Abstract
The Ecotropic viral integration site 1 (Evi1) is a zinc finger transcription factor, which is located on chromosome 3q26, over-expression in some acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS). Elevated Evi1 expression in AML is associated with unfavorable prognosis. Therefore, Evi1 is one of the strong candidate in molecular target therapy for the leukemia. MicroRNAs (miRNAs) are small non-coding RNAs, vital to many cell functions that negatively regulate gene expression by translation or inducing sequence-specific degradation of target mRNAs. As a novel biologics, miRNAs is a promising therapeutic target due to its low toxicity and low cost. We screened miRNAs which down-regulate Evi1. miR-133 was identified to directly bind to Evi1 to regulate it. miR-133 increases drug sensitivity specifically in Evi1 expressing leukemic cells, but not in Evi1-non-expressing cells The results suggest that miR-133 can be promising therapeutic target for the Evi1 dysregulated poor prognostic leukemia.
Similar content being viewed by others
Introduction
The human ecotropic viral integration site 1 (Evi1) gene is located on 3q26.2, a region frequently rearranged in acute myeloid leukemia (AML)1,2. Most patients with 3q26 rearrangements overexpress EVI12,3. In all, 5–10% of AML patients show Evi1 upregulation4. While Evi1-low patients showed >80% overall survival (OS) at 5 years, Evi1-high patients showed <60%, indicating that high expression of Evi1 correlated with poor prognosis5,6.
MicroRNAs (miRNAs) are 18–25 nt, single-stranded non-coding RNAs that are generated from primary miRNAs via pre-miRNAs. miRNAs can suppress post-transcriptional gene expression by base pairing with their target messenger RNAs (mRNAs) and inducing either translational repression or mRNA degradation7,8. miRNAs regulate a wide range of biological processes in animal development and human disease9,10.
miRNAs are promising therapeutic targets for Evi1-overexpressing AML. The advantages of miRNAs for therapy are: 1) their multiple targets; 2) low toxicity due to biological therapy; 3) and low cost due to technological innovation11. Evi1 deficiency severely affects not only hematopoietic stem cells but also other systems12. Therefore, the suppression of Evi1 is presumed to cause systemic adverse effects. miRNAs could overcome this problem because they should be expressed at an optimal dose in vivo under the control of endogenous feedback regulation and should affect the dysregulated overexpression of Evi1 in leukemic cells but not in other systems. Accordingly, Evi1-overexpressing leukemia should be a good target for miRNA-based therapy. In this study, we aimed to identify miRNAs that suppress Evi1 for therapeutic purposes. We found that miR-133, which targets Evi1, increased the drug sensitivity of Evi1-high-expressing leukemic cells, but not Evi1-non-expressing leukemic cells. This suggests that miR-133 is a promising therapeutic target for Evi1-overexpressing leukemia.
Results
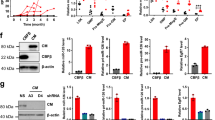
We screened for miRNAs that potentially target Evi1 to suppress its expression using computational prediction and luciferase assays. In silico prediction of Evi1 targets using the miRanda software revealed that 42 miRNAs potentially bind to the Evi1 3′UTR. (Table 1) Next, we examined whether they suppress the translation of a luciferase reporter containing the 3′UTR of the human Evi1 mRNA. The precursors for 42 miRNAs were available in our miRNA precursor library. Pre-miR™ Precursor Molecules for these 42 miRNAs were co-transfected into NIH3T3 cells with a luciferase reporter vector containing the 3′UTR region of the human Evi1 mRNA. (Figure 1a) Two miRNAs reproducibly downregulated luciferase activity: miR-133 and miR-466. (Figure 1b)
(a)The schema of the screening of miRNAs binding to the Evi1 3′UTR Synthetic miRNAs and the luciferase reporter containing the 3′-UTR of the human Evi1 mRNA were co-transfected into NIH3T3 cells to identify the miRNAs which downregulate luciferase by binding to Evi1 3′UTR. (b) The downregulated luciferase activity by binding of miR-133a,b and miR-466a-3p to Evi1 3′UTR. The longtitude axis showed the relative luciferase to the control. (c) Exogenous miR-133 decreased Evi-1 expressions in HEL cells. The sum of naïve and truncated or MDS1 bands intensity was determined by densitometry and normalized to β-actin. Three experiments were done. (*p < 0.05). (d) In Hela cells which were transfected with miR-466 or miR-3936 tough decoy (TuD), all of Evi-1 variant proteins including MDS1/Evi-1, Evi-1 and truncated Evi-1 were specifically upregulated by miR-466 suppression. The band intensity was determined as above. Three experiments were done. (*p < 0.05). (e) Expression of Evi1 in HEL, K562, HL60, U937 and THP1 cells. The expression of Evi1 was high in HEL and K562 cells (Left), while that was not detected in HL60, U937 and THP-1 cells. The band intensity was determined by densitometry and normalized to β-actin. (f) Data of real-time PCR of expression of Evi1 in HEL, K562, HL60, U937 and THP-1 cells. Expression of Evi1 was detected only in HEL and K562 cells. (*p < 0.05)
To examine whether the endogenous expression of Evi1 is affected by miR-133 and miR-466, we overexpressed miR-133 in HEL cell lines, which express high levels of Evi1. We found that miR-133 suppressed endogenous Evi1 expression in the HEL cell lines. (Figure 1c)
Overexpression of miR-466 did not affect endogenous Evi1 expression, but suppression of miR-466 activity by miR-466 TUD13 resulted in upregulation of endogenous Evi1 expression. (Figure 1d)
Since miR-133 and miR-466 both regulated Evi1, we further analyzed their functions in leukemia cell lines. Evi1 overexpression is associated with poor prognosis and shorter survival in AML, because AML shows strong drug resistance. We examined whether miR-133 and miR-466 increase sensitivity to Adriamycin (ADR), one of the key drugs in chemotherapy for AML. HEL and K562 cells derive from AML patients with high Evi1expression, while HL60, U937 and THP1 cells are from ones without Evi1 expression. (Figure 1e,f) We compared drug sensitivity between the two groups of cell lines. Ectopic expression of miR-133 in HEL cells sensitized the cells to ADR. HEL cells were transduced with a retroviral expression vector, MDH, expressing miR-466, miR-133, or no miRNA (control). Transduced HEL and K562 cells were sorted for those expressing GFP (a marker gene in the MDH vector) and were treated with ADR for 44 h. Annexin V staining was used to measure apoptosis. Approximately 3–7% of cells expressing miR-133 were Annexin V-positive (apoptotic) in the absence of ADR treatment, similar to sorted control MDH HEL, K562, HL60, U937 and THP1 cells. (Figure 2a,b) Treatment with ADR for 48 h dose-dependently increased the number of Annexin V-positive miR-133-overexpressing HEL and K562 cells, but not HL60, U937 and THP1 cells, compared to control cells. (Figure 2a,b) These results clearly show that miR-133 induces apoptosis in Evi1-overexpressing cells, but not in cells without Evi1 expression. miR-466 did not show any effect on ADR sensitivity.
Functional Analysis for miR-133.
(a) Exogenous expression of miR-133 in Evi-1high HEL cells increases the sensitivity of ADR. Annexin V positive cells including PI positive and PI negative cells were measured by FACS analysis. miR-133 overexpressing Evi-1high HEL cells and K562 cells showed more Annexin V positive cells in 4.5 and 14 μM ADR. ADR indicates Adriamycin.(*p < 005). (b) Exogenous expression of miR-133 in HL60 cells, U937 cells and THP1 cells had no effect on the sensitivity of ADR. Annexin V positive miR-133 overexpressing HL60 cells, U937cells and THP-1 cells showed no difference with the control HL60 cells, U937 cells and THP-1 cells at all the concentration of ADR. (c) Cleaved caspase 3 was increased by miR-133 in ADR dose dependent manner.
That miR-133 increases drug sensitivity in Evi1-high-expressing AML 1 cells was confirmed by caspase activation in these cells. ADR dose-dependently increased cleaved caspase-3 levels in miR-133-overexpressing cells compared with control cells. (Figure 2c) This indicates that miR-133 promoted apoptosis in the presence of ADR in Evi1-high-expressing HEL leukemic cells. (Figure 3)
The schema of the function of miR-1 and miR-133 in Evi-1high cells.
Evi1 upregulates precursor of miR-1 and miR-133. While miR-1 shows oncogenic activity, miR-133 binds 3UTR of Evi1 to downregulate Evi1, which makes “negative feedback loop”. The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see: http://www.textcheck.com/certificate/cTxRPr
Discussion
miR-1-2 and miR-133a-1 are clustered together at the same locus on chromosome 1814 suggesting that their transcription might be regulated by similar mechanisms. It was previously reported that transcription of these two miRNAs was directly regulated by Evi1, which acts as a transcription factor for them15.
Both miRNAs are upregulated by overexpression of Evi1, while only miR-1 and not miR-133, increased cell proliferation16. The function and significance of miR-133, which is transcriptionally upregulated by Evi1, needed to be explored. In this study, we demonstrated that miR-1 and miR-133 might act antagonistically, at least in Evi1-overexpressing leukemic cells. Similarly, a previous study showed that miR-1 and miR-133, which are preferentially expressed in cardiac and skeletal muscle and have been shown to regulate differentiation and proliferation of cells in these tissues, produce opposing effects: miR-1 is pro-apoptotic in cardiac cell apoptosis whereas miR-133 is anti-apoptotic. This suggests that the relative levels of miR-1 and miR-133 are more important than their absolute levels in determining the fate (apoptosis or survival) of cardiac cells. The expression difference between miR-1 and miR-133 might be governed by their biogenesis because the primary miRNA for miR-1 and miR-133 is a single transcript.
Recently, an in silico study showed that a SNP in the predicted miR-133 binding site in the 3′UTR of Evi1 predicted worse prognosis in AML. This suggests that in patients miR-133 may play a critical tumor suppressive role whose abrogation results in a worse prognosis17. Our functional assay clearly showed that miR-133 is a tumor suppressor for Evi1-overexpressing leukemic cells. The restoration of miR-133 in gastric cancer suppresses cell proliferation and induces apoptosis, indicating that miR-133 is a promising therapeutic target, consistent with our study18,19. Accordingly, regulation of miR-133 processing, chemically modified mimics of miR-133 and drug delivery systems should be further studied to better understand the function of miR-133 in Evi1-overexpressing leukemia and its therapeutic potential. Target molecules of Evi1 that induce drug resistance include ITGA6, GPR5 and ANG120,21,22.
The target genes of miR-133 include MCL1, BCLxL and IGF-1R, which have anti-apoptotic and oncogenic properties23,24,25,26,27. miR-133 may induce drug sensitivity through downregulation of Evi1 and these target genes.
In summary, we identified miR-133 as a miRNA that regulates Evi1, whose overexpression is associated with a poor prognosis in AML. Since miR-133 modulates dysregulated excess Evi1 expression but not normal expression, it could be a promising therapeutic target in Evi1-overexpressing AML patients.
Materials and Methods
Prediction of miRNAs using a computational target prediction system
To detect candidate miRNAs targeting Evi1, we first evaluated a series of miRNA precursors. To narrow the screened miRNAs to fewer than 100 miRNAs, we used a computational target prediction system (miRanda) containing updated sequences for all known miRNAs. Cutoff scores for selection of candidate miRNAs were <−20.0 for energy and >120 for binding28.
Cell culture
Five cell lines (HEL, K562, U937, HL-60 and THP1) were maintained in RPMI 1640 medium (Wako, Japan) supplemented with 10% (v/v) fetal bovine serum (FBS), 50 U/mL penicillin and 50 mg/mL streptomycin in a 10 cm dish (Corning, Inc., Corning, NY, USA). Cells were passaged twice per week.
Quantitative PCR for genes
For target gene detection, RT-PCR was performed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Inc., CA, USA) and qPCR was carried out with the Fast SYBR Green Master mix. All real-time qPCR was conducted using the StepOnePlus real-time PCR system (Applied Biosystems). Threshold cycle (CT) values were calibrated to β-actin and analyzed by the 2−ΔΔCT method. Sequences of specific primers are listed in Supplementary Table 1.
Western blotting
For Western blot analyses, cells were harvested by centrifugation and washed twice with phosphate-buffered saline (PBS). Cells (1.0 × 105) were lysed in radioimmunoprecipitation assay (RIPA) buffer for 5 min on ice. Cell lysates were centrifuged to remove debris. Protein samples were separated electrophoretically on a 5–20% SDS-polyacrylamide gel and blotted onto PVDF membranes (Bio-Rad Laboratories, Tokyo, Japan). The blots were blocked with 2% low-fat dry milk in TBST (20 mM Tris-HCl, pH 7.5, 150 mM NaCl containing 0.1% Tween 20 [Sigma, MO, USA]) for 1 h at room temperature. The blocked membrane was incubated with anti-Evi1 (CST#2593) (1:2000) or anti-βactin (1:5000) for 2 h, followed by incubation with anti-rabbit IgG (CST#7074) (1:2000) secondary antibody for 1 h.
Drug sensitivity assay
Five cell lines (HEL, K562, U937, HL-60 and THP1) were seeded in 24-well plates with 2.0 × 105 cells per well in growth medium. Adriamycin was added at specific concentrations and incubated for 48 h, before being analyzed by FACS with immunostaining for APC-Annexin V (BioLegend, Japan).
Additional Information
How to cite this article: Yamamoto, H. et al. miR-133 regulates Evi1 expression in AML cells as a potential therapeutic target. Sci. Rep. 5, 19204; doi: 10.1038/srep19204 (2015).
References
Ihle, J. N., Morishita, K., Matsugi, T. & Bartholomew, C. Insertional mutagenesis and transformation of hematopoietic stem cells. Prog Clin Biol Res 352, 329–337 (1990).
Morishita, K., Parganas, E., Parham, D. M., Matsugi, T. & Ihle, J. N. The Evi-1 zinc finger myeloid transforming gene is normally expressed in the kidney and in developing oocytes. Oncogene 5, 1419–1423 (1990).
Morishita, K. et al. Activation of EVI1 gene expression in human acute myelogenous leukemias by translocations spanning 300–400 kilobases on chromosome band 3q26. Proc Natl Acad Sci USA 89, 3937–3941 (1992).
Saito, Y. et al. CD52 as a molecular target for immunotherapy to treat acute myeloid leukemia with high EVI1 expression. Leukemia 25, 921–931 (2011).
Lugthart, S. et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood 111, 4329–4337 (2008).
Nucifora, G., Laricchia-Robbio, L. & Senyuk, V. EVI1 and hematopoietic disorders: history and perspectives. Gene 368, 1–11 (2006).
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116, 281–297 (2004).
Bagga, S. et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122, 553–563, 10.1016/j.cell.2005.07.031 (2005).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Okuyama, K., Ogata, J., Yamakawa, N., Chanda, B. & Kotani, A. Small RNA as a regulator of hematopoietic development, immune response in infection and tumorigenesis. Int J Hematol 99, 553–560, 10.1007/s12185-014-1564-4 (2014).
Esau, C. C. & Monia, B. P. Therapeutic potential for microRNAs. Adv Drug Deliv Rev 59, 101–114 (2007).
Hoyt, P. R. et al. The Evi1 proto-oncogene is required at midgestation for neural, heart and paraxial mesenchyme development. Mech Dev 65, 55–70 (1997).
Haraguchi, T., Ozaki, Y. & Iba, H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 37, e43 (2009).
Liu, N. et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA 104, 20844–20849 (2007).
Chen, J. F. et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233 (2006).
Gómez-Benito, M. et al. EVI1 controls proliferation in acute myeloid leukaemia through modulation of miR-1-2. Br J Cancer 103, 1292–1296, 10.1038/sj.bjc.6605874 (2010).
Wang, T. Y., Huang, Y. P. & Ma, P. Correlations of common polymorphism of EVI-1 gene targeted by miRNA-206/133b with the pathogenesis of breast cancer. Tumour Biol, 2213–5 (2014).
Cheng, Z. et al. miR-133 is a key negative regulator of CDC42-PAK pathway in gastric cancer. Cell Signal 26, 2667–2673 (2014).
Liu, Y. et al. Identification of miRNomes in human stomach and gastric carcinoma reveals miR-133b/a-3p as therapeutic target for gastric cancer. Cancer Lett 369, 58–66 (2015).
Yamakawa, N., Kaneda, K., Saito, Y., Ichihara, E. & Morishita, K. The increased expression of integrin α6 (ITGA6) enhances drug resistance in EVI1 (high) leukemia. PLoS One 7, e30706 (2012).
Saito, Y. et al. Maintenance of the hematopoietic stem cell pool in bone marrow niches by EVI1-regulated GPR56. Leukemia 27, 1637–1649 (2013).
Ichihara, E., Kaneda, K., Saito, Y., Yamakawa, N. & Morishita, K. Angiopoietin1 contributes to the maintenance of cell quiescence in EVI1 (high) leukemia cells. Biochem Biophys Res Commun 416, 239–245 (2011).
Bartram, I. et al. Inhibition of IGF1-R overcomes IGFBP7-induced chemotherapy resistance in T-ALL. BMC Cancer 15, 663 (2015).
Koti, M. et al. Identification of the IGF1/PI3K/NF κB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC Cancer 13, 549 (2013).
Ariës, I. M. et al. The synergism of MCL1 and glycolysis on pediatric acute lymphoblastic leukemia cell survival and prednisolone resistance. Haematologica 98, 1905–1911 (2013).
Kiprianova, I. et al. Sorafenib Sensitizes Glioma Cells to the BH3 Mimetic ABT-737 by Targeting MCL1 in a STAT3-Dependent Manner. Neoplasia 17, 564–573 (2015).
Zhou, W. et al. Inhibition of Bcl-xL overcomes polyploidy resistance and leads to apoptotic cell death in acute myeloid leukemia cells. Oncotarget 6, 21557–21571 (2015).
Oba, S. et al. A useful method of identifying of miRNAs which can down-regulate Zeb-2. BMC Res Notes 6, 470 (2013).
Acknowledgements
We thank Drs. Ryo Nasu, Kazuki Okuyama, Natsuko Yamakawa and Bidisha Chanda and the Education and Research Support Center, Tokai University, for technical assistance.
Author information
Authors and Affiliations
Contributions
H.Y., J.L., S.O. and A.K. performed research and analyzed data. A.K. designed research and wrote the paper. T.K., A.Y., N.K., K.Y., H.M., M.K., A.T., K.A., K.M. and K.K. contributed vital new reagents.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Yamamoto, H., Lu, J., Oba, S. et al. miR-133 regulates Evi1 expression in AML cells as a potential therapeutic target. Sci Rep 6, 19204 (2016). https://doi.org/10.1038/srep19204
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19204
This article is cited by
-
AKT inhibition sensitizes EVI1 expressing colon cancer cells to irinotecan therapy by regulating the Akt/mTOR axis
Cellular Oncology (2022)
-
Role of non-coding RNA networks in leukemia progression, metastasis and drug resistance
Molecular Cancer (2020)
-
Overexpression of miR-669m inhibits erythroblast differentiation
Scientific Reports (2020)
-
MicroRNAs: pivotal regulators in acute myeloid leukemia
Annals of Hematology (2020)
-
The role of zinc and its compounds in leukemia
JBIC Journal of Biological Inorganic Chemistry (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.