Abstract
Preeclampsia is a disease of pregnancy involving systemic endothelial dysfunction. However, cardiovascular consequences of preeclampsia are difficult to analyze in humans. The objective of the present study is to evaluate the cardiovascular dysfunction induced by preeclampsia by examining the endothelium of mice suffering of severe preeclampsia induced by STOX1 overexpression. Using Next Generation Sequencing on endothelial cells of mice carrying either transgenic or control embryos, we discovered significant alterations of gene networks involved in inflammation, cell cycle and cardiac hypertrophy. In addition, the heart of the preeclamptic mice revealed cardiac hypertrophy associated with histological anomalies. Bioinformatics comparison of the networks of modified genes in the endothelial cells of the preeclamptic mice and HUVECs exposed to plasma from preeclamptic women identified striking similarities. The cardiovascular alterations in the pregnant mice are comparable to those endured by the cardiovascular system of preeclamptic women. The STOX1 mice could help to better understand the endothelial dysfunction in the context of preeclampsia and guide the search for efficient therapies able to protect the maternal endothelium during the disease and its aftermath.
Similar content being viewed by others
Introduction
Preeclampsia is a complication of pregnancy characterized by gestational hypertension and proteinuria1. Preeclampsia affects ~5% of pregnant women leading to maternal an fetal morbidity and mortality2,3.
Endothelial dysfunction is a characteristic of preeclampsia4. Women affected by preeclampsia have also cardiac dysfunction and cardiac hypertrophy5. Epidemiological data report an increased risk of cardiovascular diseases later in life in women who have had a preeclamptic pregnancy6.
We recently developed a mouse model of preeclampsia by transgenic overexpression of the transcription factor STOX17. STOX1 was identified in Dutch families by a positional cloning approach and its involvement in the pathophysiology of preeclampsia was inferred8. Crossing transgenic males with WT females induces a severe preeclamptic phenotype, with gestational hypertension, proteinuria, increased plasma levels of soluble Endoglin (sENG) and soluble FMS-like tyrosine kinase 1 (sFLT1), as well as kidney histological anomalies in line with the proteinuria. In the transgenic placentas, STOX1 overexpression induces alterations of mitochondrial function and nitroso-redox imbalance, that could trigger the disease9.
A classical vision of the origin of the maternal symptoms in preeclampsia is through the release of toxic substances by the placenta that may be used for diagnostic and prognostic10, some of them such as sENG or sFLT1 being increased in the plasma of preeclamptic women even before the onset of the symptoms11. Preeclampsia remains, first and foremost a disease of the endothelium12, targeted by various placental-derived factors (antiangiogenic molecules as well as nucleic acids including miRNAs and microparticles).
Since endothelial alterations are difficult to study in human patients, we decided to evaluate thoroughly the expressional effect of a preeclamptic state on the endothelium and the heart in the STOX1 model. We used RNA-seq to examine the transcriptome of purified endothelial cells (EC) from pregnant mice bearing either WT (normal pregnancy) or STOX1 transgenic embryos (preeclamptic pregnancies). Bioinformatics analysis revealed down-regulation of genes involved in cell division and up-regulation of genes involved in fibrosis, inflammation and cardiac hypertrophy, associated with increased heart mass, abnormal heart histology and expression of markers of cardiac hypertrophy, consistently with the increasing number of reports describing links between preeclampsia and cardiovascular disease13. Overall, the alterations found in the heart and in the endothelium of wild-type females carrying transgenic embryos substantiate a model of ‘dangerous placentas’, able to induce systemic alterations in the mother. Then, we exposed an immortalized Human Umbilical Vein Endothelial Cell line (HUVEC) to plasma from human normotensive or preeclamptic pregnancies. Bioinformatics analysis revealed significant similarities between the in vivo alterations of the endothelium in the preeclamptic mice and the gene expression profile of HUVEC cells exposed to preeclamptic plasma. This suggests that our observations in mice could be extrapolated to women and paves the way to understand the molecular effects of preeclampsia in the cardiovascular system.
Methods
Animals experiments
Breedings and Ethics
The animals used in this study belonged to the FVB/N strain. WT ♀ mice were crossed with either WT or transgenic ♂ mice expressing the human STOX1 under the control of the cytomegalovirus promoter, as previously described7. Animal experiments were carried out in strict accordance with the recommendations in the guidelines of the Code for Methods and Welfare Considerations in Behavioural Research with Animals (Directive 86/609EC). And all efforts were made to minimize suffering. All experimental protocols were approved by the Local ethics committee of Jouy-en-Josas on the Ethics of Animal Experiments of the author’s institution, INRA (Permit Number 12/035 (06.29.2012)). All animal manipulations were done according to the recommendations of the French Haut Conseil aux Biotechnologies (Permit Number N°6460 (06.25.2013)). Most of the work presented here, especially the RNA-seq analysis is focused on the TgSTOX13 mice, the only strain where homozygous transgenics could be obtained. Some of the heart expression data and heart histology were carried out conjointly on females crossed with males from the other strain available (TgSTOX42). Further details are given in the Supplementary Methods.
Endothelial cells purification
Six pregnant mice carrying either WT (3) or transgenic embryos (3) were sacrificed at E16.5 . Then, muscles (from forelegs, hind legs and back) were rapidly dissected, removing fat and connective tissue. EC purification procedure is detailed in the Supplementary data., In addition to validate the gene expression modifications, three additional WT mice were sacrificed and the purified endothelium was exposed during three days either to plasma of preeclamptic mice (Carrying STOX1 overexpressing embryos) or to plasma of normal pregnant mice. The plasmas were collected at day 16.5 to 17.5 of pregnancy (normal pregnancy duration is 18.5 days in the FVB/N strain). The conditions used for treatment are the same as for the human experiment described below.
RNA-seq analysis of RNA from endothelial cells
RNA-seq was performed by the society Genotypic Technology at Bangalore (India). The data were deposited as Fastq files as project STOX1ENDO, referred to as PRJNA275924.
Immunohistochemistry
Hearts were harvested, fixed in 10% formalin and embedded in paraffin. IHC was performed as described14. The immunoreactivity was detected using the Novolink polymer detection system according to manufacturer's recommendations (Leica, Nanterre, France). Immunostaining was done using DAB and tissue sections were counterstained with Mayer’s hematoxylin solution (Merck Millipore, Guyancourt, France). Anti-CTGF antibody, rabbit polyclonal HPA031074 (Sigma Prestige antibodies, Sigma, St Louis, MO) was diluted 1:100 in PBS/ 1% BSA/0.1% Triton. IHC signals were analysed using NIH ImageJ software to quantify the staining within the heart in a 20x field of view under a microscope equipped with a DC 300F camera (digital module R, IM 1000, Leica).
qRT-PCR
Standard conditions were utilized as described previously9, using a LightCycler Roche 480 thermocycler.
Human cell experiments
Preeclamptic plasmas
The plasmas from patients were obtained at the Federative Research Institute 48 (Marseille, France) which approved the use of samples for the project (agreement 08-012); all the patients were duly informed of the use of their samples and gave an informed consent and the experiments (blood sampling) were carried out in accordance with the approved guidelines. Blood samples were collected in 4.5 ml, centrifuged for 10 min at 2000 g and 20 °C (Room Temperature). Plasma was aliquoted and kept at −80 °C. Details on the patients’ characteristics are given in the Supplementary Methods.
HUVEC and SVEC culture
HUVEC cells were grown until forming 90% confluent monolayers, washed twice in PBS and serum-starved for 12 hours. The cells were then grown in culture medium supplemented with 10% (vol/vol) plasma from either preeclamptic patients (8) or con trols (11), in 19 independent cultures. The medium was renewed every 24 hours until reaching 72 hours exposure. To note, during the experiment 5 cultures died, but no significant difference could be observed between control and preeclamptic plasmas (two from controls and three from preeclamptic plasmas, p = 0.34 by χ2, 1 df). The conditions for SVEC cultures were identical.
RNA extraction and quantitative RT-PCR conditions
Total RNA from tissues or HUVEC cells was extracted using Trizol Reagent (Invitrogen) in accordance with the manufacturer’s instructions, treated with RNase-free DNase and quantified by spectrophotometry as described previously7. Reverse Transcription and Quantitative RT-qPCR are described in Supplementary Methods, qPCR primers are listed in Table S2.
Microarray analysis of RNA from HUVEC exposed to control or preeclamptic plasmas
RNA quality was verified by Agilent bioanalyser 2100 and checked to have a RNA Integrity Number (RIN) systematically higher than 8. Two pools were hybridized on Agilent 8-plex 60 K microarrays at the Genomics Platform of the Cochin Institute according to standard validated protocols. The data were deposited at EMBL under the number E-MTAB-3348. Bioinformatics procedures are detailed in the Supplementary material.
Statistical analyses
ANOVA and post-hoc Student-Neumann-Keuls t-Tests were performed for relative expression analysis in the mouse hearts using the ΔCt values method. Differences in weight in the heart were performed using ANOVA followed by Dunnett t-test. The statistics for assessing the enrichment of group of genes or TF Binding sites in gene promoter is always based on Chi2 contingency tests that are directly performed online by DAVID, STRING or Cytoscape iRegulon tool. For basic statistics, the StatistXL add-in of Excel™ was systematically used. p < 0.05 were considered significant.
Results
Transcriptome of endothelial cells (EC) from preeclamptic mice
EC were isolated from the limbs of pregnant females and their purity was evaluated measuring the expression of endothelial markers (Vwf15, Cd3116, Vcam117 and Cdh518). The enrichment of mRNA for these markers was comprised between 6.1 fold for Cdh5 and 24 fold for Vwf (Supplemental Figure S1A) indicating efficient purification. The RNA extracted from the cells was subjected to Next Generation Sequencing analysis (RNA-seq) and the resulting reads were aligned along 94,818 transcripts of the mouse genome. Of these, 33,011 were validated, corresponding to 14,220 different genes (most significantly modified listed in Table S1). RT-qPCR was performed on independent EC samples to confirm the RNA-seq results (Figure S1B).
The complete gene expression dataset was submitted to Gene Set Enrichment Analysis (GSEA)19 to extract biological knowledge. We found 212 significantly enriched gene-sets (FDR < 0.25, p < 0.01, Table S3). Critical keywords for the gene-sets were: Mitosis, Cardiac Hypertrophy, Kidney Ageing, Fibrosis and Inflammation (Fig. 1).
Gene Set Enrichment Analysis (GSEA).
The gene expression data generated by RNA-seq was analyzed using GSEA to extract biological knowledge. Highly, significant enriched gene-sets are shown here. In every thumbnail, the green curve represents the evolution of the density of the genes identified in the RNA-seq. The False Discovery Rate (FDR) is calculated by comparing the actual data with 1000 Monte-Carlo simulations. The NES (Normalized Enrichment Score) computes the density of modified genes in the dataset with the random expectancies, normalized by the number of genes found in a given gene cluster, to take into account the size of the cluster.
Another method to analyse datasets of differentially expressed genes (DEGs) is to investigate if they coalesce into clusters of interacting genes/proteins. This analysis was performed with the 967 DEGs displaying a fold change (FC) ≥ 1.5. Cluster analysis by DAVID of these DEGs identified KEGG pathways such as Dilated Cardiomyopathy and Cell Cycle, consistently with the GSEA analysis (Figure S3). Then, using the STRING database and a high interaction confidence level (0.7), we detected 893 interactions among these DEGs while 413 were expected by chance (p < 10−100). These interactions were used to build a protein-protein interacting (PPI) network which was submitted to clustering analysis with Cytoscape to find functional modules (Fig. 2). Discrete modules were found, such as down-regulation of cell-cycle genes and up-regulation of myogenic genes. Clusters of mitochondrial genes were also identified, consistently with data obtained from the analysis of the placenta in preeclamptic pregnancies9,20. We also detected a cluster of down-regulated genes composed of interferon targets.
Protein-protein interaction network analysis.
The DEGs identified from the RNAseq analysis, with a fold change ≥1.5 were used as seeds to construct a network of interacting proteins. The interactions between the DEGs were extracted from the STRING data base and visualized using Cytoscape software (center). The network nodes represent genes and edges interactions. The gene expression levels were mapped on the network. Red indicates up-regulation and blue down-regulation. A zoom is presented on some clusters including: Interferon-induced genes, Myogenic genes, Mitochondrial modulators and genes involved in cell cycle regulation (which are in this case drastically reduced). The left-down frame presents a topological analysis of the network. The size of the nodes is proportional to the number of connections established with other genes. The color, from green to red, is a centrality measure (betweeness centrality, BC). The BC quantifies how drastically a gene influences the structure of the whole network. IL6 is the most central gene in the network and its expression is induced in the EC by placental STOX1 overexpression.
Hub genes in the network were identified by topological analysis using the Cytoscape Network Analyzer. Among these, IL6 and to a lesser extent ESR1 presented the highest centrality scores. To identify transcriptional mechanisms controlling the DEGs, the top 10 % hub genes were used to build a Minimal Essential Network, MEN (Fig. 2). Promoter analysis of the MEN genes was done with the Cytoscape iRegulon apps (Figure S4, Table S4). A significant enrichment in binding sites was detected for transcription factors involved in muscle development (SOX9), inflammation (NFKB, IRF8), unfolded protein response (ATF4), cell proliferation (P53) and cell cycle (FOXM1).
Heart weight is increased in preeclamptic mice
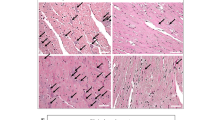
Cardiac Hypertrophy appeared systematically in the bioinformatics analyses of our dataset. Therefore, we sacrificed mice crossed with WT or transgenic males at the end of gestation, when the hypertension is the most severe and weighted their heart. An increased weight was detected in the preeclamptic mice (Figure S3). The comparative microscopic examination revealed abnormalities in heart histology, in particular fiber disorganization, fibrosis and dilated nuclei (Fig. 3A). To assess the involvement of either hypertrophy or hyperplasia leading to this increased heart weight, we evaluated the expression of three hyperplasia markers (ki67, Topo2A and Tpx2) on heart cDNA from mice that carried either WT or transgenic embryos (6 and 10, respectively). While the expression was slightly increased for ki67 and Tpx2 (+20.7 and +21.2%, respectively) these differences were not significant (data not shown). This suggests that hypertrophy (rather than hyperplasia) is the principal mechanism responsible for the observed increase of heart weight in the preeclamptic mice.
Heart histology and gene expression.
Pregnant mice crossed with WT males (n = 5), transgenic TgSTO13 (n = 5) or TgSTOX42 (n = 7) males were sacrificed at the end of gestation (E16.5-E17.5) and their hearts analyzed. (A) Heart light-field photomicrographs of Masson's trichrome-stained sections of heart collected from dams carrying WT litters (left panel) and from dams carrying transgenic litters (right panel). Arrows indicate dilated nucleus and asterisks indicate collagen deposits. (B) Relative expression of the Renin/Angiotensin System and (C). Endothelin-1 in the heart from control versus preeclamptic pregnant mice. Hearts were retrieved at the end of gestation (E16.5-E17.5) from mice crossed with wild type males (control, white bars, n = 6) or crossed with transgenic males (preeclamptic gestation, grey and black bars). The Ct were normalized by those obtained for two reference genes, Sdha and CyclophilinA and the expressions for the control gestations were then arbitrarily set to one. *p < 0,05. The third panel (C) represents the expression analysis of markers that were modified in the endothelial cells in the heart context. The genes are grouped according to their overall expression level. (D) IHC analysis of heart sections collected from dams carrying WT litters (left panel) and from dams carrying transgenic litters (right panel) for Ctgf staining. Representative data are from six animals in each group. Scale bar is on the right lower corner. The labeling of the protein confirmed the specific mRNA decrease of mice carrying transgenic embryos, a mark of cardiac hypertrophy. (E) Quantification of the IHC labeling after ImageJ treatment, transforming the image in a Black and White and measuring the labeled surfaces.
Altered gene expression profiles in the hearts from preeclamptic mice
We measured the expression of markers of the Renin/Angiotensin system (RAS), known as locally activated in case of heart hypertrophy21. We observed a significant increase of Angiotensinogen (Agt, which is the precursor of Angiotensin), Angiotensin type I receptor (Agtr1a), Angiotensinogen converting enzyme (Ace) and Ace2 mRNA in WT ♀ mice previously crossed with STOX1 transgenic ♂ compared to those crossed with WT ♂ (Fig. 3B). We also measured Endothelin-1 expression in the heart from these pregnant mice (Fig. 3B), since this factor is increased in cardiac hypertrophy22 and is known to be induced by Angiotensin II23. All these genes were induced in the heart of mice carrying transgenic embryos. This was correlated with the expression levels of the transgene (about 15 times more in TgSTOX42 than in TgSTOX13).
Since the EC of pregnant mice carrying transgenic embryos revealed deregulation of genes involved in cardiac hypertrophy, we analyzed their expression in the heart. Eleven of these genes were investigated (Ctgf, Dcn, Col1a1, Il6, Itgav, Dmd, Adcy7, Tnnc1, Myl2, Myl3, Tpm1). We observed a significant up-regulation of Myl3 and Tpm1 (Fig. 3C). However the up-regulation was mild compared to that observed in EC (more than 4 fold for Myl3 and more than 3 fold for Tpm1). Ctgf that was induced 2.9 fold in the endothelium was on the contrary down-regulated 2.4 fold in the hearts (Fig. 3C). Ctgf immunostaining of heart sections revealed strong labeling at the edges of heart muscular fibers in the mice carrying a WT litter, whereas fainter labeling is found in the heart of the mice carrying a transgenic litter. Digitalized images quantified with ImageJ, confirmed a near 3 fold down-regulation of Ctgf protein, (Fig. 3D,E).
Comparative analysis of ECs dysfunction in the STOX1 mouse model and in HUVECs exposed to preeclamptic plasma
To investigate the effects of preeclamptic plasma on EC, we exposed HUVECs to plasma from mild preeclampsia, severe preeclampsia or controls for 72 hours and analyzed the gene-expression profiles using human Agilent 60 K microarrays. This allowed identifying 3,863 DEGs (186 were modified more than twice, 96 up and 90 down). Non-supervised clustering analysis of the 100 genes that present the lowest p-value (computed by ANOVA) clearly discriminates between the cells treated with the control versus the preeclamptic plasmas (Supplementary Figure S5). The DEGs with a FC ≥ 1.5 were used as seeds to build two PPI networks describing the HUVECs response to mild or severe PE plasmas. We compared both networks with the preeclamptic-mice EC network using NeAT, a software which computes the union between two networks and assesses the statistical significance of their intersection24. The hypergeometric P-values of the intersections between the networks appeared highly significant (Table 1). However, the significance between the preeclamptic mice EC network is higher for the severe preeclampsia network than for the mild. Figure 4 shows the nodes and interactions at the intersection of these two networks. The topological analysis indicates that IL6, IL1B, TGBI and ITGB2 are the principal hub genes (Fig. 4). Biological Processes associated with this network were « blood coagulation », « chronic inflammatory response », « leukocyte migration » and « extracellular matrix » (Table S5). Finally, to evaluate in vitro the effect of the plasma of mice that had had either a preeclamptic or normal pregnancy on murine endothelial cells, we prepared primary endothelial cells from three control mice, divided them into two wells from 96-well plated and applied the two types of plasma onto the two wells. Then we prepared mRNA and cDNA and analyzed the expression of a sample of genes by qRT-PCR. We also performed the same experiment on three replicates of the SVEC mouse endothelial cell line. Overall, genes induced in the RNA-seq (in vivo experiment) were also induced in the in vitro conditions, but the correlation was much stronger with primary cells (Supplementary Figure S6).
Comparison between the transcriptomic alterations of the endothelium in the STOX1 model and HUVECs exposed to preeclamptic plasma.
HUVECs were exposed for 72 hours to mild or severe PE plasma and their transcriptome analyzed using human expression microarrays. DEGs identified in HUVECs exposed to mild or severe preeclampsia plasma were used to build networks (mild-PE-Ntw and severe_PE-Ntw)which were compared to the preeclamptic mice EC network, using the bioinformatics tool NeAT24. The figure shows the common genes and interactions resulting from the intersection between the severe-PE-Ntw and the preeclamptic mice EC network. Enrichment analysis detects sets of genes involved in inflammation, extra-cellular matrix, TGFβ cascades and coagulation processes. IL6, IL1B and ITGBI figure among the principal hub genes in the network.
Discussion
In this study, RNA-seq analysis revealed endothelial transcriptome alterations in the STOX1 mouse model of preeclampsia. In addition, cardiac hypertrophy and alterations of cardiac tissue were observed. However, our experimental design does not allow determining if the cardiac alterations are specifically due to the preeclamptic state induced by STOX1 or a consequence of the sustained increased in blood pressure.
The WT female mice cardiovascular/endothelial function and structure was systematically affected following foeto-placental expression of STOX1. The EC harbor a gene expression signature centered on IL-6 associated to inflammatory activation, a systematic down-regulation of genes involved in cell cycle and cell proliferation and an enrichment of up-regulated genes involved in fibrosis, cardiac hypertrophy and dilated cardiomyopathy. A group of down-regulated genes composed of interferon targets was also found in line with findings showing a deregulation of circulating IFN-α in lupus-affected patients25 (which have a ~4 fold increase of developing a preeclampsia relative to control patients26). In this recent study, Andrade and co-workers showed that lupus-affected pregnant patients have a higher concentration of circulating IFNα when they develop preeclampsia than when they do not. Interestingly the authors showed that sFLT1 expression is synergistically activated by co-treatment with IFNα and sFLT1, while IFNα treatment reduces the number of endometrial tubes development. Overall the results suggest that the antiangiogenic effects of sFLT1 are sensitized by IFNα. This is consistent with several studies showing that the typeI IFN system antagonizes VEGF-stimulated angiogenesis27.
The IL-6 signature of inflammation in the STOX1 mouse model is clearly reminiscent of an important amount of data obtained in humans. For instance, recently, preeclampsia was associated to gene variants present in the upstream regulator of TNFα in European Americans28. Similarly, exposition of monocytes cultures to preeclamptic plasma induced the expression of IL-6, TNFα and NFKB29, suggesting that as in humans, the pathogenesis in STOX1 mice involves aggravated inflammatory responses.
Also, comparative analysis showed that the transcriptome alterations observed in the mice endothelium are similar to that found in HUVECs exposed to preeclamptic plasma.
Down-regulation of cell-cycle genes suggests a less efficient renewal of EC, which may trigger long term alterations and ageing of the cardiovascular system. This issue may be of extreme importance to understand the reasons why women after preeclampsia are at increased risk of vascular disease as well as kidney end-stage disease, multiplied ~five-fold in women who suffered preeclampsia30. To note, one of the enriched GSEA gene-sets is linked to ageing kidney and fibrosis (Fig. 1). Our results are in line with a study showing that endothelial progenitor cells from PE are about twice as less numerous, with an increased differentiation time31.
In preeclamptic mice, heart weight was increased by ~10% and abnormal heart structure and histological features were found as well as strong alterations of the renin-angiotensin system (RAS) that may participate to cardiac hypertrophy, as observed in chronic kidney disease patients32. Resistance training-induced cardiac hypertrophy is characterized by up-regulation of the AT1R gene21. In return, RAS inhibition can reverse advanced cardiac remodeling by inducing aging in spontaneously hypertensive rats33. Endothelin-1 (ET-1) is also involved in cardiac hypertrophy induced by aging22. It is induced by Angiotensin II23. Here we have shown that several genes from the RAS system and ET-1 are increased in the hearts of mice carrying STOX1 transgenic embryos.
Cardiac hypertrophy has been described in PE women, even 6 years after the disease34. Pregnancy involves remodeling of heart tissue and increased heart blood flow, with molecular modifications of extracellular matrix (integrins, collagen, MMPs,..)35. However, this remodeling is assumed to be abnormal in preeclampsia. A link between cardiac dysfunction and preeclampsia has been substantiated by the study of Corin, a serine protease converting the pro-atrial natriuretic peptide to the active form (ANP) regulating blood pressure36. Mice lacking Corin have preeclampsia-like symptoms and mutations of the gene are found in patients with preeclampsia37,38.
Interestingly, we correlated abnormal expression of Connective Tissue Growth factor (Ctgf) with endothelial dysfunction and cardiac alterations. Ctgf (also known as IGFBP-8) is a mitogen associated with renal fibrosis39. Induction of blood pressure overload in mice that overexpress Ctgf in the heart, demonstrated that compensatory hypertrophy was limited compared to normal littermates showing that overexpression of Ctgf tends to prevent cardiac hypertrophy40. Contrastingly, inhibition of Ctgf with a monoclonal antibody has also been shown to limit the remodeling of the left ventricle following increased blood pressure41. Herein, we show that Ctgf was induced in the endothelium but repressed in the heart. In the light of the two studies described above, we conclude that down-regulation of Ctgf in the heart co-exist with cardiac hypertrophy, consistently with the results of Gravning and co-workers (2013).
A limit of our work is that we did not have the opportunity to analyze in detail heart function in the pregnant mice (cardiac rate, ejection fraction). This could be the focus on another work less centered on global analysis of gene expression. Another limit is that we did not study the long term effects of the preeclamptic pregnancy long after the gestation, which is a major issue of the health of mothers that experienced a preeclamptic pregnancy. Clearly in a further set of experiment, the STOX1 model could be used for thoroughly assessing this question.
It has been debated whether abnormal cardiovascular function was a consequence or a predisposing cause of preeclampsia42. In our model, where all the female mice are WT of the same genotype, the effects that we observed are clearly a consequence of pregnancy and not a consequence of a genetic difference between controls and affected mothers. We show that without further manipulation, STOX1 overexpression by the feto-placental unit induces cardiovascular dysfunction, suggesting that this model is a handy tool for understanding the systemic endothelial dysfunction in preeclampsia and testing therapies, to improve maternal health.
Additional Information
How to cite this article: Ducat, A. et al. Endothelial cell dysfunction and cardiac hypertrophy in the STOX1 model of preeclampsia. Sci. Rep. 6, 19196; doi: 10.1038/srep19196 (2016).
References
Sibai, B. M. Thrombophilia and severe preeclampsia: time to screen and treat in future pregnancies? Hypertension 46, 1252–3 (2005).
Firoz, T., Sanghvi, H., Merialdi, M. & von Dadelszen, P. Pre-eclampsia in low and middle income countries. Best Pract Res Clin Obstet Gynaecol 25, 537–48 (2011).
Goldenberg, R. L., Culhane, J. F., Iams, J. D. & Romero, R. Epidemiology and causes of preterm birth. Lancet 371, 75–84 (2008).
Sanchez-Aranguren, L. C., Prada, C. E., Riano-Medina, C. E. & Lopez, M. Endothelial dysfunction and preeclampsia: role of oxidative stress. Front Physiol 5, 372 (2014).
Melchiorre, K., Sutherland, G. R., Liberati, M. & Thilaganathan, B. Maternal cardiovascular impairment in pregnancies complicated by severe fetal growth restriction. Hypertension 60, 437–43 (2012).
Lykke, J. A. et al. Hypertensive pregnancy disorders and subsequent cardiovascular morbidity and type 2 diabetes mellitus in the mother. Hypertension 53, 944–51 (2009).
Doridot, L. et al. Preeclampsia-Like Symptoms Induced in Mice by Fetoplacental Expression of STOX1 Are Reversed by Aspirin Treatment. Hypertension 61, 662–668 (2013).
van Dijk, M. et al. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat Genet 37, 514–9 (2005).
Doridot, L. et al. Nitroso-Redox Balance and Mitochondrial Homeostasis Are Regulated by STOX1, a Pre-Eclampsia-Associated Gene. Antioxid Redox Signal 21, 819–34 (2014).
Sircar, M., Thadhani, R. & Karumanchi, S. A. Pathogenesis of preeclampsia. Curr Opin Nephrol Hypertens 24, 131–8 (2015).
Venkatesha, S. et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat Med 12, 642–9 (2006).
Goulopoulou, S. & Davidge, S. T. Molecular mechanisms of maternal vascular dysfunction in preeclampsia. Trends Mol Med 21, 88–97 (2015).
Chen, C. W., Jaffe, I. Z. & Karumanchi, S. A. Pre-eclampsia and cardiovascular disease. Cardiovasc Res 101, 579–86 (2014).
Marcellin, L. et al. Endometriosis also affects the decidua in contact with the fetal membranes during pregnancy. Hum Reprod 30, 392–405 (2015).
Aird, W. C., Jahroudi, N., Weiler-Guettler, H., Rayburn, H. B. & Rosenberg, R. D. Human von Willebrand factor gene sequences target expression to a subpopulation of endothelial cells in transgenic mice. Proc Natl Acad Sci USA 92, 4567–71 (1995).
DeLisser, H. M., Newman, P. J. & Albelda, S. M. Molecular and functional aspects of PECAM-1/CD31. Immunol Today 15, 490–5 (1994).
Hahne, M., Lenter, M., Jager, U., Isenmann, S. & Vestweber, D. VCAM-1 is not involved in LPAM-1 (alpha 4 beta p/alpha 4 beta 7) mediated binding of lymphoma cells to high endothelial venules of mucosa-associated lymph nodes. Eur J Cell Biol 61, 290–8 (1993).
Dejana, E. & Orsenigo, F. Endothelial adherens junctions at a glance. J Cell Sci 126, 2545–9 (2013).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102, 15545–50 (2005).
Myatt, L., Muralimanoharan, S. & Maloyan, A. Effect of preeclampsia on placental function: influence of sexual dimorphism, microRNA's and mitochondria. Adv Exp Med Biol 814, 133–46 (2014).
Barauna, V. G., Magalhaes, F. C., Krieger, J. E. & Oliveira, E. M. AT1 receptor participates in the cardiac hypertrophy induced by resistance training in rats. Am J Physiol Regul Integr Comp Physiol 295, R381–7 (2008).
Ceylan-Isik, A. F. et al. Cardiomyocyte-specific deletion of endothelin receptor A rescues aging-associated cardiac hypertrophy and contractile dysfunction: role of autophagy. Basic Res Cardiol 108, 335 (2013).
Fujita, H. et al. DPP-4 inhibition with alogliptin on top of angiotensin II type 1 receptor blockade ameliorates albuminuria via up-regulation of SDF-1alpha in type 2 diabetic patients with incipient nephropathy. Endocr J 61, 159–66 (2014).
Brohee, S. et al. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic Acids Res 36, W444–51 (2008).
Andrade, D. et al. Interferon-alpha and angiogenic dysregulation in pregnant lupus patients destined for preeclampsia. Arthritis Rheumatol 67, 977–87 (2015).
Clowse, M. E., Jamison, M., Myers, E. & James, A. H. A national study of the complications of lupus in pregnancy. Am J Obstet Gynecol 199, 127 e1–6 (2008).
Licastro, F. et al. Sharing pathogenetic mechanisms between acute myocardial infarction and Alzheimer's disease as shown by partially overlapping of gene variant profiles. J Alzheimers Dis 23, 421–31 (2011).
Harmon, Q. E. et al. Polymorphisms in inflammatory genes are associated with term small for gestational age and preeclampsia. Am J Reprod Immunol 71, 472–84 (2014).
Rahardjo, B., Widjajanto, E., Sujuti, H. & Keman, K. Different levels of IL-1alpha, IL-6, TNF-alpha, NF-kappaB and PPAR-gamma in monocyte cultures exposed by plasma preeclampsia and normotensive pregnancy. Pregnancy Hypertens 4, 187–93 (2014).
Vikse, B. E., Irgens, L. M., Leivestad, T., Skjaerven, R. & Iversen, B. M. Preeclampsia and the risk of end-stage renal disease. N Engl J Med 359, 800–9 (2008).
Hwang, H. S. et al. Increased senescence and reduced functional ability of fetal endothelial progenitor cells in pregnancies complicated by preeclampsia without intrauterine growth restriction. Am J Obstet Gynecol 199, 259 e1–7 (2008).
Raizada, V., Hillerson, D., Amaram, J. S. & Skipper, B. Angiotensin II-mediated left ventricular abnormalities in chronic kidney disease. J Investig Med 60, 785–91 (2012).
Milingos, D. et al. Insulin-like growth factor-1 isoform mRNA expression in women with endometriosis: eutopic endometrium versus endometriotic cyst. Ann N Y Acad Sci 1092, 434–9 (2006).
Ghossein-Doha, C. et al. Hypertension after preeclampsia is preceded by changes in cardiac structure and function. Hypertension 62, 382–90 (2013).
Li, J. et al. New frontiers in heart hypertrophy during pregnancy. Am J Cardiovasc Dis 2, 192–207 (2012).
Cui, Y. et al. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature 484, 246–50 (2012).
Dong, N. et al. Corin mutations K317E and S472G from preeclamptic patients alter zymogen activation and cell surface targeting. [Corrected]. J Biol Chem 289, 17909–16 (2014).
Stepanian, A. et al. Highly significant association between two common single nucleotide polymorphisms in CORIN gene and preeclampsia in Caucasian women. PLoS One 9, e113176 (2014).
Chen, X. M., Qi, W. & Pollock, C. A. CTGF and chronic kidney fibrosis. Front Biosci (Schol Ed) 1, 132–41 (2009).
Gravning, J., Ahmed, M. S., von Lueder, T. G., Edvardsen, T. & Attramadal, H. CCN2/CTGF attenuates myocardial hypertrophy and cardiac dysfunction upon chronic pressure-overload. Int J Cardiol 168, 2049–56 (2013).
Szabo, Z. et al. Connective tissue growth factor inhibition attenuates left ventricular remodeling and dysfunction in pressure overload-induced heart failure. Hypertension 63, 1235–40 (2014).
Osol, G. & Bernstein, I. Preeclampsia and maternal cardiovascular disease: consequence or predisposition ? J Vasc Res 51, 290–304 (2014).
Acknowledgements
Part of this work was supported by a ‘Who am I’ Labex grant and part by an Institut Cochin ‘Proof of Concept grant’. A.D., L.D. and R.C., are or were PhD students from the René-Descartes University under contracts by the University for the duration of their thesis.
Author information
Authors and Affiliations
Contributions
A.D., L.D. and R.C. carried out most of the experimental work and analyzed the results, C.M. carried out the heart histology experiments, J.L.V. and J.C. were in charge of the mouse work, S.B. drafted the manuscript and contributed to the q-RT, PCR experiments, as well as B.C., S.J. and F.L. carried out the transcriptome analysis of the HUVEC cells, FLG carried out with R.C. and A.D. the purification of endothelial cells from the mice, PL contributed to the RNAseq analysis, F.M. and D.V. coordinated the work, drafted the paper, carried out the bioinformatics analyses. All the authors contributed to improve the draft.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Ducat, A., Doridot, L., Calicchio, R. et al. Endothelial cell dysfunction and cardiac hypertrophy in the STOX1 model of preeclampsia. Sci Rep 6, 19196 (2016). https://doi.org/10.1038/srep19196
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19196
This article is cited by
-
The Clinical Value of Rodent Models in Understanding Preeclampsia Development and Progression
Current Hypertension Reports (2023)
-
Impaired in vitro growth response of plasma-treated cardiomyocytes predicts poor outcome in patients with transthyretin amyloidosis
Clinical Research in Cardiology (2021)
-
Pregnancy-associated cardiac dysfunction and the regulatory role of microRNAs
Biology of Sex Differences (2020)
-
Long noncoding RNAs: emerging roles in pulmonary hypertension
Heart Failure Reviews (2020)
-
FOXD1 mutations are related to repeated implantation failure, intra-uterine growth restriction and preeclampsia
Molecular Medicine (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.