Abstract
Parasitoid wasps are abundant and diverse hymenopteran insects that lay their eggs into the internal body (endoparasitoid) or on the external surface (ectoparasitoid) of their hosts. To make a more conducive environment for the wasps’ young, both ecto- and endoparasitoids inject venoms into the host to modulate host immunity, metabolism and development. Endoparasitoids have evolved from ectoparasitoids independently in different hymenopteran lineages. Pteromalus puparum, a pupal endoparasitoid of various butterflies, represents a relatively recent evolution of endoparasitism within pteromalids. Using a combination of transcriptomic and proteomic approaches, we have identified 70 putative venom proteins in P. puparum. Most of them show higher similarity to venom proteins from the related ectoparasitoid Nasonia vitripennis than from other more distantly related endoparasitoids. In addition, 13 venom proteins are similar to venoms of distantly related endoparasitoids but have no detectable venom matches in Nasonia. These venom proteins may have a role in adaptation to endoparasitism. Overall, these results lay the groundwork for more detailed studies of venom function and adaptation to the endoparasitic lifestyle.
Similar content being viewed by others
Introduction
Parasitoid wasps, being invaluable in classical and augmentative biological control of various insect pests, are among the most abundant and diverse insects on earth1. They have two basic lifestyles. Endoparasitoids lay their eggs into internal body of the host, whereas ectoparasitoids lay on the external surface of their hosts1,2. Parasitoids also injected substances into the host to ensure the successful parasitism and facilitate the successful development of their offspring, which can include venom, polydnaviruses (PDVs), virus-like particles (VLPs), ovarian fluids and teratocytes. The effects of these components depend largely on the parasitic life strategy3,4. Venoms from ectoparasitoids often induce a long-term paralysis to immobilize hosts, block their development following parasitism and also regulate their metabolism and immunity5,6. On the other hand, endoparasitoid venoms are mainly involved in temporary paralysis, host regulation by suppressing immune responses, delaying or arresting host development and synergizing the effects of PDVs/VLPs in some host-endoparasitoid systems3,4,7.
Venoms in most animals are involved in predation and/or defense4. They have been recognized as a rich source of biological active compounds8. In particular, venoms from cone snails9, snakes10, scorpions11, spiders12,13 and bees14 have received a great deal of attention. Intensive investigations have been done in these species to identify and characterize venom proteins with medical values by combining of transcriptomic, proteomic and peptidomic techniques. With an estimated number of species up to 600,000, parasitoid wasps account for around 75% of the described Hymenoptera and 10–20% of all insect species15, representing a group that dwarfs the set of venomous animals mentioned above. Venoms from parasitic Hymenoptera therefore could be an underestimated sources of valuable compounds that have potential use in pest control and pharmacy16,17. A few studies have been conducted on the compositions of several parasitoid venoms18,19,20,21,22,23,24,25,26,27. However, compared to their diversity, little is known about the composition, function and evolutionary relationship of different parasitoid venoms3,4.
Pteromalus puparum is a pupal endoparasitoid wasp that parasitizes a number of butterflies including the agricultural pest small cabbage white butterfly, Pieris rapae28. Pteromalus puparum belongs to the same subfamily Pteromalinae as the model parasitoid wasp Nasonia vitripennis. But, in contrast, N. vitripennis is an ectoparasitoid that parasitizes the pupae of various flies29. There are a number of independent origins of endoparasitoids evolving from ectoparasitoids, including in braconids, ichneumonids, chrysidoids, chalcidoids, ceraphronoids, evanioids and so on30. P. puparum represents a relatively recent evolution of endoparasitism within the subfamily Pteromalinae and thus may provide a good model for comparative studies with N. vitripennis to better understand the differences and evolutionary relationship between endo- and ectoparasitoids.
Similar to N. vitripennis, venom from P. puparum is considered to be the major maternal factor that alters both the host immunity and physiology to facilitate the development of progenies28,31. No other virulence factors such as PDV, VLP and teratocyte, have been found in P. puparum so far. Our previous studies showed that P. puparum venom could inhibit both the cellular and humoral immunity of host and regulate host development and metabolism28,32,33,34.
In this study, we investigated the P. puparum venom composition by combining both transcriptomic and proteomic approaches. Also taking differential expression and signal peptide analysis into consideration, we finally identified 70 venom gland differentially expressed secretory proteins as putative venom proteins in P. puparum. The results will help us to study the mode actions of these venom proteins and to better understand the evolution of venoms among Hymenopteran parasitoids.
Results
Assembly and analyses of P. puparum transcriptome
Three cDNA libraries were separately constructed and sequenced for transcriptome assembly: whole female adults, venom glands and female body carcasses without venom apparatus. Then all the raw data was filtered and de novo assembled to create a P. puparum transcriptome (Fig. 1). Assembly statistics showed that the N50 was 2226 bp and N80 was 825 bp (Table 1). Unigene represents a set of transcripts from the same transcription locus. Here the longest copy of redundant transcripts was regarded as a unigene. Finally, 39,738 unigenes that represented 55,958 transcripts were obtained (supplementary Figure S1). Among all unigenes, 43.73% (17,379) unigenes got matches in the nr database using blastx (e-value < 1e−5).
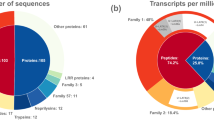
Venom gland cells from parasitoid wasps secrete venoms into the lumen of venom glands. Therefore, venom proteins are expected with secretory signal peptides in their amino acid sequences. For signal peptide analysis, transcripts with complete N terminal (subject start position of the best hit alignment = 1) were computationally translated into proteins and subjected to software SignalP. Simultaneously, their best hit sequences in nr database were also retrieved from NCBI as reference sequences for signal peptide analysis. The identity of the results by two different methods was 99% (Supplementary Table S1). Therefore, the signal peptide analysis of all unigenes was finally conducted using reference sequences. By this method, 2714 unigenes with signal peptides were identified in P. puparum combined transcriptome in total (Fig. 2A).
Identification of 70 putative venom proteins from Pteromalus puparum.
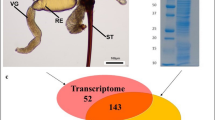
(A) Venn diagram of putative venom proteins. The orange circle indicates unigenes which were identified by proteomic approach, the purple circle indicates unigenes with signal peptides and the light green circle indicates unigenes which are differentially expressed in venom gland. (B) The SDS-PAGE (12%) analysis of venom protein. 21 gel slices are indicated by numbers on the right and shown in red boxes. The sizes and positions of molecular weight standards are indicated on the left. (C) The composition of venom proteins. DEG in VG: differentially expressed unigenes in venom gland; SignalP: signal peptide.
The expression levels of unigenes in both venom gland and carcass without venom apparatus were estimated by software eXpress35,36. To control false positive rate, the expression level cut-off was set as FPKM_VG (Venom gland) >10 and a venom gland to carcass expression ratio log2 (FPKM_VG/FPKM_Carcass) >1 and corrected P-value < 0.001 to define differentially expressed genes in venom gland. By this criterion, 2355 unigenes were identified differentially expressed in venom gland relative to carcass (whole female body minus the venom apparatus) (Fig. 2A).
Identification of venom proteins by proteomic approach
For proteomic identification, venom proteins were separated by SDS-PAGE. Several apparent bands were observed with molecular masses ranging from less than 14 kDa to more than 97 kDa (Fig. 2B). And the most abundant band was a little below 66 kDa. The SDS-PAGE gel was cut into 21 slices as the graph showed (Fig. 2B). These slices were in-gel digested by trypsin and subjected to LC-MS/MS to identify the proteins. The database for proteomic research was generated by computationally translating transcriptomic sequences into proteins according to the blastx results. Finally, 630 unigenes were identified from the venom reservoir by this approach (Fig. 2A).
Identification of putative venom proteins by combined analyses of transcriptomic and proteomic information
To identify a robust set of venom proteins, all data were analyzed under the assumption that venom proteins were secretory and differentially expressed in venom gland (Fig. 1). Combined transcriptomic and proteomic information, 70 unigenes were identified as secretory, differentially expressed in venom gland and confirmed by proteomic approach (Fig. 2A). In this study, these unigenes were defined as putative venom proteins for further analysis.
These 70 putative venom proteins were categorized into enzymes (38 records), protease inhibitors (4 records), recognition and binding proteins (4 records), others (6 records) and unknown (17 records) (Fig. 2C). The most abundant category (54%) is “enzymes”, which included esterase, serine proteases, metalloproteases, enzymes involved in DNA metabolism and so on and the second (24%) is “unknown”. These proteins are described in more detail in supplementary file 1.
Because the transcriptomic and proteomic sequencing are not replicated, gene expression in the venom gland was examined for 34 putative venom protein genes by qPCR and 8 proteins were examined for their presence in venom reservoirs by Western blotting. All 34 tested venom protein genes were differentially expressed in venom gland related to carcass (Fig. 3A), all 8 proteins were confirmed by Western blotting using antibodies to the specific venom proteins (Fig. 3B). These results showed that the putative venom proteins set in this study is reliable.
Verification of putative venom proteins by quantitative real-time PCR (qPCR) and Western blotting.
(A) qPCR verification of selected putative venom proteins. The genes and primers used for these proteins are listed in Table S1. (B) Western blotting of venom proteins from P. puparum and N. vitripennis. β-Actin was used as housekeeping protein. The accession or unigene numbers of these venom proteins are as follows. calreticulin (GenBank: ACZ68113), serine protease 22 (comp44498_c3), serine protease homolog 29 (comp44055_c7), venom protein U (comp22466_c0), GOBP-like venom protein (comp39522_c0), lipase-like venom protein (comp28596_c0), serine protease 87 (comp43143_c1), GILT-like (comp36384_c0). VA: venom apparatus; Carcass: whole body of female adult without venom apparatus; GILT-like: gamma-interferon-inducible lysosomal thiol reductase-like.
Similarity comparison of P. puparum venom proteins to N. vitripennis and other endoparasitoid venoms
Comparisons of P. puparum venoms to venom and non-venom proteins in other parastitoids were investigated by three general methods. In our initial comparisons, we performed a blastx of P. puparum venom proteins against the nr database. Excluding self matches, the large majority (68 of 70) of P. puparum venom proteins have a best hit to proteins from the ectoparasitoid, N. vitripennis (Fig. 4A, Table 2). The remaining two gave best matches to a venom protein from the parasitoid Chelonus inanitus and a non-venom protein from the bee Megachile rotundata, respectively.
Comparison of venom proteins from Pteromalus puparum to Nasonia vitripennis and other endoparasitoid venoms.
(A) Species distribution of the top BLASTX hit in the nr database for putative venom proteins from P. puparum. (B) The similarity comparison among P. puparum, N. vitripennis and other endoparasitoid venoms by BLASTP with a cutoff e-value ≤ 1e−5 and bit score ≥50. The green numbers indicate hits from P. puparum venom, the red numbers indicate hits from N. vitripennis and the blue numbers indicate hits from other endoparasitoid venoms. As numbers of similar kind of proteins can be different in different venom sets, the hits from different venoms can be different in same category. PpVen: venom proteins from P. puparum. NvVen: venom proteins from N. vitripennis. OEPVen: venom proteins from other endoparasitoid wasps.
We next specifically compared P. puparum venom proteins to venoms reported in N. vitripennis and to our database of venoms from other endoparasitoids (OEP, see methods) using blastp (Supplementary Table S2). Of these 70 venoms, 48 (68.6%) gave better e-values and 46 (65.7%) gave better bit scores to N. vitripennis venom proteins than to OEP venoms. Therefore, most P. puparum venoms are more similar to N. vitripennis than OEP venoms (e-value Wilcoxon matched signs rank (WMSR) test, p = 0.009, Supplementary Figure S2, bit score WMSR test, p = 0.001, Supplementary Figure S3). This likely reflects the closer evolutionary relationship of the endoparasitoid P. puparum to the ectoparasitoid N. vitripennis, which are in the same subfamily Pteromalinae with similar morphology (Supplementary Figure S4), than to species in the OEP, which occur in other families and superfamilies of parasitoids (e.g. Leptopilina boulardi, L. heterotoma21, Aphidius ervi20, Microplitis demolitor27, Microctonus sp24. and C. inanitus18).
Using cut-off criteria (e-value ≤ 1e−5, bit score ≥ 50, see methods), we then assigned all proteins from the three venom data sets to shared and unshared categories (Fig. 4B). Based on the criteria, 14 venom proteins were found to be unique to P. puparum, while 25 were shared only with N. vitripennis, 13 were shared only with OEP and 18 were shared among all three sets. Pteromalus puparum venom proteins are significantly more likely than are N. vitripennis venom proteins to show similarities only to OEP venoms (13/70 versus 3/79, two tailed fisher extract test, p = 0.006). Examples includ adenosine deaminase CECR1-like, protein lethal (2) essential for life-like, disulfide-isomerase A3-like, pancreatic triacylglycerol lipase-like, GILT-like, protein FAM151A-like. This set of venom proteins which only shared between P. puparum and OEP may have a role in the adaptation to endoparasitism.
Twenty-five venom proteins in P. puparum and 22 in N. vitripennis were shared in P. puparum and N. vitripennis venom only and might be Pteromalinae venom specific. Some venom proteins which were previously reported as unique in N. vitripennis were also detected in P. puparum venom, including venom protein D, G, J, L, O, U and Z. Eighteen venoms were shared among all three data sets and might present a core of venom proteins in parasitoid wasps, including venom allergen, calreticulin, serine protease, acid phosphatase, glucose dehydrogenase, gamma-glutamyltranspeptidase and so on (Supplementary Table S2).
Test of P. puparium venom antibodies against N. vitripennis venom
Antibodies against P. puparum venom proteins were tested on N. vitripennis venom to see whether they could cross detect N. vitripennis venom proteins. Antibodies against P. puparum calreticulin, GOBP-like venom protein, venom protein U, serine protease 22 and serine protease homolog 29 could also cross detect the venom proteins in N. vitripennis (Fig. 3B). The results support the view that similar proteins are present in N. vitripennis venom and share antigenic similarities. Western blotting results also showed several venom proteins were not detected in N. vitripennis venom by the antibodies against P. puparum venom proteins (Fig. 3B). GILT-like protein was absent in the venom set of N. vitripennis and as expected, couldn’t be cross detected in N. vitripennis venom by antibody against P. puparum GILT-like protein. And antibodies against P. puparum lipase-like venom protein and serine protease 87 also failed to cross detect the venom proteins in N. vitripennis. These failures might be caused by the divergence of antigens between P. puparum and N. vitripennis venom proteins, which could be sequence and/or modification differences.
Discussion
Using high throughput RNA-sequencing technology, we first assembled the transcriptome of P. puparum, which is a pupal endoparasitoid. Differential expression analysis and signal peptide analysis were conducted to search the differentially expressed secretory proteins in venom gland. In parallel, we used the shotgun proteomic approach to analyze the composition of venom. Combined the venom proteomic data with the transcriptomic information, we finally identified a robust set of putative venom proteins.
In this study, we assumed that venom proteins from P. puparum were secretory and differentially expressed in venom gland. However, proteins that were not differentially expressed in venom gland could not be totally excluded, as venom proteins, such as heat shock proteins and arginine kinases that are commonly found in parasitoid venoms. In some extreme cases, like L. boulardi21, venom proteins were even not specifically expressed in the venom gland. In P. puparum, most venom proteins are likely to be differentially expressed in the venom gland, as confirmed by qPCR in this study. In addition, many unigenes (116) from the whole body transcriptome encoded secretory proteins and were significantly more highly expressed in venom gland, but not identified by the proteomic approach. These proteins could just have local functions in the venom gland or have been missed by the proteomic approach, especially for small peptides which may not be retained by SDS-PAGE and are easy to be missed especially when there was a lack of genomic information.
Despite the rigorous filtering, the venom composition of P. puparum is still found to be quite complex. It is reasonable to believe that parasitoid venoms are much more complex than venoms from social Hymentoptera23. The parasitoid venoms must target immunity, development, metabolism and sometimes even the host nervous system to ensure successful parasitism4,23. This is quite different from the function of venoms from social Hymenoptera, which are mainly used for predation and defense.
Pteromalus puparum evolved endoparasitism from an ectoparasitoid ancestor relatively recently within the pteromalids. In the subfamily Pteromalinae, the majority of species are ectoparasitoids, such as Urolepis rufipes37, Trichomalopsis sarcophagae38, Muscidifurax raptor39, Nasonia and so on. There are also several ectoparasitoid wasps in the genus Pteromalus. For example, both P. cerealellae40 and P. sequester41 are solitary ectoparasitoids of larvae of Coleoptera. Moreover, according to the phylogenetic analysis and substitution rate results of calreticulin from parasitoid wasps, P. puparum and N. vitripennis has a relatively small evolutionary distance (supplementary Figure S5, Table S3). The evolutionary distance between P. puparum and N. vitripennis is even smaller than that between L. boulardi and L. heterotoma, which are in the same genus and have been intensively compared25,42. Thus, P. puparum and N. vitripenis provide a good model for comparative studies between endo- and ectoparasitoids and particularly to the evolutionary changes that occur when endoparasitism evolves from ectoparasitism.
As expected, most of (68/70) the identified venom proteins from the endoparasitoid P. puparum had significant similarities with proteins from the ectoparasitoid wasp N. vitripennis, which belongs to the same subfamily (Pteromalinae). Moreover, most of P. puparum venom proteins showed higher similarities to venom proteins from N. vitripennis rather than to other reported endoparasitoids. All these results are consistent with the fact that these endoparasitoids have different independent origins from ectoparasitoids30.
Although endoparasitoid wasps have different independent evolutionary origins, convergent recruiting of some similar proteins could still be expected. Strikingly, several P. puparum venoms are only shared with other reported endoparasitoids and not present in venoms of its closest sequenced relative, N. vitripennis, which is an ectoparasitoid. These venom proteins may have a role in the adaptation to endoparasitism. However, it is also possible that this pattern is caused by incomplete characterization of the venom repertoire of these species. Further investigation is therefore needed.
The development of the stinger and venoms in Hymenoptera had a single origin30. So it is expected that parasitoid wasps might contain some ancestral venom proteins. Venom antigen 5 is an example of such conservation as it is found from social Hymenoptera to parasitoid wasps (Supplementary Figure S6). In addition, different proteins have been recruited into venom for similar functions in different parasitoid wasps. These include superoxide dismutase (SOD) from Leptopilina boulardi and L. heterotoma43 and unrelated peptide Vn 4.6 from Cotesia rubecula44 which are known to inhibit the pro-phenoloxidase activation. A second example is RhoGAP (Ras homologous GTPase activating protein) from L. boulardi45, VPr3 from Pimpla hypochondriaca46,47 and SERCA (sarco/endoplasmic reticulum calcium ATPase) from Ganaspis sp.148, which are very different proteins, but all are known to alter the behavior of host hemocytes.
Taking into consideration the complexity and diversity of parasitic factors delivered to hosts (including venom, PDV and others), parasitoid wasps seem to be an untapped source of valuable molecules with agricultural and medical potential16,17. Of course, a lot of work on the compositions of parasitoid venom, functions and applications of individual venom proteins is still needed. The identification of venom composition from P. puparum in this current study is the basis for further detailed analyses of the functions of these venom proteins.
Our research revealed closer relationship of most P. puparum venom proteins to those from the pupal ectoparasitoid N. vitripennis, rather than to other reported endoparasitoid wasps. Thirteen P. puparum venom proteins show similarity to other endoparasitoid venoms but not to venom proteins of the more closely related ectoparasitoid N. vitripennis. These proteins are promising candidates for a functional role in the evolution of endopariasitism. These results will open the way to a better understanding of venom evolution in the transition from ectoparasitoids to endoparasitoids.
Methods
Insect rearing
Laboratory cultures of P. puparum and N. vitripennis were maintained at 25 °C with a photoperiod of 14: 10 h (light: dark) as described previously28,31 and used in all experiments. Once emerged, the wasp females were collected and held in glass containers, fed ad lib on 20% (v/v) honey solution to lengthen life span.
Venom gland preparation and RNA isolation
Mated female wasps aged 0–7 days were anaesthetized in −70 °C refrigerator for 5 min and dissected in Ringer’s saline (KCl 182 mM; NaCl 46 mM; CaCl2 3 mM; Tris-HCl 10 mM) with 1 unit/μl RNAase inhibitor (TOYOBO, Osaka, Japan) on the ice plate under a stereoscope (Olympus). Venom glands and carcasses without venom apparatus were collected into Trizol reagent (Invitrogen, USA), respectively. The total RNA was extracted using Trizol reagent according to the manufacture’s protocol.
Construction and sequencing of cDNA library
The construction and sequencing of cDNA library were done by Beijing Genomics Institute (BGI, Shenzhen, China). Briefly, the isolated RNA was purified using Sera-mag Magnetic Oligo (dT) beads (Illumina), then transcribed using N6 primers followed by synthesis of second cDNA strand. After end pair processing and ligation of adaptor, RNA was amplified by PCR and purified using QIAquick Gel extraction Kit (Qiagen, Germany). The cDNA library of whole female adult was sequenced on Illumina Hiseq 2000 with paired-end reads of 100 bp. The cDNA libraries of venom gland and carcass were sequenced on 1G Illumina Genome Analyzer (Illumina, San Diego, USA) with paired-end reads of 75 bp.
Analysis of transcriptomic data
The transcriptomic raw data was assembled using Trinity v2013-02-1649. All unigenes were annotated by blastx search against NCBI nr database (March, 2013) with a cutoff of 1e−5. The expression level was estimated by software eXpress v1.3.335,36. Differential expression analysis between venom gland and carcass was performed using the R package DEGSeq v1.2.250. The p-values were adjusted using the Benjamini & Hochberg method. Corrected p-value < 0.001, log2 (FPKM_VG/FPKM_Carcass) >1 and FPKM_VG (Venom gland) >10 were set as the threshold for significantly differential expression in venom gland. Presence of signal peptides was analyzed by software SignalP 4.151. The putative venom unigenes were manually checked using blastx on NCBI website and categorized into enzymes, protease inhibitors, recognition and binding proteins, others and unknown based on their blast results and domain information.
Comparison of P. puparum venom proteins to N. vitripennis and other endoparasitoid venoms
For similarity comparison, blastp were performed between three different venom data sets, P. puparum venom, N. vitripennis venom23 and a database of other endoparasitoid venoms generated here. The other endoparasitoid venom database was manually generated, including venom proteins from Leptopilina boulardi, L. heterotoma21, Aphidius ervi20, Microplitis demolitor27, Microctonus sp24. and Chelonus inanitus18. All the nucleotide acid sequences from P. puparum venom and other endoparasitoid venoms were translated into proteins by OrfPredictor40 (http://proteomics.ysu.edu/tools/OrfPredictor.html). And venom proteins in other endoparasitoid venom database were further clustered by CD-HIT41 with sequence identity cutoff=0.5 (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index.cgi?cmd=cd-hit) to remove redundancy. As bit score is independent on database size and more suitable than e-value for comparing similarity scores from different searches (http://www.ncbi.nlm.nih.gov/BLAST/tutorial/), we analyzed a range of bit scores to determine how these criteria affect similarity scores among the different venom protein sets (Supplementary Table S4). Criteria that incorporated a bit score criterion (e-value ≤ 1e−5, bit sore ≥ 50) was used for the further analyses.
Multiple amino acid sequence alignments were performed using MUSCLE v3.852. Phylogenetic analysis was conducted by MEGA 5 using maximum likelihood algorithm53. Pairwise substitution rates were calculated by CodeML in PAML v4.854.
Extraction of venom proteins
Mated female wasps aged 0–7 days of P. puparum and N. vitripennis, were anaesthetized in −70 °C refrigerator for 5 min as mentioned above and then dissected in Ringer’s saline (KCl 182 mM; NaCl 46 mM; CaCl2 3 mM; Tris-HCl 10 mM) with 1 mM phenylmethanesulfonyl fluoride (PMSF) (Sigma, St. Louis, MO) on the ice plate under a stereoscope (Olympus). The venom reservoirs were washed for several times and then transferred to an Eppendorf tube. After centrifugation at 16,000 g and 4 °C for 1 min, the supernatant was filtered with 0.22 μm Millipore filter and stored at −70 °C until use. The concentration of venom protein was determined using Bradford method55.
Mass spectrometric venom protein identification by LC-MS/MS
Pteromalus. puparum venom sample containing 100 μg proteins dissolved in 20 μl rehydration solution (7 M urea, 2 M thiourea, 4% CHAPS, 0.5% Triton X-100, 65 mM DTT, 0.5% Bio-Lyte and 0.001% bromophenol blue) were separated by SDS-PAGE and stained with Coomassie Brilliant Blue R-250 (Bio-Rad, USA). The gel was excised into 21 slices, depending on the molecular masses of protein bands. Each gel slice was digested by trypsin and lyophilized separately followed by 1DLC-LTQ-Velos (Thermo Finnigan, San Jose, CA). In this study, samples were desalted on Zorbax 300 SB-C18 (Agilent Technologies, Wilmington, DE) and then separated on a RP-C18 column (150 μm i.d., 150 mm length) (Column technology Inc., Fremont, CA). The buffer A was water with 0.1% formic acid, buffer B was 84% acetonitrile with 0.1% formic acid and the gradient was from 4% buffer B to 50% buffer B in 1h. The charge-to-mass ratios of peptides and fractions of peptides were collected 20 times after every full scan. The resulting MS/MS spectra were searched against the translated P. puparum transcriptome using Sequest search algorithm56. Carbamidomethyl of cysteine and oxidation of methionine were set as fixed and variable modifications, respectively. Delta CN (≥0.1) and cross-correlation scores (Xcorr, one charge ≥1.9, two charges ≥2.2, three charges ≥3.75) were used to filter the peptide identification. This part was done by Shanghai Applied Protein Technology Co., Ltd (Shanghai, China).
Quantitative real-time PCR (qPCR)
cDNA from venom glands and carcasses without venom apparatus was synthesized, respectively, using TransScript one-step gDNA Removal and cDNA Synthesis SuperMix (TransGen, China) with random primers. All the primer sequences (Supplementary Table S5) used were designed on website Primer 357 and synthesized commercially (Sangon, Chnia). The PCR reaction was run in ABI 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) using SsoFast EvaGreen Supermix with Low Rox (Bio-Rad, USA) according to the manufacture’s protocol. The cycling conditions for qPCR were as follows: enzyme activation at 95 °C for 30 sec, followed by 40 cycles with denaturation at 95 °C for 5 sec, annealing at 60 °C for 34 sec. Relative expression level of putative venom proteins was normalized to reference gene (18S rRNA) using 2−ΔΔCT method58. Statistical analysis was performed using Student’s t test. Unigenes with log2(Expression ratio venom gland/carcass) >1 and p-values < 0.05 were considered differentially expressed in venom gland.
Western blotting
The antibodies against different P. puparum venom proteins were prepared as previously described59. Recombinant venom protein GOBP was expressed in the pGEX-4T-2 vector with a GST tag, others were expressed in pET-28a vector with a His tag. The primary antibody against β-actin was bought commercially (Huabio, China). The venom and carcass proteins of P. puparum and N. vitripennis were separated by 12% SDS-PAGE and then transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, USA) using Mini-ProTEAN Tetra system (Bio-Rad, Hercules, CA) at 16 V for 16 h. The PVDF membrane was blocked and washed. Anti-venom protein antibodies (diluted from 1: 500 to 1: 2000, depending on the antibody) and anti-actin antibody (diluted 1: 5000) were respectively used as the primary antibody. And goat anti-rabbit IgG-horseradish peroxidase (HRP) conjugate (Sigmae Aldrich, Taufkirchen, Germany; diluted 1: 5000) was used as the secondary antibody. The PVDF membranes were detected using ECL Western Blotting Substrate (Promega, Madison, WI, USA) and imaged in Chemi Doc-ItTM 600 Imaging System (UVP, Cambridge, UK).
Availability of supporting data
All RNA-seq raw data have been deposited at the NCBI Sequence Read Archive under accession number SRP055738. This Transcriptome Shotgun Assembly project has been deposited at GenBank under the accession GECT00000000. The version described in this paper is the first version, GECT01000000.
Additional Information
How to cite this article: Yan, Z. et al. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. 6, 19604; doi: 10.1038/srep19604 (2016).
References
Quicke, D. L. Parasitic Wasps, (Chapman and Hall, 1997).
Pennacchio, F. & Strand, M. R. Evolution of developmental strategies in parasitic hymenoptera. Annu. Rev. Entomol. 51, 233–258 (2006).
Asgari, S. Venom proteins from polydnavirus-producing endoparasitoids: their role in host-parasite interactions. Arch. Insect Biochem. Physiol. 61, 146–156 (2006).
Asgari, S. & Rivers, D. B. Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 56, 313–335 (2011).
Mrinalini et al. Parasitoid venom induces metabolic cascades in fly hosts. Metabolomics 11, 350–366 (2015).
Martinson, E. O. et al. Nasonia vitripennis venom causes targeted gene expression changes in its fly host. Mol. Ecol. 23, 5918–5930 (2014).
Schmidt, O., Theopold, U. & Strand, M. Innate immunity and its evasion and suppression by hymenopteran endoparasitoids. Bioessays 23, 344–351 (2001).
Pimenta, A. M. C. & De Lima, M. E. Small peptides, big world: biotechnological potential in neglected bioactive peptides from arthropod venoms. J. Pept. Sci. 11, 670–676 (2005).
Lewis, R. J., Dutertre, S., Vetter, I. & Christie, M. J. Conus venom peptide pharmacology. Pharmacol. Rev. 64, 259–298 (2012).
Koh, D. C. I., Armugam, A. & Jeyaseelan, K. Snake venom components and their applications in biomedicine. Cell. Mol. Life Sci. 63, 3030–3041 (2006).
de la Vega, R. C. R., Schwartz, E. F. & Possani, L. D. Mining on scorpion venom biodiversity. Toxicon 56, 1155–1161 (2010).
King, G. F. & Hardy, M. C. Spider-venom peptides: structure, pharmacology and potential for control of insect pests. Annu. Rev. Entomol. 58, 475–496 (2013).
Sanggaard, K. W. et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat. Commun. 5 (2014).
Peiren, N. et al. The protein composition of honeybee venom reconsidered by a proteomic approach. Biochim. Biophys. Acta 1752, 1–5 (2005).
Heraty, J. In Insect Biodiversity: Science and Society (eds Peter H. A. & Robert G. F. ) 445–462 (Wiley-Blackwell, 2009).
Moreau, S. J. M. & Guillot, S. Advances and prospects on biosynthesis, structures and functions of venom proteins from parasitic wasps. Insect Biochem. Mol. Biol. 35, 1209–1223 (2005).
Danneels, E. L., Rivers, D. B. & de Graaf, D. C. Venom proteins of the parasitoid wasp Nasonia vitripennis: recent discovery of an untapped pharmacopee. Toxins 2, 494–516 (2010).
Vincent, B. et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genomics 11, 693 (2010).
Parkinson, N. M. et al. Towards a comprehensive view of the primary structure of venom proteins from the parasitoid wasp Pimpla hypochondriaca. Insect Biochem. Mol. Biol. 34, 565–571 (2004).
Nguyen, T. T. A. et al. Early presence of an enolase in the oviposition injecta of the aphid parasitoid Aphidius ervi analyzed with chitosan beads as artificial hosts. J. Insect Physiol. 59, 11–18 (2013).
Goecks, J. et al. Integrative approach reveals composition of endoparasitoid wasp venoms. Plos One 8 (2013).
Doremus, T. et al. Venom gland extract is not required for successful parasitism in the polydnavirus-associated endoparasitoid Hyposoter didymator (Hym. Ichneumonidae) despite the presence of numerous novel and conserved venom proteins. Insect Biochem. Mol. Biol. 43, 292–307 (2013).
de Graaf, D. C. et al. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 19, 11–26 (2010).
Crawford, A. M. et al. The constituents of Microctonus sp. parasitoid venoms. Insect Mol. Biol. 17, 313–324 (2008).
Colinet, D. et al. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: the case of Leptopilina parasitoids of Drosophila. Insect Biochem. Mol. Biol. 43, 601–611 (2013).
Colinet, D. et al. Identification of the main venom protein components of Aphidius ervi, a parasitoid wasp of the aphid model Acyrthosiphon pisum. BMC Genomics 15, 342 (2014).
Burke, G. R. & Strand, M. R. Systematic analysis of a wasp parasitism arsenal. Mol. Ecol. 23, 890–901 (2014).
Cai, J., Ye, G. Y. & Hu, C. Parasitism of Pieris rapae (Lepidoptera: Pieridae) by a pupal endoparasitoid, Pteromalus puparum (Hymenoptera: Pteromalidae): Effects of parasitization and venom on host hemocytes. J. Insect Physiol. 50, 315–322 (2004).
Werren, J. H. et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science 327, 343–348 (2010).
Whitfield, J. B. Phylogeny and evolution of host-parasitoid interactions in hymenoptera. Annu. Rev. Entomol. 43, 129–151 (1998).
Zhang, Z., Ye, G. Y., Cai, J. & Hu, C. Comparative venom toxicity between Pteromalus puparum and Nasonia vitripennis (Hymenoptera: Pteromalidae) toward the hemocytes of their natural hosts, non-target insects and cultured insect cells. Toxicon 46, 337–349 (2005).
Fang, Q. et al. Pteromalus puparum venom impairs host cellular immune responses by decreasing expression of its scavenger receptor gene. Insect Biochem. Mol. Biol. 41, 852–862 (2011).
Fang, Q. et al. Venom of parasitoid, Pteromalus puparum, suppresses host, Pieris rapae, immune promotion by decreasing host C-type lectin gene expression. Plos One 6 (2011).
Zhu, J. Y., Ye, G. Y., Dong, S. Z., Fang, Q. & Hu, C. Venom of Pteromalus puparum (Hymenoptera: Pteromalidae) induced endocrine changes in the hemolymph of its host, Pieris rapae (Lepidoptera: Pieridae). Arch. Insect Biochem. Physiol. 71, 45–53 (2009).
Roberts, A. & Pachter, L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10, 71–U99 (2013).
Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. & Pachter, L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 12 (2011).
Stenseng, L., Skovgård, H. & Holter, P. Life table studies of the pupal parasitoid Urolepis rufipes (Hymenoptera: Pteromalidae) on the house fly Musca domestica (Diptera: Muscidae) in Denmark. Environ. Entomol. 32, 717–725 (2003).
Floate, K. Field trials of Trichomalopsis sarcophagae (Hymenoptera: Pteromalidae) in cattle feedlots: a potential biocontrol agent of filth flies (Diptera: Muscidae). Can. Entomol. 135, 599–608 (2003).
Geden, C. J., Johnson, D. M., Kaufman, P. E. & Boohene, C. K. Competition between the filth fly parasitoids Muscidifurax raptor and M. raptorellus (Hymenoptera: Pteromalidae). J. Vector Ecol. 39, 278–287 (2014).
Howard, R. W. & Baker, J. E. Morphology and chemistry of dufour glands in four ectoparasitoids: Cephalonomia tarsalis, C. waterstoni (Hymenoptera: Bethylidae), Anisopteromalus calandrae and Pteromalus cerealellae (Hymenoptera: Pteromalidae). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 135, 153–167 (2003).
Gurreaa, M. P., Canoa, J. M. & Fisher, J. R. The ability of an artificial diet to sustain larvae of Exapion ulicis (Coleoptera: Apionidae) and the Occurrence of Pteromalus sequester (Hymenoptera: Pteromalidae) from field-collected larvae in Oregon. Fla. Entomol., 89, 405–406.
Schlenke, T. A., Morales, J., Govind, S. & Clark, A. G. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 3, 1486–1501 (2007).
Colinet, D., Cazes, D., Belghazi, M., Gatti, J.-L. & Poirie, M. Extracellular superoxide dismutase in insects: characterization, function and interspecific variation in parasitoid wasp venom. J. Biol. Chem. 286, 40110–40121 (2011).
Asgari, S., Zareie, R., Zhang, G. & Schmidt, O. Isolation and characterization of a novel venom protein from an endoparasitoid, Cotesia rubecula (Hym: Braconidae). Arch. Insect Biochem. Physiol. 53, 92–100 (2003).
Labrosse, C. et al. A RhoGAP protein as a main immune suppressive factor in the Leptopilina boulardi (Hymenoptera, Figitidae)-Drosophila melanogaster interaction. Insect Biochem. Mol. Biol. 35, 93–103 (2005).
Richards, E. H. & Dani, M. P. Biochemical isolation of an insect haemocyte anti-aggregation protein from the venom of the endoparasitic wasp, Pimpla hypochondriaca and identification of its gene. J. Insect Physiol. 54, 1041–1049 (2008).
Dani, M. P. & Richards, E. H. Cloning and expression of the gene for an insect haemocyte anti-aggregation protein (VPr3), from the venom of the endoparasitic wasp, Pimpla hypochondriaca. Arch. Insect Biochem. Physiol. 71, 191–204 (2009).
Mortimer, N. T. et al. Parasitoid wasp venom SERCA regulates Drosophila calcium levels and inhibits cellular immunity. Proc. Natl. Acad. Sci. USA 110, 9427–9432 (2013).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Wang, L., Feng, Z., Wang, X., Wang, X. & Zhang, X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010).
Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 1–19 (2004).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Untergasser, A. et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Wang, L. et al. Inhibition of host cell encapsulation through inhibiting immune gene expression by the parasitic wasp venom calreticulin. Insect Biochem. Mol. Biol. 43, 936–946 (2013).
Acknowledgements
This research was supported by grants from China National Program on Key Basic Research Projects (973 Program, 2013CB127600), National Natural Science Foundation of China (Grant no. 31272098, 31472038), National Science Fund for Innovative Research Groups of Biological Control (Grant no. 31321063) and China National Science Fund for Distinguished Young Scholars (Grant no. 31025021). Work by John Werren is supported by US NIH (RO1GM098667). We thank Ellen Martinson and Mrinalini for access to data analyses in Nasonia.
Author information
Authors and Affiliations
Contributions
G.Y., J.H.W. and Q.F. conceived and designed the research; Z.Y., L.W., Y.Z. and F.W. perfomed the experiments; Z.Y., J.L. and F.L. analyzed the data. G.Y., J.H.W., Q.F. and Z.Y. wrote and revised the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yan, Z., Fang, Q., Wang, L. et al. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci Rep 6, 19604 (2016). https://doi.org/10.1038/srep19604
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19604
This article is cited by
-
Genome of the parasitoid wasp Cotesia chilonis sheds light on amino acid resource exploitation
BMC Biology (2022)
-
Shrew's venom quickly causes circulation disorder, analgesia and hypokinesia
Cellular and Molecular Life Sciences (2022)
-
An integrated transcriptomic and proteomic approach to identify the main Torymus sinensis venom components
Scientific Reports (2021)
-
Venomics of the ectoparasitoid wasp Bracon nigricans
BMC Genomics (2020)
-
Identification and characterization of serine protease inhibitors in a parasitic wasp, Pteromalus puparum
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.