Abstract
Plant microRNAs (miRNAs) play pivotal roles in many biological processes. Although many miRNAs have been identified in various plant species, the functions of these miRNAs remain largely unknown due to the shortage of effective genetic tools to block their functional activity. Recently, miRNA target mimic (TM) technologies have been applied to perturb the activity of specific endogenous miRNA or miRNA families. We previously reported that Tobacco rattle virus (TRV)-based TM expression can successfully mediate virus-based miRNA silencing/suppression (VbMS) in plants. In this study, we show the Potato virus X (PVX)-based TM expression causes strong miRNA silencing in Nicotiana benthamiana. The PVX-based expression of short tandem target mimic (STTMs) against miR165/166 and 159 caused the corresponding phenotype in all infected plants. Thus, a PVX-based VbMS is a powerful method to study miRNA function and may be useful for high-throughput investigation of miRNA function in N. benthamiana.
Similar content being viewed by others
Introduction
Plant microRNAs (miRNAs) are a class of endogenous non-coding small RNAs with 20–24 nucleotides1,2. Plant miRNAs mainly act as negative regulators of gene expression at the post-transcription level by triggering endonuclease cleavage or by promoting translation repression of target mRNAs3. They therefore play essential roles in various biological processes including growth and development4,5, metabolism6,7, hormone signaling8,9 and responses to biotic10,11,12,13 and abiotic14,15,16 stresses. The functions of plant miRNAs can be investigated by enhancing their activity through ectopic- or over- expression of the miRNA gene9,17 or by blocking miRNA function through expression of a miRNA-resistant target that has uncleavable silent mutations in the mRNA sequence18. Despite great progress in predicting and identifying miRNAs in many plants, characterization of their biological roles has largely lagged behind, especially in non-model plant species. This is for two reasons: on the one hand, traditional loss-of-function approaches are difficult to apply due to the extensive genetic redundancy of miRNA genes in plant genomes19,20; on the other hand, the use of miRNA resistant targets can only partially reveal the function of those miRNAs that regulate multiple targets21,22,23.
miRNA function is naturally regulated by target mimics (TMs). The TM molecules have a mismatch bulge at the position corresponding to nucleotides 10–11 of the relative miRNA, the cleavage site of target mRNAs, thus blocking the cleavage of target mRNAs by miRNA-Argonaute proteins24,25. Artificially designed TM transcripts based on Arabidopsis INDUCED BY PHOSPHATE STARVATION 1 (IPS1) are highly complementary to specific miRNAs but contain extra nucleotides that can form a mismatching loop at the cleavage site in the miRNA-TM duplex, thus preserving TMs from cleavage and sequestrating miRNA functions25. A short tandem target mimic (STTM), consisting of two TMs separated by a short linker sequence, suppressed miRNA activity more efficiently than the regular TM in transgenic plants26,27,28. Thus miRNA TM technology provides an effective method to manipulate the activity of endogenous miRNAs and dissect their function(s)27,29,30.
We previously developed a plant virus-based miRNA expression system, in which a Tobacco rattle virus (TRV)-based vector was used to express artificial TMs and cause virus-based miRNA silencing/suppression (VbMS)31,32. Several research groups have developed VbMS systems based on different plant viruses, including Cotton leaf crumple virus (CLCrV)33 and Cucumber mosaic virus (CMV)34,35 and have demonstrated good miRNA suppression. VbMS enables a prompt elucidation of miRNA function and has proved to be effective in Nicotiana benthamiana31,34, N. tabacum34, Solanum lycopersicum31,34, Arabiopsis32,35 and Gossypium hirsutum33. However, the efficiency of VbMS caused by currently available TRV vectors is not strong enough and weak phenotypes may be overlooked31,32.
The precise mode of action of TMs is unknown, but TM molecules induce small RNA degradation by pairing with target miRNA molecules27 and the levels of TM transcripts inversely correlate with those of the targeted miRNAs25,27,36. Thus viral vectors which provide higher accumulations of TM transcripts could cause higher VbMS efficiency. PVX derived vectors can be used to mediate high expression of foreign genes37,38 and so might be modified to deliver improved VbMS.
In this study, we found that PVX-based VbMS causes very efficient silencing of miRNAs in N. benthamiana and so may be a useful tool for high-throughput investigation of miRNA function in plants.
Results
Development of PVX-based VbMS vector
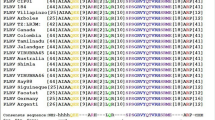
To perform high-throughput cloning of TMs, we generated a modified PVX vector PVX-LIC (Fig. 1) by cloning a ligation independent cloning (LIC) cassette into pSPDK65839. We showed previously that STTM-mediated VbMS suppressed miRNA more strongly than regular IPS1-based VbMS31. Therefore, we used the PVX-based vector to express STTMs under the control of the coat protein (CP) sub-genomic promoter (sgP) to inhibit miRNA function in this study (Fig. 1). The designed STTM sequences can be cloned into PVX-LIC by the high throughput LIC method.
Diagram of the Potato Virus X based PVX-LIC vector for VbMS.
The PVX-LIC vector was generated by introducing the LIC cassette into a T-DNA PVX vector. Artificially designed TM sequences can be cloned into PVX-LIC via the LIC reaction and expressed under the control of the CP subgenomic promoter. LB: T-DNA left border, RB: T-DNA right border, 35S: Cauliflower mosaic virus 35S promoter, NOSt: nopaline synthase terminator, RdRP: RNA dependent RNA polymerase, 25K: PVX 25K protein, 16K: PVX 16K protein, 8K: PVX 8K protein, CP: coat protein.
PVX-based VbMS of miR165/166
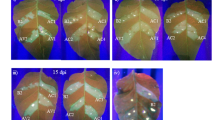
In N. benthamiana, miR165/166 is predicted to target the homeodomain-leucine zipper (HDZip) gene family. In an earlier study, TRV-based expression of STTM against Nbe-miR165/166 (STTM165/166) led to reduced apical dominance and only in extreme cases caused generation of an ectopic leaf from the midrib31. PVX-based expression of STTM165/166 (Fig. 2a) caused a strong silencing phenotype (Figs 2 and 3) with pleiotropic developmental defects. The most general defect was ectopic leaf outgrowths from veins on the leaf abaxial surface (Fig. 2b). A leaf lamina appeared on the back side of the leaf rib with opposite symmetry to the normal leaf (Fig. 2b). In some cases, alteration of the adaxial-abaxial organization led to the formation of abnormal bilateral symmetric leaves with a double layered lamina split by the mid vein (Fig. 3a) and cup-shaped leaves (Fig. 3b), reflecting an alteration of the leaf primordia caused by blockage of miR165/16640. Occasionally, the midrib grew out from the lamina (Fig. 3c) and in certain extreme cases, trumpet-shaped leaves were generated along the rib of the adaxial side of some leaves in addition to the terminally extended mid vein (Fig. 3d). At the nodes, leaves grew out of the stem from ectopically generated axillary meristems near leaf insertion sites (Fig. 3e), comparable to the strong phenotypes of the miR165/166 resistant PHAVOLUTA (NsPHAV) in Nicotiana sylvestris40.
Suppression of miR165/166 by PVX-STTM165/166 in Nicotiana benthamiana.
(a) Scheme of the base pairing pattern of STTM165/166 and miR165/166. -: denotes no nucleotide at this position. 48 nts: the 48-nt long stem-loop linker. (b) Leaf phenotypes of plants inoculated with PVX-STTM165/166. The abaxial side of leaves from the PVX control and PVX-STTM165/166 plants were photographed 15 days post infiltration (dpi). Magenta arrows denote the ectopically generated leaf tissues in the leaf veins, orange arrow heads indicate the larger leaf lamina formed from the midrib. Bars represent 1 cm. (c) Relative miR165/166 levels as measured by stem-loop RT-PCR in plants inoculated with PVX or PVX-STTM159. (d) Relative expression levels of the miR165/166 target HDZIP like gene TC21810. Data are means of 3 independent real-time RT-PCR experiments. Error bars show the standard deviation (±SD).
Varied severe phenotypes of miR165/166 blockage by PVX-STTM165/166 in Nicotiana benthamiana.
(a) Abnormal bilateral symmetric leaf with double layered lamina split by the mid vein. (b) Cup-shaped leaf with adaxial surface on the outside and abaxial surface inside. (c) Midrib growing out from the lamina and forming rod like terminus (arrow). (d) Trumpet-shaped leaves generated along the midrib (arrow). (e) Ectopic axillary meristems at the vicinity of leaf insertion sites; arrow heads indicate outgrowth of leave tissues from the stem surface. Bars represent 1 cm.
All PVX-STTM165/166 infiltrated plants displayed obvious leaf developmental defects, implicating a great efficiency of VbMS. Real time RT-PCR analysis verified that the mRNA levels of the predicted Nbe-miR165/166 target TC21810, a HD-Zip gene, were elevated greatly in PVX-STTM165/166 infiltrated plants, (Fig. 2c) whereas Nbe-miR165/166 levels were highly reduced (Fig. 2d).
PVX-based VbMS of miR159
In Arabidopsis, miR159 targets a set of transcription factor genes containing the MYB domain that are involved in vegetative development and flowering41,42. Deep sequencing data show that miR159 is conserved in N. benthamiana43,44,45 and it is predicted that Nbe-miRNA159 cleaves several MYB like transcription factor genes (http://plantgrn.noble.org/psRNATarget/, N. benthamiana genome version 1.0.1). The exact biological function of miR159 in N. benthamiana development has not been studied but it is reported to have a role in the pathogenesis of severe CMV strains34,35.
In our experiments, PVX-STTM159 inoculated plants were stunted and compact with downward-curled leaves, darker green pigmentation, smaller leaf area and shorter petioles than PVX control plants (Fig. 4a), indicating that functional blockage of miR159 triggers serious leaf morphogenesis defects. In addition, the internodes of PVX-STTM159 plants were shorter and the leaves were clustered together at the shoot apex (Fig. 4b). Compared to the PVX control plants, the mRNA levels of the miR159 target, MYB-like transcription factor (NbMYBL1, N. benthamiana genome version 1.0.1, Niben101Scf01383g07027.1), were increased in PVX-STTM159 plants while the miR159 levels decreased greatly (Fig. 4c,d). These results indicate that the PVX VbMS method can inhibit different endogenous miRNAs and provide an indication of miRNA/target mRNA function.
PVX-based VbMS of miR159 by PVX-STTM159 in Nicotiana benthamiana.
(a) Diagrammatic representation of PVX-STTM159. -: denotes no nucleotide at this position. 48 nts: the 48-nt long stem-loop linker. (b) Plants infiltrated with PVX control or PVX-STTM159 photographed at 20 dpi. Top row, top view; bottom row, side view. Bar represents 1 cm. (c) Relative miR159 levels as measured by stem-loop RT-PCR in plants inoculated with PVX or PVX-STTM159. (d) Relative mRNA levels of miR159 target NbMYBL1 in the PVX control and PVX-STTM159 plants. Values are means ± SD from 3 independent experiments.
Discussion
In this study, we have shown that the modified PVX vector can be used to express TM molecules and successfully inhibit the function of several endogenous miRNAs in N. benthamiana. There was a very high efficiency of miRNA silencing in the plants. All PVX-STTM165/166 infiltrated plants exhibited ectopic leaf outgrowths, similar to those observed in the gain-of-function HD-ZIP III phv1 mutants in N. sylvestris40 and in plants with CMV-based VbMS of miR165/16634. By comparison, in N. benthamiana plants infected with TRV containing STTM-165/166 (TRV-STTM165/166), ectopic leaf outgrowths on the leaf middle vein were observed only in certain extremes31. All PVX-STTM159 N. benthamiana plants had a leaf developmental defect (Fig. 4) and resembled the phenotypes induced by CMV-based VbMS of miR159 as well as a severe CMV strain Fny-CMV. However, we did not observed any leaf phenotype in TRV-STTM159 plants. These results suggest that the PVX based VbMS is highly effective in blocking the function of targeted miRNAs in N. benthamiana.
PVX (in this study) or CMV34 seems to have a higher efficiency of VbMS than our previously reported TRV system for the miRNAs expressed in leaves of N. benthamiana. It is possible that TRV is a better VIGS vector and that VIGS may cause a lower level of viral RNA containing TM. Since TRV has a wide host range and also infects the meristems of plants, whereas PVX and CMV do not, TRV-based VbMS is still very useful for dissecting the function of many miRNAs expressed in the meristems and in some important host plants.
In the previous report, the success of miRNA silencing depended on the expression levels of the TM transcripts. Highly expressed TMs may trigger more effective silencing effects on targeted miRNAs and greater enhancement of miRNA target mRNA levels. Meanwhile, different designed TMs may have differing effectiveness in plant cells25,27,36.
Manipulation of the sequence composition of the TM molecule, by inserting 1–3 nucleotides to produce central mismatches between the cleavage site or at other recognition sites, provides subtle regulation of target miRNA inactivation36,46. It has been reported that no single design can guarantee the most dramatic silencing of different miRNA families29. However, it appears that the most efficient TMs are obtained quite empirically; the most important factor is to avoid the formation of another new cleavage site as previous reported29.
In addition to N. benthamiana, PVX-mediated gene expression has been reported to function in N. clevelandii, N. tabacum, Solanum tuberosum and S. lycopersicum in either local or systemic tissues47,48. The high levels of foreign gene products synthesized and the long term expression produced by PVX makes the system suitable for miRNA TM expression throughout the duration of plant development30,49. Thus PVX can be utilized as a general vector for various plants. As PVX can tolerate extra larger insertions and repetitive regions with high homology37,50, it may be feasible to insert multiple TM segments into the PVX vector to promote expression of heterogenic TM molecules. The PVX VbMS may provide an excellent system for engineering different miRNA families involved in related or unrelated processes in plants. Therefore, the modified PVX-based VbMS described in this report has great potential for the characterization of functions of miRNAs.
Materials and Methods
Plant Growth
N. benthamiana plants were grown in a growth room at 24 °C with a 16 hr/8 hr light/dark photoperiod cycle, under white light at 100 μM m−2 s−1. Four week old plants with 7–8 leaves were used for PVX VbMS infiltration.
TM molecular design
The 3~4 nt insertions in STTM molecules were designed empirically at the site corresponding to nts 10–11 of the targeted miRNA. All PCR reactions for cloning were amplified by using the synthesized 48 nt oligonucleotide as template with EasyPfu polymerase (Transgene). All primers used for cloning are listed in Supplementary Table S1.
Plasmid Construction
PVX-LIC was generated by inserting the LIC cassette containing the ccdB and chloramphenicol-resistant genes into the PVX T-DNA vector39 by replacing the original multiple cloning site and was maintained and propagated in E. coli strain DB3.1. Before LIC cloning, PVX-LIC was digested with SmaI and treated with T4 DNA polymerase in the presence of dTTP (0.5 mM) and DTT (1 mM) for 30 min at 37 °C to produce the 14 nt sticky 5′ end. After the polymerase was inactivated at 75 °C for 20 min, the sticky-ended PVX-LIC vector was purified by phenol extraction and ethanol precipitation. TM PCR products were treated with T4 DNA polymerase in the presence of dATP (0.5 mM) and DTT (1 mM), followed by a 75 °C inactivation step and purified by ethanol precipitation. For LIC cloning, equal volumes of PVX-LIC and PCR product treated with T4 DNA polymerase were mixed and incubated at 37 °C for 30 min and then transformed into E. coli strain DH5α. The resulting constructs were verified by sequencing and transformed to Agrobacterium tumefaciens strain GV3101 (or GV2260).
Agrobacteria infiltration
Agrobacterium strain GV3101 (or GV2260) carrying the PVX derivatives was grown at 28 °C overnight, then harvested by centrifugation at 5,000 g and re-suspended in infiltration buffer (10 mM MES, 10 mM MgCl2 and 200 μM acetosyringone) to a final OD600 of 1.0. The suspensions were incubated at 25 °C for 2.5 ~ 4 hrs and infiltrated into leaves of N. benthamiana with needleless syringes. Each experiment was performed three times with at least 6 plants for each construct. Symptom development was monitored starting from 10 days after infiltration. Plants exhibiting phenotypes were then photographed and tissues were collected for subsequent analyses.
RNA extraction and Real time RT-PCR analysis
Total RNA was isolated using TRNzol-A+Reagent (Tiangen Biotech), treated with DNase I (Sigma Aldrich) to eliminate genomic DNA and converted into cDNA using the QuantScript reverse transcription Kit (Tiangen Biotech). Real-time PCR primers were designed with Primer Express 3.0. Real-time PCR was performed using SYBR Green PCR Master Mix (Life technologies) with sequence-specific primers. Stem loop RT-PCR was performed to analyze mature miRNA levels as described previously51. N. benthamiana eukaryotic Initiation Factor-4A (eIF4A) was used as an internal control and each assay was replicated at least three times. The expression data illustrated by the quantification cycle (Cq) were collected, statistically processed by the ΔΔCq algorithm and plotted using Origin 2015 software. The primers used in RNA analysis are shown in Supplemental Table S1.
Additional Information
Accession codes: NbAP2L1 (Genbank Accession number CK287095); eIF4a (Dana-Farber Cancer Institute N. benthamiana Gene Index TC19454).
How to cite this article: Zhao, J. et al. An efficient Potato virus X -based microRNA silencing in Nicotiana benthamiana. Sci. Rep. 6, 20573; doi: 10.1038/srep20573 (2016).
References
Rogers, K. & Chen, X. Biogenesis, Turnover and Mode of Action of Plant MicroRNAs. The Plant Cell 25, 2383–2399, 10.1105/tpc.113.113159 (2013).
Cuperus, J. T., Fahlgren, N. & Carrington, J. C. Evolution and Functional Diversification of MIRNA Genes. The Plant Cell 23, 431–442, 10.1105/tpc.110.082784 (2011).
Li, S. et al. microRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574, 10.1016/j.cell.2013.04.005 (2013).
Bhogale, S. et al. MicroRNA156: A Potential Graft-Transmissible MicroRNA That Modulates Plant Architecture and Tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 164, 1011–1027, 10.1104/pp.113.230714 (2014).
Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025, 10.1126/science.1088060 (2004).
Kettles, G. J., Drurey, C., Schoonbeek, H.-j., Maule, A. J. & Hogenhout, S. A. Resistance of Arabidopsis thaliana to the green peach aphid, Myzus persicae, involves camalexin and is regulated by microRNAs. The New Phytologist 198, 1178–1190, 10.1111/nph.12218 (2013).
Jones-Rhoades, M. W. & Bartel, D. P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799, 10.1016/j.molcel.2004.05.027 (2004).
Guo, H.-S., Xie, Q., Fei, J.-F. & Chua, N.-H. MicroRNA Directs mRNA Cleavage of the Transcription Factor NAC1 to Downregulate Auxin Signals for Arabidopsis Lateral Root Development. The Plant Cell 17, 1376–1386, 10.1105/tpc.105.030841 (2005).
Li, W. et al. Transcriptional Regulation of Arabidopsis MIR168a and ARGONAUTE1 Homeostasis in Abscisic Acid and Abiotic Stress Responses. Plant Physiol. 158, 1279–1292, 10.1104/pp.111.188789 (2012).
Du, P. et al. Viral Infection Induces Expression of Novel Phased MicroRNAs from Conserved Cellular MicroRNA Precursors. PLoS Path. 7, e1002176, 10.1371/journal.ppat.1002176 (2011).
Navarro, L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439, 10.1126/science.1126088 (2006).
Li, Y. et al. Identification of MicroRNAs Involved in Pathogen-Associated Molecular Pattern-Triggered Plant Innate Immunity. Plant Physiol. 152, 2222–2231, 10.1104/pp.109.151803 (2010).
Zhai, J. et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553, 10.1101/gad.177527.111 (2011).
Fujii, H., Chiou, T. J., Lin, S. I., Aung, K. & Zhu, J. K. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15, 2038–2043, 10.1016/j.cub.2005.10.016 (2005).
Liu, H.-H., Tian, X., Li, Y.-J., Wu, C.-A. & Zheng, C.-C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843, 10.1261/rna.895308 (2008).
Zhou, L. et al. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 61, 4157–4168, 10.1093/jxb/erq237 (2010).
Turner, M. et al. Ectopic Expression of miR160 Results in Auxin Hypersensitivity, Cytokinin Hyposensitivity and Inhibition of Symbiotic Nodule Development in Soybean. Plant Physiol. 162, 2042–2055, 10.1104/pp.113.220699 (2013).
Xue, X.-Y. et al. Interaction between Two Timing MicroRNAs Controls Trichome Distribution in Arabidopsis. PLoS Genet. 10, e1004266, 10.1371/journal.pgen.1004266 (2014).
Sieber, P., Wellmer, F., Gheyselinck, J., Riechmann, J. L. & Meyerowitz, E. M. Redundancy and specialization among plant microRNAs: role of the MIR164 family in developmental robustness. Development 134, 1051–1060, 10.1242/dev.02817 (2007).
Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell. Dev. Biol. 25, 21–44, 10.1146/annurev.cellbio.042308.113417 (2009).
Bartel, D. P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 136, 215–233, 10.1016/j.cell.2009.01.002 (2009).
Huang, D., Koh, C., Feurtado, J. A., Tsang, E. W. T. & Cutler, A. J. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics 14, 140–140, 10.1186/1471-2164-14-140 (2013).
Jeong, D.-H. et al. Parallel analysis of RNA ends enhances global investigation of microRNAs and target RNAs of Brachypodium distachyon. Genome Biology 14, R145–R145, 10.1186/gb-2013-14-12-r145 (2013).
Wu, H.-J., Wang, Z.-M., Wang, M. & Wang, X.-J. Widespread Long Noncoding RNAs as Endogenous Target Mimics for MicroRNAs in Plants. Plant Physiol. 161, 1875–1884, 10.1104/pp.113.215962 (2013).
Franco-Zorrilla, J. M. et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037, 10.1038/ng2079 (2007).
Tang, G. et al. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods 58, 118–125, 10.1016/j.ymeth.2012.10.006 (2012).
Yan, J. et al. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427, 10.1105/tpc.111.094144 (2012).
Jia, X. et al. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta 242, 283–293, 10.1007/s00425-015-2305-5 (2015).
Todesco, M., Rubio-Somoza, I., Paz-Ares, J. & Weigel, D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6, e1001031, 10.1371/journal.pgen.1001031 (2010).
Haraguchi, T., Ozaki, Y. & Iba, H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 37, e43, 10.1093/nar/gkp040 (2009).
Sha, A. et al. Virus-based microRNA silencing in plants. Plant Physiol. 164, 36–47, 10.1104/pp.113.231100 (2014).
Yan, F. et al. A virus-based miRNA suppression (VbMS) system for miRNA loss-of-function analysis in plants. Biotech J. 9, 702–708, 10.1002/biot.201300523 (2014).
Gu, Z., Huang, C., Li, F. & Zhou, X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649, 10.1111/pbi.12169 (2014).
Liao, Q., Tu, Y., Carr, J. P. & Du, Z. An improved cucumber mosaic virus-based vector for efficient decoying of plant microRNAs. Sci. Rep. 5, 13178, 10.1038/srep13178 (2015).
Du, Z. et al. Using a viral vector to reveal the role of microRNA159 in disease symptom induction by a severe strain of cucumber mosaic virus. Plant Physiol. 164, 1378–1388, 10.1104/pp.113.232090 (2014).
Ivashuta, S. et al. Regulation of gene expression in plants through miRNA inactivation. PloS one 6, e21330, 10.1371/journal.pone.0021330 (2011).
Dickmeis, C., Fischer, R. & Commandeur, U. Potato virus X-based expression vectors are stabilized for long-term production of proteins and larger inserts. Biotechnol J 9, 1369–1379, 10.1002/biot.201400347 (2014).
Chapman, S., Kavanagh, T. & Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 2, 549–557 (1992).
Liu, Y., Schiff, M., Serino, G., Deng, X.-W. & Dinesh-Kumar, S. P. Role of SCF Ubiquitin-Ligase and the COP9 Signalosome in the N Gene–Mediated Resistance Response to Tobacco mosaic virus. The Plant Cell 14, 1483–1496, 10.1105/tpc.002493 (2002).
McHale, N. A. & Koning, R. E. MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16, 1730–1740, 10.1105/tpc.021816 (2004).
Palatnik, J. F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263, 10.1038/nature01958 (2003).
Allen, R. S. et al. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104, 16371–16376, 10.1073/pnas.0707653104 (2007).
Yin, K., Tang, Y. & Zhao, J. Genome-wide characterization of miRNAs involved in N Gene-mediated Immunity in response to tobacco mosaic virus in Nicotiana benthamiana. Evol. Bioinform. 1–11, 10.4137/EBO.S20744 (2015).
Amin, I., Patil, B. L., Briddon, R. W., Mansoor, S. & Fauquet, C. M. A common set of developmental miRNAs are upregulated in Nicotiana benthamiana by diverse begomoviruses. Virology Journal 8, 143–143, 10.1186/1743-422X-8-143 (2011).
Xiao, B. et al. A diverse set of miRNAs responsive to begomovirus-associated betasatellite in Nicotiana benthamiana. BMC Plant Biol. 14, 60–60, 10.1186/1471-2229-14-60 (2014).
Reichel, M., Li, Y., Li, J. & Millar, A. A. Inhibiting plant microRNA activity: molecular SPONGEs, target MIMICs and STTMs all display variable efficacies against target microRNAs. Plant Biotechnol. J., 10.1111/pbi.12327 (2015).
Wang, Y. et al. Development of new potato virus X-based vectors for gene over-expression and gene silencing assay. Virus Res. 191, 62–69, 10.1016/j.virusres.2014.07.018 (2014).
Takken, F. L. W. et al. A functional cloning strategy, based on a binary PVX-expression vector, to isolate HR-inducing cDNAs of plant pathogens. Plant J. 24, 275–283, 10.1046/j.1365-313x.2000.00866.x (2000).
Giritch, A. et al. Rapid high-yield expression of full-size IgG antibodies in plants coinfected with noncompeting viral vectors. Proc Natl Acad Sci USA 103, 14701–14706, 10.1073/pnas.0606631103 (2006).
Chapman, S., Kavanagh, T. & Baulcombe, D. Potato virus X as a vector for gene expression in plants. Plant J. 2, 549–557, 10.1046/j.1365-313X.1992.t01-24-00999.x (1992).
Varkonyi-Gasic, E. & Hellens, R. P. Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol Biol 744, 145–157, 10.1007/978-1-61779-123-9_10 (2011).
Acknowledgements
This work was supported, in whole or in part, by the National Natural Science Foundation of China (31270182, 31530059, 31470254 and 31300134), the National Basic Research Program of China (2014CB138400), the Special Fund for Agro-scientific Research in the Public Interest of China (201303028), the National Transgenic Program of China (2014ZX08010, 2014ZX0800104B), the China Postdoctoral Science Foundation (2014M550049), the Initial Funding of Zhejiang Academy of Agricultural Sciences and the Cultural Funding for Youth Talent of Zhejiang Academy of Agricultural Sciences (2015R21R08E03). We thank Professor Michael J. Adams at Rothamsted Research, Stevenage, UK for help in correcting the English of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.L. and J.Z. designed the experiment. J.Z., Q.L., P.H., Q.J., N.L. and K.Y. performed experiment, data acquisition and analysis. J.Z. and Y.L. wrote paper. J.C., F.Y. and Y.C. were involved in discussions.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhao, J., Liu, Q., Hu, P. et al. An efficient Potato virus X -based microRNA silencing in Nicotiana benthamiana. Sci Rep 6, 20573 (2016). https://doi.org/10.1038/srep20573
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep20573
This article is cited by
-
iTRAQ based protein profile analysis revealed key proteins involved in regulation of drought-tolerance during seed germination in Adzuki bean
Scientific Reports (2021)
-
Function identification of miR482b, a negative regulator during tomato resistance to Phytophthora infestans
Horticulture Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.