNF-α activates NF-κB to transactivate the expression of miR-155, which in turn suppresses TNF-α production through directly targeting the 3′-UTR of CARHSP1. Therefore, miR-155 is a key component of a negative feedback loop to prevent atherosclerosis-associated foam cell formation by repressing CARHSP1.
A notable limitation of this study is that the importance of the miR-155-CARHSP1 axis in atherosclerosis needs to be reinforced by transgenic animal data. Another limitation is that beyond macrophages there are other types of cells contributing to the elevated plasma miR-155 levels in AS patients.
To summarize, we found that miR-155 is overexpressed in the plasma and atherosclerotic lesions of patients with AS and its induction by oxLDL in human macrophages relieves inflammation and foam cell formation by depressing CARHSP1-mediated stimulation of TNF-α expression. We therefore conclude that miR-155 plays a key role in the anti-inflammation activity of macrophages, which thereby attenuates atherosclerosis-associated foam cell formation.
Methods
Patient characteristics
We enrolled 70 patients with AS, who were diagnosed by clinical symptoms and angiography from the Central Hospital of Wuhan between January 2014 and February 2015. Fifty-five healthy volunteers were recruited as the control group. We also identified 17 pairs of atherosclerotic lesions and normal veins from the same patients. Patients were excluded who had the following: 1) Any disease that activates monocytes, such as arthritis, bronchial asthma and infectious diseases caused by bacteria or viruses. 2) Any serious cardiovascular disease, such as acute myocardial infarction or heart failure. Informed consent was obtained from all human subjects. This study was approved by the Medical Ethics Committee of The Central Hospital of Wuhan and all human subject research was performed in accordance with institutional, national and Declaration of Helsinki requirements.
Plasma collection and storage
Peripheral blood plasma from patients and the control group were collected in EDTA tubes and processed within 4 hours by centrifuging at 1000 x g at 4 °C for 10 minutes, gently transferring the plasma to a fresh RNase/DNase-free 1.5-mL EP tube (Axygen, Union City, CA, USA) while avoiding pipetting of the middle layer of white blood cells and centrifuging again at 16,000 x g at 4 °C for 10 minutes. The supernatant was then transferred to fresh RNase/DNase-free tubes and stored at −80 °C.
Cell culture and establishing the foam cell model
The human monocytic cell line THP-1 and human embryonic kidney (HEK)293T cells were purchased from the American Type Culture Collection (ATCC). THP-1 cells were cultured in RMPI-1640 medium (Gibco, Carlsbad, CA, USA) and HEK-293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco). Both media were supplied with 10% fetal bovine serum (Gibco), 100 μg/mL streptomycin and 100 IU/mL penicillin (Thermo Fisher Scientific, Rockford, IL, USA). Cells were incubated in a humidified incubator at 37 °C with 5% CO2. To drive monocyte differentiation into macrophages and ultimately establish a foam cell model, THP-1 cells were first seeded at 1 × 106 per mL with 100 nM propylene glycol monomethyl ether acetate (PMA, Sigma-Aldrich, St. Louis, MO, USA) for 12 hours to differentiate them into adherent macrophages18,50. The THP-1 macrophages were then stimulated with 50 μg/ml oxLDL (Luwen, Shanghai, China) for 12 hours to establish the foam cell model51.
Oil Red O staining
Macrophages derived from THP-1 cells were transfected with a miR-155 mimic or a miR-155 inhibitor at a working concentration of 100 nM using Lipofectamine® LTX and Plus reagent (Invitrogen, Carlsbad, CA, USA). After 24 h, the cells were washed three times with 1 × phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 minutes, then stained with Oil Red O stain (Beisuo, Zhuhai, Shenzhen, China) in a 6:4 ratio of 0.5% Oil Red O solution with ddH2O for 20 minutes, followed by destaining with 60% isopropanol for 1 minute. The foam cells were then photographed under a microscope (Olympus, Tokyo, Japan) at 400 × magnification.
RNA isolation and qRT-PCR
Plasma miRNA from AS patients and healthy volunteers was extracted using the mirVana PARIS kit (Ambion, Carlsbad, CA, USA) according to the manufacturer’s instructions. miRNA reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Carlsbad, CA, USA) and the miRNAs were analyzed by qRT-PCR using the TaqMan Small RNA Assay kit and TaqMan universal PCR master mix (Applied BioSystems) in an ABI StepOne Plus qPCR instrument (Applied Biosystems). Total RNA from THP-1 macrophages and atherosclerotic lesions was isolated using TRIzol reagent (Invitrogen). Total RNA was measured using SYBR Green (Applied BioSystems) in an ABI StepOne Plus qPCR instrument and cDNA for target gene detection was synthesized from 1 μg total RNA. The following primers were used: CARHSP1 sense (5′-CCTGCACATCTCTGATGTGGA-3′) and antisense (5′-TGGTGCCAGGTGAGTGATGA-3′); TNF-α sense (5′-GTAGCCCATGTTGTAGCAAACCCTC-3′) and antisense (5′TGAAGAGGACCTGGGAGTAGAT3′); NF-κB p65 sense (5′-TCCCATCTTTGACAATCGTGCCC-3′)and antisense (5′-TCTGCACCTTGTCACACAGTAG-3′). PCR results were normalized between samples with U6 as an internal control and expressed as 2–(CT[miRNA]–CT[U6]), with CT denoting the threshold cycle52.
Western blot
Cells were lysed in RIPA buffer containing protease inhibitors (Beyotime, Shanghai, China) and the protein concentration was measured using a BCA protein assay kit (Thermo Fisher Scientific). Soluble lysate was mixed with loading buffer and boiled for 10 min. Lysates were separated via 12% SDS-PAGE electrophoresis, then transferred to a 0.22-μm PVDF membrane and blocked with 5% nonfat milk. The membranes were then incubated with primary antibodies against β-actin (1:1000, Santa Cruz, Delaware, CA, USA), CARHSP1 (1:1000, Santa Cruz), or TNF-α (1:1000, R&D Systems, Minneapolis, MN, USA) overnight at 4 °C. The membranes were then incubated with the HRP-conjugated secondary antibody (1:10,000, Santa Cruz) and the protein level detected by chemiluminescence.
Enzyme-linked immunosorbent assay (ELISA) for cytokine detection
Secreted pro-inflammatory cytokines (TNF-α) were detected in aliquots of THP-1 macrophage supernatants and quantitated by ELISA (R&D Systems). The entire procedure was performed according to the manufacturer’s instructions.
Luciferase reporter assay
The prediction of miRNA target genes was performed using online software packages, including MirDB, Targetscan and PicTar. A fragment of the CARHSP1 3′-UTR containing a putative miR-155 binding site was amplified from human genomic DNA from THP-1 cells by PCR using specific primers (sense, 5′-TATCTAGAGGAGGAGGCAGCAGACACT-3′; antisense, 5′-ATGCGGCCGCAAAGGCAACATCCGTCCAAT-3′). A mutant 3′-UTR lacking the miR-155 seed sequence was also amplified with specific primers (sense 5′-TATCTAGAGGAGGAGGCAGCAGACACT-3′; antisense, 5′-AATGCTTTTGGGCGGCTTCAGATGGTGAGAGG-3′). The amplified products were restricted and ligated to the XbaI and NotI (New England Biolabs, NEB, Ipswich, MA, England) sites of the pRL-TK vector (Promega, Madison, WI, USA) and the reporter plasmids were confirmed by DNA sequencing. Approximately 1 × 104 HEK-293T cells were seeded into 96-well plates 24 hours before transfection and the cells were then co-transfected with pRL-TK-CARHSP1-3′UTR or pRL-TK-CARHSP1-3′UTR mutant vectors (100 ng), 10 ng of pGL3 control (Promega) and 10 pmol miR-155 mimic or miR-155 inhibitor using Lipofectamine® LTX and Plus reagent (Invitrogen). After a 48-hour transfection, luciferase activity was detected using the Dual-Glo luciferase reporter assay system (Promega), according to the manufacturer’s protocol. miR-155 mimic and miR-155 inhibitor were purchased from GenePharma (Suzhou, Jiangsu, China). The mimic is a chemically synthesized double-stranded RNA oligonucleotide that mimics the effects of endogenous miR-155. The inhibitors are chemically synthesized miRNA hairpin inhibitors that effectively inhibit endogenous miR-155 function.
Transfection of miR-155 promoter constructs and luciferase assay
The human miR-155 promoter region containing NF-κB p65 binding sites, extending from –1 bp to –2000 bp relative to the transcription start site, was generated by PCR using the following primers: sense, 5′-TATCTAGAGCGAGTTATATTGGCTGGGGTGG-3′; antisense, 5′-ATGCGGCCGC TGTGATACAAGCAATGGAGGT-3′. The genomic DNA of THP-1 cells was isolated for use as template. The purified fragment was digested using XbaI and NotI and inserted into the XbaI- and NotI-cut pRL-TK luciferase reporter vector. The promoter region was sequenced and compared with sequences in Genbank. The miR-155 promoters mutated at the p65 binding site 1 (–1108 bp to –1099 bp), site 2 (–978 bp to –969 bp) and site 3 (–714 bp to –704 bp) were generated by site-directed mutagenesis using a special polymerase contained in the PhusionTM High-Fidelity PCR kit (NEB).
Chromatin immunoprecipitation (ChIP) assay
To detect which putative binding site within the miR-155 promoter binds to NF-κB p65, chromatin immunoprecipitation assays were performed using the Magna ChIP™ HiSens kit (Millipore, Temecula, CA, USA), according to the manufacturer’s instructions. In brief, proteins and DNA were first cross-linked using 37% formaldehyde for 10 minutes and the cells were then lysed in lysis buffer for 15 minutes to release cross-linked protein–DNA complexes. In the next step, the isolated chromatin was sonicated on wet ice to shear the DNA, yielding chromatin fragments of 200–1000 bp and the final step was immunoprecipitation (IP) of cross-linked protein–DNA complexes using p65 antibody (4 μg, Millipore) after incubation overnight at 4 °C. The ChIP results were semi-quantified by agarose gel electrophoresis.
CARHSP1 overexpression or knockdown
The open reading frame (ORF) sequence of CARHSP1 was searched using the UCSC and Ensemble genome browsers and the ORF clone was amplified by PCR in order to append cloning sites to the 5′ and 3′ ends of the sequence using the following primers: sense, 5′-GAGGCGATCGCCATGTCATCTGAGCCTCCCCC-3′; antisense, 5′-GCGACGCGTGGAGCTGATGACATGTCCAGACCA-3′. The RNA was first isolated from THP-1 cells and the full-length cDNA clone used as the template for CARHSP1 ORF cloning using the Phusion TM High-Fidelity PCR kit (NEB). The size of the amplification product was confirmed by agarose gel electrophoresis. The amplification product was digested using Sgf I and MIu I (NEB) and inserted into the Sgf I- and MIu I-cut pCMV6-AC-GFP vector (Origene, Rockville, MD, USA). The CARHSP1 overexpression plasmid sequences were confirmed by DNA sequencing and the transfection efficiency was determined by qPCR. CARHSP1 siRNA was purchased from Santa Cruz Biotechnology and 50 pmol/ml siRNA was transfected into THP-1 cells according to the manufacturer’s instructions.
mRNA stability assay
OxLDL stimulated THP-1 cells were treated with actinomycin D (ActD) (5 μg/ml) to inhibit transcription after transfection with CARHSP1 siRNA or control. Total RNAs were extracted at different time points after ActD administration and TNF-α mRNA was measured by qRT-PCR. RNA half-life (t1/2) was calculated by linear regression analysis53,54.
Statistical analysis
Student’s t-test was performed for comparison between two groups and the data was also analyzed by one-way analysis of variance (ANOVA ) among multiple groups using GraphPad Prism software. All measurements were performed at least three times independently. P value < 0.05 were considered to be a statistically significant difference. *P < 0.05, **P < 0.01, ***P < 0.001.
Additional Information
How to cite this article: Li, X. et al. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci. Rep. 6, 21789; doi: 10.1038/srep21789 (2016).
References
Glass, C. K. & Witztum, J. L. Atherosclerosis. the road ahead. Cell 104, 503–516 (2001).
Moore, K. J. & Tabas, I. Macrophages in the pathogenesis of atherosclerosis. Cell 145, 341–355 (2011).
Tuttolomondo, A. et al. Atherosclerosis as an inflammatory disease. Curr Pharm Des 18, 4266–4288 (2012).
Hansson, G. K. Inflammation, atherosclerosis and coronary artery disease. N Engl J Med 352, 1685–1695 (2005).
Moore, K. J. & Tabas, I. Macrophages in the Pathogenesis of Atherosclerosis. Cell 145, 341–355 (2011).
Ross, R. Atherosclerosis is an inflammatory disease. Am Heart J 138, S419–420 (1999).
Weber, C. & Noels, H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med 17, 1410–1422 (2011).
Ouimet, M. & Marcel, Y. L. Regulation of lipid droplet cholesterol efflux from macrophage foam cells. Arterioscler Thromb Vasc Biol 32, 575–581 (2012).
Ambros, V. microRNAs: tiny regulators with great potential. Cell 107, 823–826 (2001).
Williams, A. E. Functional aspects of animal microRNAs. Cell Mol Life Sci 65, 545–562 (2008).
Eulalio, A., Huntzinger, E. & Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 132, 9–14 (2008).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Devaux, Y. et al. MicroRNAs: new biomarkers and therapeutic targets after cardiac arrest? Crit Care 19, 54 (2015).
Wang, G. K. et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 31, 659–666 (2010).
Eis, P. S. et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA 102, 3627–3632 (2005).
Faraoni, I., Antonetti, F. R., Cardone, J. & Bonmassar, E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta 1792, 497–505 (2009).
Ma, X., Ma, C. & Zheng, X. MicroRNA-155 in the Pathogenesis of Atherosclerosis: A Conflicting Role? Heart, Lung and Circulation 22, 811–818 (2013).
Zhu, J. et al. Regulation of microRNA-155 in atherosclerotic inflammatory responses by targeting MAP3K10. PLoS One 7, e46551 (2012).
Nazari-Jahantigh, M. et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J Clin Invest 122, 4190–4202 (2012).
Wistow, G. Cold shock and DNA binding. Nature 344, 823–824 (1990).
Lindquist, J. A., Brandt, S., Bernhardt, A., Zhu, C. & Mertens, P. R. The role of cold shock domain proteins in inflammatory diseases. J Mol Med 92, 207–216 (2014).
Schafer, C., Steffen, H., Krzykowski, K. J., Goke, B. & Groblewski, G. E., CRHSP-24 phosphorylation is regulated by multiple signaling pathways in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol 285, G726–734 (2003).
Hou, H. et al. Structure-functional analyses of CRHSP-24 plasticity and dynamics in oxidative stress response. J Biol Chem 286, 9623–9635 (2011).
Yan, G., You, B., Chen, S. P., Liao, J. K. & Sun, J. Tumor necrosis factor-alpha downregulates endothelial nitric oxide synthase mRNA stability via translation elongation factor 1-alpha 1. Circ Res 103, 591–597 (2008).
Zhang, T., Kruys, V., Huez, G. & Gueydan, C. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans 30, 952–958 (2002).
Pfeiffer, J. R., McAvoy, B. L., Fecteau, R. E., Deleault, K. M. & Brooks, S. A. CARHSP1 Is Required for Effective Tumor Necrosis Factor Alpha mRNA Stabilization and Localizes to Processing Bodies and Exosomes. Mol Cell Biol 31, 277–286 (2010).
Zhu, G. et al. miR-155 inhibits oxidized low-density lipoprotein-induced apoptosis of RAW264.7 cells. Mol Cell Biochem 382, 253–261 (2013).
Lindquist, J. A., Brandt, S., Bernhardt, A., Zhu, C. & Mertens, P. R. The role of cold shock domain proteins in inflammatory diseases. J Mol Med 92, 207–216 (2014).
De Simone, V. et al. Th17-type cytokines, IL-6 and TNF-alpha synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene 34, 3493–3503 (2015).
Dvoriantchikova, G. & Ivanov, D. Tumor necrosis factor-alpha mediates activation of NF-κB and JNK signaling cascades in retinal ganglion cells and astrocytes in opposite ways. Eur J Neurosci 40, 3171–3178 (2014).
Weber, C. & Noels, H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med 17, 1410–1422 (2011).
Schober, A. et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat Med 20, 368–376 (2014).
Tabet, F. et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun 5, 3292–3303 (2014).
Menghini, R. et al. MicroRNA 217 Modulates Endothelial Cell Senescence via Silent Information Regulator 1. Circulation 120, 1524–1532 (2009).
Donners, M. M. et al. Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice. PLoS One 7, e35877 (2012).
Ceppi, M. et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci USA 106, 2735–2740 (2009).
Zhu, N. et al. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 215, 286–293 (2011).
Wu, X., Fan, W., Fang, R. & Wu, G. Regulation of microRNA-155 in endothelial inflammation by targeting nuclear factor (NF)-κB P65. J Cell Biochem 115, 1928–1936 (2014).
Huang, R. S., Hu, G. Q., Lin, B., Lin, Z. Y. & Sun, C. C. MicroRNA-155 Silencing Enhances Inflammatory Response and Lipid Uptake in Oxidized Low-Density LipoproteinYStimulated Human THP-1 Macrophages. J Investig Med 58, 961–967 (2010).
Sun, H. X. et al. Essential Role of MicroRNA-155 in Regulating Endothelium-Dependent Vasorelaxation by Targeting Endothelial Nitric Oxide Synthase. Hypertension 60, 1407–1414 (2012).
Zhang, R. N. et al. Tongxinluo inhibits vascular inflammation and neointimal hyperplasia through blockade of the positive feedback loop between miR-155 and TNF-α. AJP: Heart and Circulatory Physiology 307, H552–562 (2014).
Tian, F. J. et al. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc Res 103, 100–110 (2014).
Groblewski, G. E. et al. Purification and characterization of a novel physiological substrate for calcineurin in mammalian cells. J Biol Chem 273, 22738–22744 (1998).
Hou, H. et al. Structure-functional analyses of CRHSP-24 plasticity and dynamics in oxidative stress response. J Biol Chem 286, 9623–9635 (2011).
Vargova, K. et al. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood 117, 3816–3825 (2011).
Yin, Q., Wang, X., McBride, J., Fewell, C. & Flemington, E. B-cell Receptor Activation Induces BIC/miR-155 Expression through a Conserved AP-1 Element. J Biol Chem 283, 2654–2662 (2008).
Li, P. et al. Signal transducer and activator of transcription-3 induces microRNA-155 expression in chronic lymphocytic leukemia. PLoS One 8 e64678 (2013).
Thompson, R. C., Vardinogiannis, I. & Gilmore, T. D. Identification of an NF-kappaB p50/p65-responsive site in the human MIR155HG promoter. BMC Mol Biol 14, 24 (2013).
Gatto, G. et al. Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF- B pathway. Nucleic Acids Res 36, 6608–6619 (2008).
Daigneault, M., Preston, J. A., Marriott, H. M., Whyte, M. K. & Dockrell, D. H. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One 5, e8668 (2010).
Bao, Y. et al. Salvianolic acid B inhibits macrophage uptake of modified low density lipoprotein (mLDL) in a scavenger receptor CD36-dependent manner. Atherosclerosis 223, 152–159 (2012).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101–1108 (2008).
Subbaram, S. et al. Integrin 3 1 controls mRNA splicing that determines Cox-2 mRNA stability in breast cancer cells. J Cell Sci 127, 1179–1189 (2014).
Xu, T. P. et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene 34, 5648–5661 (2015).
Acknowledgements
This project was supported by grants from the National Natural Sciences Foundation of China (No. 81071921 and 81372325), the National High Technology Research and Development Program of China (863 Program; 2011AA02A111), the Key project of Natural Sciences Foundation of Hubei Province (2015CFA078), Science and technology support program (foreign scientific and technological cooperation) of Hubei Province (2015BHE022), the Yellow Crane Talent Plan Foundation, Research Fund of Wuhan Public Health Bureau (WX11A03, WX14C13 and WX14B10) and Youth Foundation of Wuhan Central Hospital (YQ14A01 and YQ15A03).
Author information
Authors and Affiliations
Contributions
X.L., D.K. and H.C. carried out the molecular biology analysis, participated in the design of the study and the clinical specimen collection and drafted the manuscript. S.L., H.H., T.W., W.C., J.W. and Y.N. carried out the clinical specimen collection, participated in the data analysis and performed the statistical analysis. Z.L. and Y.L. conceived of and designed the study and participated in the data analysis and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Li, X., Kong, D., Chen, H. et al. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci Rep 6, 21789 (2016). https://doi.org/10.1038/srep21789
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep21789
This article is cited by
-
Microglia specific deletion of miR-155 in Alzheimer’s disease mouse models reduces amyloid-β pathology but causes hyperexcitability and seizures
Journal of Neuroinflammation (2023)
-
miR-155 activates the NLRP3 inflammasome by regulating the MEK/ERK/NF-κB pathway in carotid atherosclerotic plaques in ApoE−/− mice
Journal of Physiology and Biochemistry (2022)
-
Inflammatory Modulation of miR-155 Inhibits Doxorubicin-Induced Testicular Dysfunction via SIRT1/FOXO1 Pathway: Insight into the Role of Acacetin and Bacillus cereus Protease
Applied Biochemistry and Biotechnology (2022)
-
Genome-wide CRISPR/Cas9 screening identifies CARHSP1 responsible for radiation resistance in glioblastoma
Cell Death & Disease (2021)
-
Atherosclerotic-nephropathy: an updated narrative review
Journal of Nephrology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.