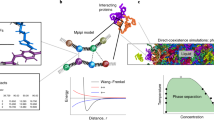
Abstract
In order to investigate the contribution of individual amino acids to protein and peptide solubility, we carried out 100 ns molecular dynamics (MD) simulations of 106 Å3 cubic boxes containing ~3 × 104 water molecules and 27 tetra-peptides regularly positioned at 23 Å from each other and composed of a single amino acid type for all natural amino acids but cysteine and glycine. The calculations were performed using Amber with a standard force field on a special purpose MDGRAPE-3 computer, without introducing any “artificial” hydrophobic interactions. Tetra-peptides composed of I, V, L, M, N, Q, F, W, Y and H formed large amorphous clusters and those containing A, P, S and T formed smaller ones. Tetra-peptides made of D, E, K and R did not cluster at all. These observations correlated well with experimental solubility tendencies as well as hydrophobicity scales with correlation coefficients of 0.5 to > 0.9. Repulsive Coulomb interactions were dominant in ensuring high solubility, whereas both Coulomb and van der Waals (vdW) energies contributed to the aggregations of low solubility amino acids. Overall, this very first all-atom molecular dynamics simulation of a multi-peptide system appears to reproduce the basic properties of peptide solubility, essentially in line with experimental observations.
Similar content being viewed by others
Introduction
Protein solubility and consequently aggregation, is a critical issue in several areas of biochemical and biopharmaceutical research, such as the production and maintenance of protein pharmaceuticals or industrial enzymes1,2. However except for general trends, the physico-chemical aspects of protein solubility are not well understood and a hydrophobic/hydrophilic model has been traditionally applied to analyze solubility data. In general, hydrophobic proteins are aggregation prone3, whereas proteins displaying charged residues on their surfaces are hydrophilic and thus highly soluble4,5. However, the initial purpose of the hydrophobic/hydrophilic model was to describe the equilibrium between an amino acid’s solubility in a non-polar and a polar or aqueous environment6,7.Thus, although this model properly describes the structural stabilization of globular proteins by approximating its interior as non-polar and its exterior as polar8, this does not warrant its suitability for describing protein solubility or aggregation tendency.
Experimental scales for describing amino acid’s solubility and aggregation tendencies independently from hydrophobic scales have been reported. Typically, the solubility of individual amino acids have been measured in the 1970’s and standard values for amino acid solubility have been compiled9. However, such solubility measurements are subject to artifacts because they are carried out at extremely high peptide (or amino acid) concentrations, which can cause jellification, poor phase separation, sticky fluids, pH shifts, etc10.
In order to alleviate issues arising from high molecule’s concentration, two recent studies have measured the contribution of amino acids to the relative solubility change of a model protein using a systematic mutational analysis. In one of them, the solubility of Ribonuclease Sa was examined by systematically mutating Thr76, which is located on its molecular surface, to all of the 20 natural amino acids11. In the second example, 5-residue peptide tags, composed of a single amino acid type, were fused to a model protein, a simplified BPTI variant12 and the amino acid’s contribution to protein’s solubility was determined by measuring the corresponding BPTI variant’s solubility13,14. Overall, these two studies suggest that systematic mutational analysis measuring relative solubility changes, could yield a solubility propensity scale, which might provide an estimate for the relative solubility of a poly-peptide from its amino acid sequence alone.
Molecular dynamics (MD) simulation is a powerful method for investigating protein dynamics as well as its interaction with substrates at atomic level15,16. Recently, it has also been applied to analyzing the molecular interactions at the early stage of self-assembly involving the formation of seed (nucleus) and fibrils in amyloid forming peptides, which are difficult to investigate experimentally15,17,18,19. Realistic simulations of multi-peptide systems are computationally demanding and thus time consuming and implicit solvent20 as well as coarse-grained models of amino acids are often used21. However, given the key role of peptide-water interaction in the solvation or aggregation of peptides, MD simulations with explicit solvent models are required for characterizing peptide solubility from a genuine physico-chemical view point.
The purpose of this study is to shed light into the molecular determinants of peptide and protein solubility associated to amorphous aggregation (aggregation hereafter) using all atom molecular dynamics (MD) simulations of 106 Å3 cubic systems containing approximately 3 × 104 water molecules and 27 tetra-peptides for all 20 amino acids but Gly and Cys. To date, this is a large system for an all-atom MD simulation and the closest attempt reported so far, is an analysis of amyloidogenic aggregation with systems containing merely 2 to 8 peptides22,23. The large scale calculation was made possible by intensive use of a state-of-the-art special-purpose computer system for MD simulations24 and its scale enabled a systematic and detailed statistical analysis of the results, which strongly suggested that amorphous aggregation properties is quantitatively reproducible using first principle interactions. In particular, the ranking of both the fraction of monomers and the mean cluster size agreed with several, but not all, experimentally derived amino acids’ solubility and hydrophobicity scales. Furthermore, MD simulations indicated that peptides that were soluble at low concentration did aggregate at higher concentration, which is very much in line with experimental observation. Markov state model (MSM) analysis revealed that small clusters containing 2 peptides, which may correspond to seeds, accumulated at the initial stage of aggregation18 and then extend to larger ones. Finally, the calculation indicated that high solubility was due to repulsive electrostatic interactions, whereas clusters were stabilized essentially through van der Waals (vdW) interactions, but also accessorily through H-bond and T-stacking interactions. This study is the first of its kind and it shows that simulations based on solely physico-chemical first principles without introducing artificial parameters can indeed reproduce peptide’s solubility roughly in line with experimental observations.
Methods
Model Systems
We performed MD simulations for 18 kinds of tetra-peptides composed of the same four amino acids in this study (e.g. KKKK, RRRR, DDDD, NNNN, etc.), except cysteine and glycine. To remove the effect of the terminal charge and isolate the contribution of the sidechain onto the peptide’s solubility, all the tetra-peptides were capped by an acetyl group (CH3CO-) at the N-terminus and N-methyl (-NHCH3) group at the C-terminal. Asp and Glu were not protonated (denoted as ASP and GLU in AMBER). On the other hand, we performed the calculation for His both protonated and non-protonated at its epsilon nitrogen (named in AMBER as HIP and HIE, respectively).
The initial configuration was prepared by placing 27 tetra-peptides, with parallel orientation, on a 23.0 Å spaced grid (Fig. S1). Sodium and chloride ions were added using the leap module of Amber 8.0 software package25 for neutralizing the net charge of the system. The cubic dimensions, about 106 Å3, were chosen to give a peptide concentration of about 40 mM. The total number of water molecules in each system was approximately 30,00026.
MD Simulation
MD simulations were performed using the Amber 8.0 software package25 on a personal computer (Xeon 3.2 GHz) equipped with special-purpose computer boards for MD simulations, MDGRAPE-327,28. The all-atom point-charge force-field ff9929 was used to represent the peptides. All bond-lengths were constrained to equilibrium lengths using the SHAKE module30. A 2 fs time integration step was used in all simulations. Long range Coulomb interactions were treated with the Particle Mesh Ewald (PME) method31, wherein the real-space component was calculated using MDGRAPE-3, while the host computer calculated the wave number-space component and the bonded-interactions. We used a cut-off radius of 14 Å for the real-space component in order to optimize the balance between the components’ calculation times.
First, energy minimization was performed using the steepest descent protocol, followed by conjugated gradient. After a 5,000 steps energy minimization, the systems were gradually heated from 0 K to 300 K at a heating rate of 6 K/ps. Subsequently, the temperature and pressure were maintained constant at 300 K and 1 atm, respectively, with a coupling constant of 1.0 ps. The MD simulation was performed for 100 ns and data were saved at 10 ps intervals for analysis. Each 100 ns run took approximately 6 months to complete and produced 66 Gbyte of data.
Analysis of Clusters
The solubility of the peptides was quantitatively characterized by analyzing the formation of clusters (or aggregates) and their sizes in the MD trajectories. In this study, a “cluster” was defined when the distance between two atoms belonging to different tetra-peptide chains was less than the sum of their vdW radius. We defined a “cluster size” (CS) as the number of peptides forming a cluster and calculated a mean cluster size (MCS) as MCS = ( ) /N. CS i, t means the cluster size to which peptide i belongs at time t and N is the total number of peptides in the system (27 in this study).
) /N. CS i, t means the cluster size to which peptide i belongs at time t and N is the total number of peptides in the system (27 in this study).
Energy Calculation
VdW energies and electrostatic energies were calculated using, respectively, the molecular mechanics and generalized Born method32, wherein water molecules were replaced with implicit solvent. In the generalized Born calculation, the dielectric constants inside and outside the molecule were set to 1.0 and 78.5, respectively. The H-bonds were calculated with HBPLUS33.
Markov State Model Analysis
We investigated the time dependent cluster’s size distribution using a Markov state model (MSM) and computed a 27 × 27 transition matrix T(Si, Sj) describing the transition among cluster size states. The transition matrix was calculated by counting the total number of peptides that undergo the i to j transition: T(Si, Sj) = ∑t = 0(Si,t → Sj,t + 1), where Si, t and Sj,t+1 represent respectively a cluster size state i and j at time t and t + 1.
The MSM describes the dynamics among the cluster states using a transition matrix P, where its element P(i, j) is the transition’s probability for a peptide, currently in state i (i-mer), to move to state j (j-mer) in the next step. The transition probability P is calculated by normalizing the transition matrix elements with the sum of elements contained in the corresponding row as follows: P(Si, Sj) = T(Si, Sj) / ∑j = 127 (Si,t → Sj,t + 1). The fraction of trajectories in each state (N-mer) after n propagation steps is thus computed as the row vector π (n) = π (0) Pn, where π (0) is a row vector containing the starting fractional populations.
Results and Discussion
Cluster Analysis
We analyzed the formation of clusters during the MD trajectories, as we anticipated that the cluster’s size could be related to the solubility (or aggregation propensity) of an amino acid. From the 100 ns MD simulations, we found that the tetra-peptides made of I, V, L, N, Q, F, M, H, W and Y formed large size amorphous clusters, which consisted of 23 ~ 27 peptides (85 ~ 100% of all peptides in the system), whereas those containing E, D, R and K did not cluster at all (Fig. 1 and Table 1, Fig. S2). The tetra-peptide made of T formed medium size amorphous clusters, consisting of ≤20 peptides (less than 74% of all peptides) and tetra-peptides containing A, P and S formed small clusters containing less than 10 peptides (less than 37% of all peptides). The above observations are generally in line with our expectations drawn from experimental solubility and hydrophobicity data including our own ones14,34. Similar conclusions were reached using the fraction of monomers (Fig. 1B) or monomers and dimers (Fig. S2), which was expected since the monomer fraction correlates well with the MCS. Additionally, we did not observe β sheets or bridges in our clusters (Fig. 2B), as expected since the tetra peptides are not amyloid forming peptides, but perhaps also because the force field do not favor beta- strands formation in contrast with many amyloid MD simulations19,22,23. Overall, the analysis indicated that the results of a full atom molecular dynamics simulation of a multiple peptide system is indeed be related to experimentally determined “solubility” values.
Time-dependence of the mean cluster size (MCS) and number of monomer
(B) for 18 simulated systems.(A) MCS and (B) the number of monomers are, respectively, the mean number of peptides forming cluster and the number of monomeric peptides, as defined using the default cutoff parameters given in the Method section.
Time-dependent number of inter-peptide H-bonds formed
(A) H-bonds between mainchains of Ile4 and Asn4. H-bonds between mainchain and sidechain and sidechain and sidechain are also shown for Asn4 (A). (B) Secondary structures, i.e. coil, strand and helix, formed in Ile4 and Asn4 as calculated using DSSP. The helix faction overlaps with the bottom horizontal axis and is not visible.
Robustness of the Results
To assess the robustness of our results we first examined the influence of the starting configuration on the results. To this end, we repeated the above calculation with 27 randomly disposed Ile4 and Arg4 peptides. Arginine did not cluster in agreement to the above results obtained with peptides disposed at equal intervals on a grid. Similarly, randomly disposed Ile4 clustered in line with our above results (Fig. 3A). Hence, these calculations confirmed that the initial configuration did not significantly influence the aggregation states reported thereafter.
Effect of parameter variation on the MCS analysis.
Time-dependent MCSs obtained from MD simulations starting with a set of 27 tetra-peptides placed randomly (A). Though the MCS of Ile4 after a 100 ns simulation decreased slightly from 27 to 23, the trajectories clearly indicate that the final size will eventually reach the same value. (B) MCSs as a function of time calculated by adding 0.7, 1.4, 2.1 and 2.8 Å to the interatomic vdW distance used for identifying a cluster. (C) MCSs as a function of time calculated by varying the number of atoms pairs for defining a cluster from 1 to 3.
Additionally, we examined possible influence of the initial peptide’s conformation on the aggregation states as reported thereafter. To this end, we performed MD simulations of single tetra-peptides (Ile4 and Arg4) and compared their conformation using the Phi/Psi dihedral angles with those adopted during the initial 10 ns of the 27-tetra-peptides systems (Fig. S3). In brief, the peptides in both the 27-tetra-peptide and isolated tetra-peptide systems adopted, during the initial stage of the simulation, all of the allowed Phi/Psi angle region including the α-helix and the extended (β) structures, strongly suggesting that the initial conformation of the peptides had no or a minimal influence on our calculation.
We next examined the influence of the parameters, namely, the inter-atomic distances and the number of atom pairs, used for defining the clusters. Although the use of stringent cutoff values slightly reduced the cluster sizes, the trends remained the same (Fig. 3B,C), strongly suggesting that the trends are independent from parameters used to define the clusters. In order to investigate why hydrophobic Ala4 did not form a large cluster we increased the number of peptides of Ala4 and Arg4 in the simulation boxes (54 peptides). Ala4 formed a larger cluster at higher concentration (Fig. S4) but Arg4 did not form any cluster. We concluded that, in contrast to the other peptides, the hydrophobicity of Ala4 was observed at higher concentration. We also confirmed the dependency of Ala4 aggregation on peptide-concentration in this result.
Insights into the Molecular Mechanisms of Cluster Formation
Repulsive electrostatic interaction appeared to hamper the formation of clusters for charged tetra-peptides (D, E, R, K; Fig. 1A)14. This view was confirmed by calculating the Coulomb energies of tetra-peptides in clustered and non-clustered states (Fig. 4A). On the other hand, uncharged amino acids, including polar ones, did not exhibit any repulsive effect as expected. Further, both vdW and Coulomb energies showed good correlation with MCS with correlation coefficients of 0.7 ~ 0.9 for systems with MCS > 10.0 (Figs S5, S6 and Table S1).
Time-dependent differences of Coulomb and vdW interaction energies for clustered and non-clustered systems of charged residue peptides.
(A) Coulomb interaction energies (dielectric constant = 1.0) for clustered systems of charged residue peptides; The electrostatic interaction energies of clustered charged peptides, which did not cluster during the MD simulation, were calculated by using the backbone structures of clustered Ile4 peptides and mutating the sidechains to those of the charged amino acids after removal of potential atomic clashes arising from the mutations by energy-minimization. (B) vdW energies; and Coulomb interaction energies computed using a dielectric constant of 80.0 (C) and 1.0 (D).
For the purpose of discussion, let us consider the above results with regard to the physical forces that compose a “solvation energy”, which is used to describe a molecule’s solubility and aggregation in thermodynamic terms but are not explicitly included in all-atom MD simulations25. Overall, the aforementioned results indicate that both vdW and Coulomb interactions contribute to the clustering of the peptides, but their roles are different. The solubility of charged tetra-peptides originated from inter-molecular repulsive electrostatic forces, whereas no repulsion occurred between neutral amino acids, which cluster through vdW interaction (Fig. 4) and H-bonds (see next paragraph). Noteworthy, the strength of repulsive electrostatic interaction between charged residues is much larger than the vdW interaction stabilizing the clusters of neutral amino acids (Fig. 4B,D). Similarly, polar amino acids clustered through vdW interaction but the clusters disintegrated when the electrostatic interaction between partial charges become destabilizing. Though these observations are in line with one’s intuitive anticipation, this is the first systematic and quantitative analysis of the physical forces that drive the solubility of peptides from an atomistic viewpoint.
We also examined the contribution of H-bonds to the formation of clusters. The mainchains of clustered hydrophobic peptides formed a substantial number of H-bonds (Fig. 2A), which was approximately the same as the number of mainchain-mainchain H-bonds in polar peptides (Fig. 2A). Among polar amino acids, the MCS of Thr4 and Ser4 were smaller than those of Gln4 and Asn4 (Fig. 1), but the numbers of mainchain-mainchain H-bonds, both water mediated and non mediated ones, were approximately the same in all four peptides (Table 2). To date, mainchains H-bonds were mostly intermolecular and no secondary structures were formed as assessed by DSSP35 (Fig. 2B).
We further examined H-bonds formed by the sidechains of polar amino acids, as they contain electron acceptors and donors for H-bonding. Sidechains H-bonds exhibited different properties among the polar amino acids: Thr4 and Ser4 formed virtually no direct and very few water mediated H-bonds between their sidechains as they have no electron donors (Table 2). Similarly, Thr4 and Ser4 formed fewer direct sidechain-mainchain H-bonds than Gln4 and Asn4, but almost the same number of water mediated sidechain-mainchain H-bonds were observed in all four tetra-peptides (Table 2). All in all, these results are fully in line with the fact that the sidechains of Asn and Gln, which have both a donor and an acceptor, have a stronger ability to form H-bonds than Ser and Thr sidechains, which have only an acceptor.
Tetra-peptides made of aromatic residues (Y, W, F) formed large clusters (Fig. 1), where several π − π interactions were present (Fig. S7A). Tyrosine’s hydroxyl groups (−OηH) were mostly solvent exposed and formed H-bonds with water molecules, but a few formed a H-bond with a mainchain’s carbonyl group or a -OηH (Figs. S7B,C). Comparison with Phe4 indicated that the presence of −OηH caused only a minor solubility change (Fig. 1), as assessed both by MCS and the fraction of monomers. Although many CH-π stackings were observed in both Tyr4 and Phe4 clusters, face-to-face stacking were present only in Tyr4, not in Phe4 clusters, possibly due to the presence of −OηH in Tyr (Fig. 5). Thus, the phenol ring adopted specific configurations including π- and T- stacking when the rings were close to each other, which is generally in line with previous simulation and PDB database analysis36.
Spacial orientation of closely located aromatic rings.
The distribution of θ and γ angle for pairs with Rcen < 5.5 Å were calculated for Phe4 and Tyr4. A schematic explanation of the parameters (Rcen, θ and γ) describing the ring’s conformation is given at the bottom of the figure. Rcen is the distance between the two geometrical centers of the aromatic rings. θ is the angle between the normal vector to the first ring and the vector relying the two ring centers. γ is the angle between the two ring’s surface normal vectors.
Buried Water Molecules
As mentioned above, several buried peptide groups formed H-bonds, some of which were formed with buried water molecules, which motivated us to further characterize them. We defined buried water as molecules located within 75% of the radius of gyration from the center of gravity. Buried water molecules were observed in all tetra-peptide clusters, including Thr4 and Ser4’s clusters despite their smaller MCS. Clusters of peptides composed of hydrophilic residues tended to contain more buried water molecules than those made of hydrophobic amino acids. Peptide clusters made of aromatic amino acids had high water contents and among all peptides Tyr4 had the highest one (Table 3).
Relationship between Experimental Solubility/Hydrophobicity scales and Cluster Formation
Our calculation uses force field derived from physico-chemical first principles. Aggregation or low solubility is usually considered to be caused by the so called “hydrophobic” interactions37, which, in our calculation originates from the absence of repulsive electrostatic forces, the absence of H-bonds with water molecules, the formation of inter-peptide H bonds and the occurrence of attractive short range vdW interaction as discussed in the above subsection. We thus assessed potential correlation between our results and selected experimental hydrophobicity/solubility scales (Figs 6 and S8). The relative amino acid’s solubility and hydrophobicity were overall well reproduced in our calculation. Namely, hydrophobic and aromatic residues formed clusters and the MCS ranked as A ≪ V < Y < F~W~I~L which is roughly reflected in the hydrophobicity scales (A < V < L < Y~I~F < W; Table 1). Similarly, the MCS of polar tetra-peptide and Met4 ranked as S < T < N < Q ≪ M which also reflected the ranking of their hydrophobicities (S~N~Q < T ≪ M). Finally charged residues (R, D, E, K) remained monomeric (MCS ~ 1) which would correspond to their high solubilities or low hydrophobicities. Similar patterns of correlations were observed with the hydropathy scale, which correlates strongly correlate to hydrophobicity (Table 1). Finally, the present results roughly reproduced a recent amino acid’s hydrophilicity scale as well as a recently determined amino acid contribution scale to proteins’ solubility (Fig. S8; R = 0.87 calculated without Asn).
Comparison of calculated solubility parameters and experimental solubility/hydrophobicity scales.
In both panels, the calculated parameters are represented by black bars. (A) Averaged mean cluster size (black bar) vs hydropathy (circle)8 and hydrophobicity (triangle)41,42. The number of monomers and mean cluster sizes are values averaged over the last 10 ns of the MD simulations. (B) The number of monomer (black bar) averaged over the last 10 ns is shown vs hydrophobicity (triangle) and the contribution to protein solubility (circle) that we reported in an earlier study14. (C) Correlation between the averaged number of monomer and hydrophobicity. Rtot indicates the correlation coefficients of all residues.
The lowest correlation was found with the individual amino acid’s solubilities as tabulated in the CRC Handbook of Chemistry and Physics (R = −0.38 excluding Pro)9, where proline has a reported solubility of 1600 g/L and Lysine’s solubility is low at 5 g/L. These two figures, which are not reproduced in our calculation, contradict our intuitive anticipation and are also at odd with several solubility/hydrophobicity scales (Table 1). Thus, given such moderate correlation among the various experimental solubility and hydrophobicity scales, correlation coefficients of up to ~0.9 between the fraction of monomer as well as the MCS and some of the experimental scales is surprisingly good and one may hope that they could serve as a scale for estimating an amino acid’s contribution to a peptide or protein solubility.
MSM Analysis
We analyzed the MD trajectories using MSM, which provides a convenient way to model kinetic networks between different conformational states38,39. MSM is appropriate for analyzing the MD results because of the relatively large size of our 27-peptide system, which was not the case for much smaller systems used in previous studies22,23. We calculated the transition matrix P whose components are the fraction of i-mers undergoing a transition to a j-mer at a given propagation step. In order to analyze the initial stage for the aggregation mechanism, we used the first 20 ns trajectories for constructing the transition matrix. MSM revealed that small clusters (2~4-mer) rapidly accumulated, with dimmers constituting up to 18% of the oligomers (Fig. 7; Tyr), before growing or consolidating into medium (5~9-mer) and large size amorphous clusters (greater than 10-mer). The small size clusters might correspond to seeds playing a key role at the initial stage of amorphous aggregation18,40.
Conclusion
We reported a systematic and in-depth analysis of amino acid’s contribution to protein/peptide solubility using a molecular dynamics simulation of multi-peptide systems with explicit solvents. Eighteen 100 ns MD simulations of tetra-peptides corresponding to all amino acids, but Gly and Cys, were carried out. The results were overall in line with previously reported experimental amino acid’s solubility and hydrophobicity scales. To our knowledge, this is the very first study of this kind and it was surprising that the solubility of amino acids is fairly well reproduced using standard MD simulation methods, without a need to introduce artificial attractive or repulsive forces. A finer analysis of the calculation indicated that the high solubility of charged residues originated from repulsive Coulomb energies, whereas the lower solubility of uncharged residues had various origins. To date, “hydrophobic” residues, such as Ile, Leu and Val, which lack repulsive electrostatic interaction, clustered predominantly through vdW interactions and accessorily through mainchain H-bonds. Markov state model analysis suggested that small clusters consisting of 2 ~ 4 peptides accumulated before growing or merging into larger clusters.
Additional Information
How to cite this article: Kuroda, Y. et al. All-atom molecular dynamics analysis of multi-peptide systems reproduces peptide solubility in line with experimental observations. Sci. Rep. 6, 19479; doi: 10.1038/srep19479 (2016).
References
Fowler, S. B. et al. Rational design of aggregation-resistant bioactive peptides: reengineering human calcitonin. Proc. Natl. Acad. Sci. USA 102, 10105–10 (2005).
Ricci, M. S. & Brems, D. N. Common structural stability properties of 4-helical bundle cytokines: possible physiological and pharmaceutical consequences. Curr. Pharm. Des. 10, 3901–11 (2004).
Niwa, T. et al. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. USA 106, 4201–6 (2009).
Lawrence, M. S., Phillips, K. J. & Liu, D. R. Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc. 129, 10110–2 (2007).
Simeonov, P., Berger-Hoffmann, R., Hoffmann, R., Strater, N. & Zuchner, T. Surface supercharged human enteropeptidase light chain shows improved solubility and refolding yield. Protein Eng. Des. Sel. 24, 261–8 (2011).
Nozaki, Y. & Tanford, C. The solubility of amino acids and related compounds in aqueous urea solutions. J. Biol. Chem. 238, 4074–81 (1963).
Baldwin, R. L. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA 83, 8069–72 (1986).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–32 (1982).
Dawson, R. M. C. et al. in CRC Handbook of Chemistry and Physics (eds David, R. L. et al.) Sec. 8, 92–121 (CRC Press, 2009).
Kramer, R. M., Shende, V. R., Motl, N., Pace, C. N. & Scholtz, J. M. Toward a molecular understanding of protein solubility: increased negative surface charge correlates with increased solubility. Biophys. J. 102, 1907–15 (2012).
Trevino, S. R., Scholtz, J. M. & Pace, C. N. Amino acid contribution to protein solubility: Asp, Glu and Ser contribute more favorably than the other hydrophilic amino acids in RNase Sa. J. Mol. Biol. 366, 449–60 (2007).
Kuroda, Y. & Kim, P. S. Folding of bovine pancreatic trypsin inhibitor (BPTI) variants in which almost half the residues are alanine. J. Mol. Biol. 298, 493–501 (2000).
Kato, A. et al. Mutational analysis of protein solubility enhancement using short peptide tags. Biopolymers 85, 12–8 (2007).
Islam, M. M., Khan, M. A. & Kuroda, Y. Analysis of amino acid contributions to protein solubility using short peptide tags fused to a simplified BPTI variant. Biochim. Biophys. Acta 1824, 1144–50 (2012).
Akiyama, S. et al. Experimental identification and theoretical analysis of a thermally stabilized green fluorescent protein variant. Biochemistry 51, 7974–82 (2012).
Suenaga, A. et al. Molecular dynamics simulations reveal that Tyr-317 phosphorylation reduces Shc binding affinity for phosphotyrosyl residues of epidermal growth factor receptor. Biophys. J. 96, 2278–88 (2009).
Colombo, G., Daidone, I., Gazit, E., Amadei, A. & Di Nola, A. Molecular dynamics simulation of the aggregation of the core-recognition motif of the islet amyloid polypeptide in explicit water. Proteins 59, 519–27 (2005).
Oosawa, F. & Kasai, M. A theory of linear and helical aggregations of macromolecules. J. Mol. Biol. 4, 10–21 (1962).
Lin, Y. S., Bowman, G. R., Beauchamp, K. A. & Pande, V. S. Investigating how peptide length and a pathogenic mutation modify the structural ensemble of amyloid beta monomer. Biophys. J. 102, 315–24 (2012).
Roux, B. & Simonson, T. Implicit solvent models. Biophys. Chem. 78, 1–20 (1999).
Urbanc, B., Borreguero, J. M., Cruz, L. & Stanley, H. E. Ab initio discrete molecular dynamics approach to protein folding and aggregation. Methods Enzymol. 412, 314–38 (2006).
Nguyen, P. H., Li, M. S., Stock, G., Straub, J. E. & Thirumalai, D. Monomer adds to preformed structured oligomers of Abeta-peptides by a two-stage dock-lock mechanism. Proc. Natl. Acad. Sci. 104, 111–116 (2006).
Itoh, S. G. & Okumura, H. Dimerization Process of Amyloid-β(29–42) Studied by the Hamiltonian Replica-Permutation Molecular Dynamics Simulations. J. Phys. Chem. B 118, 11428–11436 (2014).
Ohmura, I., Morimoto, G., Ohno, Y., Hasegawa, A. & Taiji, M. MDGRAPE-4: a special-purpose computer system for molecular dynamics simulations. Philos. Trans. A. Math Phys. Eng. Sci. 372, 10.1098/rsta.2013.0387 (2014).
Case, D. A. et al. Amber 8.0 (2004).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926 (1983).
Narumi, T., Yasuoka, K., Taiji, M. & Hofinger, S. Current performance gains from utilizing the GPU or the ASIC MDGRAPE-3 within an enhanced Poisson Boltzmann approach. J. Comput. Chem. 30, 2351–7 (2009).
Kikugawa, G. et al. Application of MDGRAPE-3, a special purpose board for molecular dynamics simulations, to periodic biomolecular systems. J. Comput. Chem. 30, 110–8 (2009).
Duan, Y. et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 24, 1999–2012 (2003).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089 (1993).
Onufriev, A., Bashford, D. & Case, D. A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55, 383–94 (2004).
McDonald, I. K. & Thornton, J. M. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238, 777–93 (1994).
Khan, M. A., Islam, M. M. & Kuroda, Y. Analysis of protein aggregation kinetics using short amino acid peptide tags. Biochim. Biophys. Acta Proteins and Proteomics 1834, 2107–2115 (2013).
Joosten, R. P. et al. A series of PDB related databases for everyday needs. Nucleic Acids Res. 39, D411–9 (2011).
Chelli, R., Gervasio, F. L., Procacci, P. & Schettino, V. Stacking and T-shape competition in aromatic-aromatic amino acid interactions. J. Am. Chem. Soc. 124, 6133–43 (2002).
Baldwin, R. L. Gas-liquid transfer data used to analyze hydrophobic hydration and find the nature of the Kauzmann-Tanford hydrophobic factor. Proc. Natl. Acad. Sci. 109, 7310–7313 (2012).
Singhal, N., Snow, C. D. & Pande, V. S. Using path sampling to build better Markovian state models: predicting the folding rate and mechanism of a tryptophan zipper beta hairpin. J. Chem. Phys. 121, 415–25 (2004).
Jayachandran, G., Vishal, V. & Pande, V. S. Using massively parallel simulation and Markovian models to study protein folding: examining the dynamics of the villin headpiece. J. Chem. Phys. 124, 164902 (2006).
Caspar, D. L. & Klug, A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 27, 1–24 (1962).
Wolfenden, R. V., Cullis, P. M. & Southgate, C. C. Water, protein folding and the genetic code. Science 206, 575–7 (1979).
Wolfenden, R., L. Andersson, P. M. Cullis & C. C. B. Southgate . Affinities of amino acid side chains for solvent water. Biochemistry 20, 849–855 (1981).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–32 (1982).
Nozaki, Y. & Tanford, C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J. Biol. Chem. 246, 2211–7 (1971).
von Heijne, G. & Blomberg, C. Trans-membrane translocation of proteins. The direct transfer model. Eur. J. Biochem. 97, 175–81 (1979).
Acknowledgements
We thank Mr. Kenta Zushi for preliminary calculations, members of the Kuroda’s Laboratory for discussion and Dr. Gentaro Morimoto and RIKEN RICC for their computational support. This work was supported by a JSPS grant-in-aid for scientific research (KAKENHI:21300110) to Y.K.
Author information
Authors and Affiliations
Contributions
Y.K. designed the project, Y.K. and A.S. supervised the data analysis, Y.S. and S.K. performed the analysis and prepared the figures, Y.K., A.S., S.K. and Y.K. wrote the manuscript, M.T. provided computational facility and technical expertise. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kuroda, Y., Suenaga, A., Sato, Y. et al. All-atom molecular dynamics analysis of multi-peptide systems reproduces peptide solubility in line with experimental observations. Sci Rep 6, 19479 (2016). https://doi.org/10.1038/srep19479
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19479
This article is cited by
-
Designing a new bispecific tandem single-chain variable fragment antibody against tumor necrosis factor-α and interleukin-23 using in silico studies for the treatment of rheumatoid arthritis
Journal of Molecular Modeling (2020)
-
Calculation of absolute binding free energies between the hERG channel and structurally diverse drugs
Scientific Reports (2019)
-
Large-scale all-atom molecular dynamics alanine-scanning of IAPP octapeptides provides insights into the molecular determinants of amyloidogenicity
Scientific Reports (2019)
-
Theoretical analysis on thermodynamic stability of chignolin
Scientific Reports (2019)
-
Tracing whale myoglobin evolution by resurrecting ancient proteins
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.