Abstract
The transcription factors HSF1 and p53 both modulate the stress response, thereby protecting and facilitating the recovery of stressed cells, but both have the potential to promote tumor development. Here we show that a p53 target gene, IER5, encodes an activator of HSF1. IER5 forms a ternary complex with HSF1 and the phosphatase PP2A and promotes the dephosphorylation of HSF1 at numbers of serine and threonine residues, generating a novel, hypo-phosphorylated active form of HSF1. IER5 is also transcriptionally upregulated in various cancers, although this upregulation is not always p53-dependent. The IER5 locus is associated with a so-called super enhancer, frequently associated with hyperactivated oncogenes in cancer cell lines. Enhanced expression of IER5 induces abnormal HSF1 activation in cancer cells and contributes to the proliferation of these cells under stressed conditions. These results reveal the existence of a novel IER5-mediated cancer regulation pathway that is responsible for the activation of HSF1 observed in various cancers.
Similar content being viewed by others
Introduction
The p53 tumor suppressor gene is one of the most frequently mutated genes in human cancer and the loss of functional p53 is a prerequisite for oncogenesis in many cancers1. p53 functions as a transcriptional activator that induces various genes involved in the suppression of tumorigenesis2,3,4. These target genes modulate growth arrest, DNA repair, cell death, metabolism, cellular homeostasis and a variety of other functions. Under conditions of severe stress, p53 induces apoptosis and/or senescence to eliminate cells that are irreparably damaged. However, under conditions of mild stress, p53 instead elicits a survival response, induces genes that are involved in cell-cycle arrest, DNA repair and regulation of metabolism and thereby acts to protect cells and facilitate their recovery from stress5. This p53-mediated survival response may suppress tumorigenesis in normal cells, but may have the potential to promote tumor development in cells that otherwise would not recover or be repaired. For example, among the p53 target genes are several that adapt cells to metabolic changes such as nutrient deprivation and ROS. This function could enable cancer cells to survive under harsh conditions and thereby contribute to cancer progression4.
HSF1 is a constitutively expressed transcriptional activator, a master regulator of the heat shock response and is post-translationally activated following heat shock6. Under non-stressed conditions, HSF1 exists as a monomer in complex with HSP90, which negatively regulates its activity. Heat shock induces the dissociation of HSF1 from HSP90, allowing HSF1 to multimerize into a trimer, which can accumulate in the nucleus and bind DNA. Activated HSF1 induces a number of HSF1 target genes including the HSP family of genes, which allow the cell to adapt and recover from stress. Moreover, it has been recently reported that HSF1 can promote tumorigenesis. HSF1 is constitutively activated in cancer tissues and higher HSF1 activity is related to poorer prognosis of cancer patients7. HSF1 has also been shown to transactivate a variety of genes involved in tumor progression8. A study of HSF1-deficient mice showed that tumorigenesis driven by oncogenic ras or mutant p53 is HSF1-dependent9. The ability of HSF1 to promote tumor development is critically dependent on its ability to upregulate HSF1 target genes, including the HSP family of genes, which facilitate the survival of cancer cells and their adaptation to hostile conditions8. Thus, HSF1 resembles p53 in its ability to both protect stressed cells and in its potential to promote tumor formation.
Previously, we searched for novel genes regulating tumorigenesis by analyzing p53 target genes10. We introduced a temperature-sensitive p53 mutant into the p53-null Saos2 cell line and looked for genes that were induced upon temperature shift to the permissive temperature. In addition, to identify genes modulated directly by p53, we performed Chip-seq analysis using HCT116 cells, which contain wild-type p53. From these analyses, we identified several putative p53 target genes, i.e., genes that are both induced by p53 and to which p53 binds11,12. Here, we report that one of these p53 target genes, IER5, is a hitherto unknown activator of HSF1 that regulates HSF1 activity via a novel mechanism. We show that IER5 facilitates the dephosphorylation of several serine and threonine residues in HSF1, generating a hypo-phosphorylated active form of HSF1 that is distinct from that induced by heat shock. The IER5 gene has been known as an immediate-early gene induced by various growth-promoting stimuli and is overexpressed in various cancers13,14. We observe that depletion of IER5 in cancer cells results in decreased HSF1 activity. Furthermore, IER5-mediated activation of HSF1 is required for anchorage-independent cell growth of cancer cells. These results collectively indicate that IER5 has oncogenic potential and is responsible for the activation of HSF1 in cancer.
Results
The IER5 gene is a p53 target gene
In order to discover potentially novel, cancer associated genes, we previously undertook a comprehensive effort to identify p53 target genes and subsequently analyzed several whose functions were unknown10,11,12,15,16. This study focuses on one of these genes, IER5. IER5 mRNA (Fig. 1A–C) and protein (Fig. 1D,E) are induced by DNA-damaging reagents such as 5-FU, γ-ray and Adriamycin (doxorubicin). In addition, induction of IER5 mRNA and protein by these treatments is p53-dependent (Fig. 1A,B,D).
The IER5 gene is a p53 target gene.
(A–C) IER5 expression was analyzed by Northern blotting. HCT116 p53 (+/+), HCT116 p53 (−/−) were subjected to γ-ray irradiation (20 Gy) (A). Normal human fibroblasts (MRC5 cells) transfected with control lentivirus or lentivirus expressing p53 shRNA were subjected to γ-ray irradiation (30 Gy) (B). Cell lines carrying wild-type p53 were treated with Adriamycin (1 μM) for 24 hrs (MCF7, U2OS and MRC5) or 19 hrs (A549) (C). (D) HCT116 p53 (+/+), HCT116 p53 (−/−) were treated with 5-FU (0.038 mM) for 24 hrs. Expression of IER5, p53 and p21 (representative p53 target gene) were analyzed by Western blotting. (E) Cell lines carrying wild-type p53 were treated with Adriamycin (1 μM) for 24 hrs (MRC5, MCF7, U2OS) or 19 hrs (A549). Expression of IER5 and p53 were analyzed by Western blotting. (F) Genomic locus of IER5 is shown together with the ChIP-seq and 4C-seq results. HCT116 p53 (+/+) cells treated with or without 5-FU were used for ChIP-seq analysis. ChIP-seq analyses were performed using antibodies against p53, H3K27ac, H3K4me1, H3K4me3 and phospho-RNAP II. Two p53 binding sites RE1 (11 kb downstream) and RE2 (46 kb downstream) were identified. Chromatin interaction was detected by 4C-seq analysis in HCT116 p53+/+ and HCT116 p53−/− cells with or without 5-FU. Primers were designed around RE2 and the genomic regions interacting with RE2 were determined. (G) The positions and nucleotide sequences of RE1 and RE2. (H) Double-stranded synthetic oligonucleotides containing RE1, RE2 or mutant RE2 (the sequences are shown in G) were inserted into the luciferase reporter plasmid containing a minimal promoter. The assay was performed 24 hrs post-transfection. The experiment was run in triplicate and data are represented as the mean-fold activation ±SD.
The IER5 gene promoter region was analyzed by ChIP-seq analysis of 5-FU treated and untreated HCT116 p53+/+ cells. We detected H3K4 tri-methylation surrounded by H3K4 mono-methylation at the IER5 gene promoter region, indicating that IER5 is actively transcribed in both unstressed and stressed conditions (Fig. 1F). Furthermore, we detected phospho-RNA polymerase II binding to the IER5 promoter and this binding increased upon 5-FU treatment, consistent with enhanced transcription of IER5. ChIP-seq analysis identified two p53 binding sites 11 kb (RE1) and 46 kb (RE2) downstream of the IER5 gene (Fig. 1F). p53 binding to RE2 was strong, while binding to RE1 was relatively weak. In addition, H3K27 acetylation was detected at RE2 and was strongly increased by 5-FU treatment, showing that RE2 is an active enhancer that is highly activated upon DNA damage. In contrast, we could not detect H3K27 acetylation at RE1, suggesting that it is not an active enhancer, even though it does bind p53. We found sequences highly similar to the p53 consensus binding sequence at the center of RE2 (TRANSFAC match score; 0.94) but not in RE1 (TRANSFAC match score; 0.74, Fig. 1G). We generated DNA oligonucleotides representing RE1 and RE2 and created a version of RE2 mutated in the p53 consensus sequence (RE2 mut; sequences shown in Fig. 1G) and cloned each upstream of a luciferase reporter gene containing a minimal promoter. As shown in Fig. 1H, p53 strongly activated a reporter containing RE2, but not RE2 mut or RE1. The lack of RE1 enhancer activity is consistent with the weak binding of p53 and the lack of H3K27 acetylation at RE1 seen with ChIP-seq, indicating that RE1 is not an active enhancer.
We further performed 4C-seq analysis to identify chromatin regions that might interact with RE2. As shown in Fig. 1F, we detected chromatin interaction between RE2 and RE1 and between RE2 and the IER5 gene promoter region, in both HCT116 p53 +/+ and −/− cells. The interactions between the RE2 and the IER5 promoter region seem to be stabilized by 5-FU treatment in HCT116 p53 +/+ cells but not in HCT116 p53 −/− cells. In contrast, the interactions between RE2 and RE1 seem to be stronger in HCT116 p53 +/+ cells compared to HCT116 p53 −/− cells, but are unchanged by 5-FU treatment. The RE2 region also interacts with several regions that are positive for H3K4me1. Collectively, these results show that RE2 is localized in the vicinity of the IER5 promoter when a higher order chromatin structure is formed and identifies RE2 as the p53-responsive element of the IER5 gene. In addition, since RE1 and RE2 were constitutively associated in the absence of 5-FU treatment in HCT p53 +/+ cells, p53 binding to both RE1 and RE2 may facilitate the formation of higher-order chromatin structure at this locus.
IER5 transcription is driven by super-enhancers in cancer cells, leading to enhanced IER5 expression
Originally, IER5 was reported as a gene induced by growth-promoting stimuli and that is highly expressed in cervical cancers13,14. Consistent with this, we observed that IER5 could be induced by various mitogenic stimuli such as PMA, serum and ionomycin (Fig. 2A and S1A). In addition, IER5 upregulation and overexpression has been observed in a number of cancers (Fig. 2B and S1B). These results collectively suggest that various growth-promoting and oncogenic signals can induce expression of IER5.
IER5 mRNA expression is increased in various cancers and IER5 is required for anchorage-independent cancer cell proliferation.
(A) Mouse embryonic fibroblasts and NIH3T3 cells were treated with TPA (160 ng/ml) for 30 min. NIH3T3 cells were also cultured with 0.5% FBS DMEM medium overnight and then stimulated by 10% FBS DMEM for 2 hrs. Expression of IER5 mRNA was analyzed by quantitative RT-PCR. (*p < 0.05, **p < 0.01). (B) IER5 mRNA expression levels were analyzed in the indicated cancers and their corresponding normal tissues using the Gene Logic SCIANTIS database. (*p < 0.05, ***p < 0.001). (C) The IER5 gene is associated with super-enhancers in various cancer cell lines. The locus of the IER5 gene is shown with both of the human genome reference versions, Hg19/Build37 (upper) and Hg18/Build36 (lower). The regions identified as containing super enhancers by Hnisz et al. are shown by yellow lines17. (D) IER5 mRNA expression in various cancer cell lines. Significance of difference was calculated between non-super enhancer cell lines and super enhancer cell lines. (**p < 0.01, ***p < 0.001, #p < 0.0001). (E–G) OE33 cells were plated in adherent (E) or suspension (F) 96-well culture plates and control or IER5-targeting siRNAs were introduced. Cell growth assays were performed at the indicated days. Mean relative cell numbers ±SD are shown. IER5 mRNA expression was analyzed by quantitative RT-PCR 48 hrs post-transfection (G). (**p < 0.01, #p < 0.0001). (H–J) Anchorage-independent growth in soft agar was analyzed. Control or IER5-targeting siRNAs were introduced into OE33 cells. The representative images (H) and the mean numbers of colonies ±SD are shown (I). Expression of IER5 (J) was analyzed by quantitative RT-PCR 48 hrs post-transfection. (*p < 0.05, **p < 0.01). (K,L) Cell growth assays were performed using the indicated cell lines. Cells were plated in suspension culture plates and analyzed as in (F). Cell growth was analyzed 7 days (HCC1954, MCF7 and HeLa cells) or 8 days (MDA-MB-231 cells) post-transfection (K). IER5 mRNA expression was analyzed by quantitative RT-PCR 48 hrs post-transfection (L). (*p < 0.05, **p < 0.01).
Recently, it was reported that many cancer cells acquire so-called “super-enhancers” at key driver oncogenes and other genes important for tumor pathogenesis17,18. Since enhanced IER5 expression is observed in many cancers, we asked whether the IER5 gene locus might also be associated with a super-enhancer in cancer cells. A comprehensive analysis of super-enhancers in 6 blood cancer-derived cell lines and 12 solid cancer-derived cell lines has been recently reported17. The IER5 gene was included in this analysis and as shown in Fig. S1C, there was no association of the IER5 gene with a super enhancer in the 6 blood cancer-derived cell lines we examined, but there was an association in 5 of the 12 solid cancer-derived cell lines, regardless of p53 status. These super enhancers were localized around the IER5 gene locus including the RE1 and RE2 regions (Fig. 2C). This remarkably high incidence of super-enhancer association with the IER5 gene in cancer cell lines supports the idea that IER5 may be an important gene in tumor pathogenesis.
IER5 is required for anchorage-independent cancer cell growth
In order to analyze the function of IER5 in tumorigenesis, we examined the effect of IER5 expression on the growth of cancer cells. First, we analyzed the expression levels of IER5 in several cancer cell lines including cell lines identified as having or not having IER5-associated super enhancers: HCC1954, HeLa, MCF7, LNCaP and HepG2 (Fig. 2D). Cell lines that have super enhancers at the IER5 locus had higher IER5 mRNA expression compared to cell lines without them. We took the OE33 esophageal cancer cell line, as it expressed relatively high levels of IER5 and analyzed the effect of IER5 knockdown under both adherent (Fig. 2E) and suspension (Fig. 2F) cell culture conditions. IER5 knockdown was confirmed as shown in Fig. 2G. We found that decreased IER5 expression had no effect on growth in adherent cultures, but had a significant effect in suspension cultures. In addition, IER5 knockdown significantly decreased colony formation in soft agar (Fig. 2H,I). IER5 knockdown was confirmed as shown in Fig. 2J. We next analyzed the effect of IER5 knockdown in four of the other cancer cell lines and saw that IER5 had a significant effect on the growth in each line under suspension culture conditions (Fig. 2K). IER5 knockdown was confirmed as shown in Fig. 2L. These data collectively indicate that IER5 has an important function in anchorage-independent proliferation of cancer cells.
IER5 induces transcription of HSP family genes
We next undertook a comprehensive gene expression analysis in cells expressing IER5 using gene microarrays. As shown in Fig. 3A, several genes from the HSP family were modestly or highly induced by IER5 expression. These results were further confirmed by Northern blotting (Fig. 3B) and found to require intact N- and C- termini of IER5 (Fig. S2). Upregulation of HSP family mRNA expression by IER5 was accompanied by markedly enhanced HSP family protein expression (Fig. 3C). Conversely, siRNA suppression of endogenous IER5 resulted in decreased HSP family mRNA and protein expression (Fig. 3D–F).
HSP family genes are induced by IER5.
(A) H1299 or 293T cells were transfected with control vector or an IER5 expression vector. Cells were harvested 21 hrs or 27 hrs post-transfection and microarray expression analysis was performed. The table shows the HSP family genes, among the genes induced by IER5. (B) H1299 cells were transfected with control, IER5-Flag or mutant IER5-Flag expression vectors (representative image of mut 1 is shown in Fig. S1). Cells were harvested 27 hrs post-transfection and mRNA expressions of the HSP family genes were analyzed by Northern blotting. (C) H1299 and 293T cells were transfected with control vector or IER5-Flag expression vector and cells were harvested 24 hrs post-transfection. Expressions of the HSP family proteins were analyzed by Western blotting. (D–F) Control or IER5-targeting siRNAs were introduced into OE33 cells. Cells were harvested 52 hrs post-transfection. Expression of IER5 (D,F) and HSPA1A (E,F) were analyzed by quantitative RT-PCR (D,E) and Western blotting (F). (**p < 0.01). (G) The promoter regions of HSPA1A, HSPA1B and HSPA6 were inserted into the luciferase reporter plasmid containing a minimal promoter and assayed 24 hrs post-transfection. Experiments were run in triplicate and data are represented as the mean-fold activation ±SD. (H) Serially deleted regions of the HSPA1A promoter were analyzed as in (G). Numbers indicate the position of the 5′ most nucleotide relative to the transcription initiation site. A heat shock element (HSE), to which HSF1 binds, was found between positions −132 and −109.
We next employed a luciferase promoter reporter assay to analyze whether IER5 activates the promoters of HSP family genes. As shown in Fig. 3G, the HSPA1A, HSPA1B and HSPA6 gene promoters were strongly activated by IER5. In order to identify the IER5-responsive element(s), we generated a series of serially deleted HSPA1A promoter regions and cloned these upstream of the luciferase reporter gene. This analysis indicated that the region between −310 and −100 relative to the transcription initiation site of the HSPA1A gene mediates IER5 responsiveness (Fig. 3H). Analysis of the sequences within this region revealed the presence of HSF1 transcription factor binding sites, so-called heat shock elements (HSEs).
IER5 activates HSF1
HSF1 binds to HSEs within the promoters of HSP family genes and induces their transcription in response to stresses such as heat stress19. As we found an HSE within the IER5-responsive region of the HSPA1A gene promoter, we analyzed whether induction of HSP family genes by IER5 requires HSF1. As shown in Fig. 4A–C, siRNA suppression of endogenous HSF1 abrogated the induction of these HSP family genes by IER5, showing that IER5 upregulation of HSP family genes is dependent on HSF1. HSF1 activation can be regulated at two steps: HSF1 oligomerization and nuclear localization. It has been reported that the association of HSF1 with HSP90 keeps HSF1 in an inactive state by inhibiting HSF1 oligomerization. Accordingly, the dissociation of HSP90 from HSF1 allows oligomerization and activation of HSF16. As shown in Fig. 4D,E, expression of IER5 resulted in decreased association of HSP90 with HSF1 and increased HSF1 oligomerization. Furthermore, expression of wild-type IER5, but not mutant IER5, resulted in a greater accumulation of HSF1 in the nucleus (Fig. 4F). Finally, we examined DNA binding by HSF1 in cell extracts using biotin-tagged DNA probes containing an HSE and bound to streptavidin-coated beads. Binding of HSF1 to HSE was strongly increased in cells overexpressing IER5 (Fig. 4G). These data collectively indicate that IER5 induces HSF1 activation and transactivation of the HSP family of genes.
IER5 is a novel activator of HSF1.
(A–C) Control or HSF1-targeting siRNAs were introduced into H1299 cells. Subsequently, cells were transfected with control vector or an IER5 expression vector 24 hrs post-siRNA transfection. Cells were harvested 24 hrs post-DNA transfection. Expression of HSPA1A (A), HSPA6 (B) and HSF1 (C) mRNAs were analyzed by quantitative RT-PCR. (#p < 0.0001). (D) 293T cells were transfected with HA-HSF1 and control vector or IER5-Flag. Cells were harvested 24 hrs post-transfection. Cell lysates were crosslinked by DSP (1 mg/ml) and immunoprecipitated using anti-HA antibody. HSP90 binding to HA-HSF1 was detected by Western blotting. (E) H1299 cells were transfected with control vector or IER5-Flag expression vector. Cells were harvested 25 hrs post-transfection. Whole cell lysates were crosslinked with EGS. HSF1 and IER-Flag expression was analyzed by Western blotting. (F) H1299 cells were transfected with control vector, IER5-Flag, mut 1-Flag expression vector. Cells were harvested 24 hrs post-transfection. Subcellular localizations of endogenous HSF1 (detected using anti-HSF1 antibody), IER5-Flag and mut 1 (detected using anti-Flag antibody) were analyzed. HSF1 and IER5 expression levels were quantitated and shown at the bottom. (G) HSF1 binding to Heat Shock Elements (HSEs) was analyzed by streptavidin pull-down assay. Biotinylated control or HSE-containing DNA probes were bound to streptavidin-coated beads and mixed with control or IER5-expressing cell lysates. Bead-bound HSF1 was denatured in Laemmli buffer and analyzed by Western blotting.
IER5 dephosphorylates HSF1 at multiple Ser and Thr residues, including sites involved in the repression of HSF1 activity
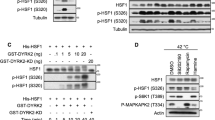
HSF1 activation is regulated by various post-translational modifications, especially phosphorylation. For example, phosphorylation at Ser 230, 320 and 326 has been reported to increase HSF1 transactivation20,21,22. We therefore asked if HSF1 phosphorylation is affected by expression of IER5. We observed that HSF1 shifts to a higher electrophoretic mobility in cells expressing IER5 (Fig. 5A). Conversely, ablation of endogenous IER5 expression resulted in decreased HSF1 electrophoretic mobility (Fig. 5B). When HSF1 protein was treated with phosphatase prior to electrophoresis, no difference in HSF1 migration between IER5-expressing and control cells was observed (Fig. 5C). These results show that IER5 dramatically changes the post-translational modification of HSF1, in particular causing HSF1 dephosphorylation. Heat shock, which activates HSF1, induced strong phosphorylation of serines known to be involved in HSF1 activation: Ser230, 320 and 326 (Fig. 5D). In contrast, HSF1 phosphorylation at these sites was relatively unaffected by IER5 expression (Fig. 5E).
IER5 dephosphorylates HSF1 at multiple Ser and Thr residues including sites involved in repression of HSF1 activity.
(A–E) Whole cell extracts and immunoprecipitated samples were analyzed by Western blotting. (A) 293T cells were transfected with HSF1-Flag together with control vector or an IER5 expression vector. Cells were harvested 21 hrs post-transfection. (B) 293T cells were transfected with control vector or HSF1-Flag expression vector, together with control or IER5-targetting siRNAs. Cells were harvested 49 hrs post-transfection. (C) 293T cells were transfected with HSF1-Flag and control vector or IER5. Cells were harvested 27 hrs post-transfection. Cell lysates were immunoprecipitated using anti-Flag antibody and incubated with or without CIAP for 30 min. Total HSF1 and p-Ser320 were detected. (D) 293T cells were transfected with control vector or HA-HSF1 expression vector. Cells were treated with or without heat shock at 42 °C for 3 hrs and harvested 24 hrs post-DNA transfection. (E) 293T cells were transfected with HA-HSF1 and control or IER5-Flag expression vector. Cells were harvested 24 hrs post-DNA transfection. (F) HSF1 modification was analyzed by LC-MS/MS. The experiment was performed nine times and the numbers of analysis that detected each phosphorylation are shown. Significant reductions in phosphorylation at 5 residues (S121, S307, S314, T3232 and T367) were detected. Phosphorylation sites previously reported to be involved in the repression of HSF1 activity (S121 and S307) are shown in blue. (G) 293T cells were transfected with the indicated plasmids and harvested 24 hrs post-transfection. HSPA1A mRNA expression was analyzed by quantitative RT-PCR and other proteins were detected by Western Blotting. (***p < 0.001, #p < 0.0001). (H,I) 293T cells were transfected with control vector or IER5-Flag expression vector. Cells were treated with or without heat shock at 42 °C for 3 hrs and harvested 24 hrs post-DNA transfection. Endogenous HSF1, IER5-Flag protein expression was analyzed by Western blotting (H) and HSPA1A, HSPA6 and HSPA1B mRNA expression was analyzed by quantitative RT-PCR (I). (*p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001).
In order to comprehensively identify the modifications induced by IER5, we analyzed HSF1 in IER5-expressing versus control cell extracts by LC-MS/MS. We found that expression of IER5 was associated with decreased phosphorylation levels at multiple sites (Fig. 5F). In particular, significant reductions in phosphorylation were observed at 5 residues (S121, S307, S314, T323 and T367). We also observed a reduction in the acetylation of E113 and K118 (Fig. S3). Interestingly, the amino acids exhibiting these modification differences reside within the regulatory domain (RD, a.a. 221–383) or the flexible linker region (a.a. 110–130) of HSF1. The RD is located between the heptad repeat (HR)-A/B and the HR-C domain and is involved in the repression of HSF1 activity. It has been reported that various post-translational modifications in the RD modulate its ability to repress HSF1 function. For example, phosphorylation of Ser320 and 326 increases HSF1 activity, while phosphorylation of Ser303 and 307 represses HSF1 activity23,24,25. In addition, the flexible linker region that connects the HSF1 DNA binding domain and the HR-A/B domain has been reported to be involved in the regulation of HSF1 activity and to influence HSF1 trimerization6. For example, phosphorylation of Ser121 within the linker region promotes HSP90 binding, which in turn represses HSF1 trimerization26. IER5 expression significantly reduces phosphorylation of Ser121 and 307, which are involved in the negative regulation of HSF1 activity (Fig. 5F). We therefore next analyzed whether HSF1 activation by IER5 involves the dephosphorylation of S121, S307, S314, T323 and/or T367. As shown in Fig. 5G, endogenous HSF1 induced expression of HSPA1A, an indicator of HSF1 activation, in a manner dependent on IER5. HSPA1A was not induced by ectopic expression of HSF1 alone, but was strongly induced when IER5 was expressed together with HSF1. We next constructed several mutant versions of HSF1 containing phosphorylation mimic mutants of these sites, as shown in Fig. 5G. We observed that IER5-dependent HSPA1A induction was significantly lower than wild-type when any of these mutants (2S/D, 3S/D, 4S/D and 5S/D) was expressed (Fig. 5G). In particular, we note that the 5S/D mutant dramatically reduces HSPA1A induction (22% activity compared to ectopic wild-type HSF1). These results suggest that dephosphorylation of S121, S307, S314, S323 and T367 are essential for HSF1 activation.
IER5 converts HSF1 to a hypo-phosphorylated active form, while heat shock converts HSF1 to a hyper-phosphorylated active form. To investigate whether IER5-mediated dephosphorylation of HSF1 can further activate HSF1 under conditions of heat stress, we subjected control or IER5-expressing cells to heat shock treatment (Fig. 5H,I). Induction of HSP family genes following heat shock was normal in control cells, but dramatically enhanced in cells expressing IER5. An increase in HSF1 electrophoretic mobility was also observed in heat-shocked cells expressing IER5 compared to HSF1 in heat-shocked cells in which IER5 was not expressed (Fig. 5H). Dephosphorylation of HSF1 by IER5 can therefore further activate HSF1 above the levels of activation induced by heat shock alone.
IER5 regulates dephosphorylation of HSF1 by PP2A
Our results indicate that IER5 modulates HSF1 activity by inducing dephosphorylation at serine and threonine residues, suggesting the potential involvement of a Ser/Thr protein phosphatase. To test this idea, we treated the cells with NaF, a general inhibitor of Ser/Thr protein phosphatases and found that IER5-activated, HSF1-dependent induction of two HSP family genes was significantly reduced (Fig. 6A,B). Moreover, NaF treatment caused HSF1 to be hyper-phosphorylated, indicating the involvement of a Ser/Thr protein phosphatase that constitutively dephosphorylates HSF1 (Fig. 6C). One potential candidate involved in this response is PP2A, a regulatory subunit of which was reported to associate with IER5 and which was reported to interact with the HSF family protein HSF227,28. PP2A is a phosphatase involved in various biological phenomena, including tumorigenesis29,30. To examine whether PP2A is the phosphatase responsible for the dephosphorylation and activation of HSF1 by IER5, we treated cells expressing IER5 and HSF1 with okadaic acid, an inhibitor of PP2A. As shown in Fig. 6D, okadaic acid treatment resulted in the decreased electrophoretic mobility of HSF1 and reduced induction of HSPA1A. Okadaic acid also dramatically enhanced the phosphorylation of Ser121 and 303/307, suggesting that PP2A may dephosphorylate these residues (Fig. 6E). Lastly, we knocked down PP2A expression in IER5-expressing cells and found that this caused a decrease in HSF1 electrophoretic mobility, as well as a decrease in HSPA1A and HSPA6 induction (Fig. 6F–H). These results collectively indicate that PP2A is required for HSF1 activation and HSF1 dephosphorylation by IER5.
IER5 induces PP2A-dependent dephosphorylation and activation of HSF1.
(A–C) 293T cells were transfected with HA-HSF1 together with control vector or IER5-Flag. Cells were treated with 20 mM NaF acid for 4 hrs and harvested 24 hrs post-transfection. Expression of HSPA1A and HSPA6 were analyzed by quantitative RT-PCR (A,B) and expression of IER5-Flag and HA-HSF1 were detected by Western Blotting (C). (**p < 0.01, ***p < 0.001). (D) 293T cells were transfected with HA-HSF1 together with control vector or IER5-Flag. Cells were treated with 500 nM okadaic acid for 8 hrs and harvested 24 hrs post-transfection. Expression of HSPA1A was analyzed by quantitative RT-PCR and expression of IER5-Flag and HA-HSF1 were detected by Western Blotting. (***p < 0.001). (E) 293T cells were transfected as in (D). Cells were treated with 250 nM okadaic acid for 4 hrs and harvested 24 hrs post-transfection. Cell lysates were immunoprecipitated using anti-HA antibody. Total HSF1 and HSF1 phosphorylated at Ser121 or p-Ser303, 307 were detected by Western Blotting. (F–H) Control or PPP2CA (PP2A catalytic subunit)-targeting siRNAs were introduced into 293T cells. Subsequently, cells were transfected with HA-HSF1 and control vector or IER5-Flag expression vector 72 hrs post-siRNA transfection. Cells were harvested 24 hrs post-DNA transfection. IER5, HSF1 and PPP2CA protein levels were analyzed by Western blotting and HSPA1A (G) and HSPA6 (H) mRNAs were analyzed by quantitative RT-PCR. (**p < 0.01). (I,J) 293T cells were transfected with HA-HSF1 together with control vector, IER5-Flag or IER5-Flag mut 1-Flag and harvested 24 hrs post-transfection. IER5-Flag (I) or HA-HSF1 (J) was immunoprecipitated using anti-Flag or anti-HA antibodies. Association of HSF1, PP2A catalytic subunit PPP2CA and PP2A regulatory subunit B55 with IER5 was analyzed by Western blotting in (I) and association of IER5, PPP2CA and B55 with HSF1 was analyzed in (J). (K) Endogenous IER5 in OE33 cells was immunoprecipitated using anti-IER5 antibody. Association of HSF1 and PPP2CA with IER5 was analyzed. (L) PP2A phosphatase activities were analyzed using the indicated amounts of cell lysates. 293T cells were treated with or without Okadaic acid (500 nM) for 8 hrs. Control or IER5-Flag expression vector were transfected and harvested 24 hrs post-transfection.
IER5 forms a ternary complex with HSF1 and PP2A
We next analyzed the mechanism by which IER5 and PP2A activate HSF1. As shown before in Fig. 4F, expression of IER5 results in HSF1 accumulation in the nucleus and co-localization with IER5, suggesting a possible physical association of these two factors. To test this, we transfected 293T cells with Flag-tagged wild-type- or mutant IER5 and HA-tagged HSF1. Next, IER5 and HSF1 were immunoprecipitated with anti-Flag or anti-HA antibody, respectively. Figure 6I,J show that HSF1 associates with wild-type IER5, but not with mut 1, a mutant of IER5 incapable of activating HSF1. In addition, we observed that IER5 was associated with the PP2A catalytic subunit PPP2CA and regulatory subunit B55 (Fig. 6I), confirming a previous study that comprehensively identified PP2A-interacting proteins27. Similar associations were observed between endogenous IER5 and HSF1 and between endogenous IER5 and PP2A (Fig. 6K). We could not detect an association of PPP2CA with HSF1 under the conditions employed (Fig. 6J). Since HSF1 is a substrate of PP2A, it is possible that PP2A dissociates from HSF1 when HSF1 is dephosphorylated and therefore this association may be difficult to detect. Furthermore, PP2A phosphatase activity was not affected by IER5 expression (Fig. 6L). Collectively, these results suggest that IER5 functions as a scaffold to bring HSF1 and PP2A in proximity to one another, thereby allowing PP2A to dephosphorylate and activate HSF1.
The IER5/HSF1 axis is required for cancer cell proliferation
We have shown that IER5 is required for the anchorage-independent growth of cancer cells (Fig. 2) and is an activator of HSF1. We speculate that IER5-mediated HSF1 activation is especially required for the protection of cells under stress, such as that which occurs when cells are placed in suspension cultures. We therefore next examined the potential involvement of HSF1 in anchorage-independent cell growth. As shown in Fig. 7A,B, knockdown of HSF1 suppressed cell growth similar to knockdown of IER5 in OE33 cells. Growth suppression was minor for adherent cells, but was significant for cells growing under conditions of stress caused by loss of anchorage. We confirmed that IER5 and HSF1 were indeed depleted under these experimental conditions (Fig. 7C,D). Knockdown of HSF1 or IER5 in HeLa cells caused similar suppression of cell proliferation (Fig. S4). Furthermore, we took a constitutively active form of HSF1 (caHSF1), which has the ability to induce HSF1 target genes independently of HSF1 activating signal31 and expressed this in OE33 cells. These cells no longer required IER5 for HSP family expression and HSP family proteins were constitutively expressed regardless of IER5 status (Fig. 7E). Subsequently, we treated caHSF1-expressing cells with siRNA targeting IER5 in addition to the parental OE33 cells. While the parental OE33 cells required IER5 for proliferation from day 5 (Fig. 7B), caHSF1 expression rescued the proliferation of cells in which IER5 is knocked down completely up to day 5 (Fig. 7F). Theses results show that HSF1 activation downstream of IER5 is important for proliferation of cancer cells.
IER5/HSF1/HSP family gene axis in cancer and in stressed cells.
(A–D) OE33 cells (2 × 103 cells) were plated in adherent (A) or suspension (B) 96 well culture plates and control, IER5-targeting or HSF1-targeting siRNAs were introduced. Cell growth assays were performed on the indicated days. Relative cell numbers were analyzed with CellTiter-Glo reagents from four wells and the mean cell numbers ±SD are shown. IER5 and HSF1 mRNA expression was analyzed by quantitative RT-PCR 48 hrs post-transfection (C,D). (**p < 0.01, #p < 0.0001). (E) OE33 cells were stably transfected with caHSF1 and the indicated siRNAs were introduced. Cell lysates were prepared 48 hrs post-transfection and expression of caHSF1-Flag, IER5, HSPA1A/1B were analyzed by Western blotting. (F) Cell growth assays were performed as in (B) using OE33 cells expressing caHSF1. (**p < 0.01). (G,H) Expression of IER5 (G) and HSPA6 (H) and prognosis in cancer patients. Disease-specific survival of patients with bladder cancer (Transitional cell carcinoma, dataset GSE13507) was analyzed using the PrognoScan database. (I–K) Expression of IER5 (I), HSPA6 (J) and HSPA1A (K) mRNA in cells treated with Adriamycin. Cell lines carrying wild-type p53 were treated with Adriamycin (1 μM) for 24 hrs (MRC5, MCF7, U2OS) or 19 hrs (A549). (*p < 0.05, **p < 0.01). (L) Expression of IER5, p53 and HSPA1A protein level in U2OS cells. Control, p53 and IER5 targeting siRNAs were introduced. Cells were treated with Adriamycin (1 μM) for 24 hrs and harvested 48 hrs post siRNA-transfection. (M) IER5 is transiently induced downstream of p53 and activates HSF1 in stressed cells, while IER5 is overexpressed and constitutively activates HSF1 in cancer cells.
IER5 is responsible for HSF1 activation in cancers and high expression of IER5 is associated with poor prognosis of cancer patients
Search of a publicly available cancer microarray database (PrognoScan;32) revealed that higher IER5 expression is associated with poorer prognosis in bladder, breast and brain cancer patients (Fig. 7G and S5). Interestingly, higher HSPA6 (the gene most strongly induced by IER5 expression, shown in Fig. 3A) expression is also related to poorer prognosis and we observed overall similar patterns of survival among bladder and brain cancer patients exhibiting high versus low IER5 and HSPA6 expression (Fig. 7H and S5). The correlation between IER5 and HSPA6 expression is also quite strong (Fig. 7G and S5, r = 0.47 and p < 0.0001 in bladder cancer samples, r = 0.63 and p < 0.0001 in breast cancer samples and r = 0.2896 and p < 0.005 in brain cancer samples). These results suggest that the IER5-HSF1-HSP family axis of regulation may be involved in cancer progression. The association of IER5 and HSPA6 expression with cancer progression may be related to the in vitro observation that IER5 and HSF1 are required for the proliferation of cancer cells under stressed conditions.
It has been reported that when HSF1 is activated in cancers, it specifically induces expression of cancer-associated HSF1 target genes in addition to the HSP family of genes8. We therefore analyzed the correlation between IER5 mRNA levels and the expression of cancer-associated HSF1 target genes in cancer patients. As shown in Fig. S5A, expression of IER5 and cancer-associated HSF1 target genes (RBM23 and EIF4A2) showed a significant association in bladder cancer (Fig. S5A, r = 0.64 and p < 0.0001 for RBM23 and r = 0.54 and p < 0.0001 for EIF4A2). In addition, an even higher association of IER5 and RBM23 and EIF4A2 were found in breast cancer patients (Fig. S5B, r = 0.75 and p < 0.0001 for RBM23 and r = 0.65 and p < 0.0001 for EIF4A2). These results collectively indicate that IER5 is responsible for HSF1 activation in cancer.
The HSP family of genes is induced by DNA damage in a p53- and IER5-dependent manner
As described earlier, IER5 gene is a target of p53 and its expression is induced under conditions of cellular stresses. Therefore, we examined if HSP family gene expression can be induced by DNA damage. As shown in Fig. 7I–K, Adriamycin treatment resulted in the concomitant upregulation of IER5, HSPA6 and HSPA1A mRNAs in several cell lines containing wild-type p53. In addition, HSPA1A protein upregulation by Adriamycin treatment was dependent on both p53 and IER5 (Fig. 7L). Thus, our data show that HSP family proteins are induced by p53 and IER5 upon DNA damage and may play a role in the recovery and protection of normal cells, as well as of cancer cells having wild-type p53.
Discussion
p53-dependent and -independent IER5 gene induction
We previously showed by microarray expression analysis that IER5 is a p53-inducible gene10 and we now have shown by ChIP-seq analysis that the IER5 is directly regulated by p53. We also have shown that the RE2 motif, a p53 binding site 46 kb downstream of the IER5 gene, has all the properties of an active enhancer and can function as an enhancer of the IER5 gene. Recently, Melo et al. performed a comprehensive genome-wide analysis of p53 binding sites and reported that the IER5 gene is regulated by p5333. They further reported that the IER5 promoter region interacts intrachromosomally with the same region that we identified as RE2. We have shown that there is a weak, but detectable IER5 promoter-RE2 interaction under non-stressed conditions. This interaction was weaker but still detectable in HCT116 p53−/− cells. We also have shown that IER5 is expressed in a p53-independent manner and that the region containing the RE2 motif functions as a super-enhancer in various cancer cell lines, regardless of p53 functional status. It will be interesting to analyze whether this IER5 promoter-RE2 interaction is required for p53-independent IER5 gene expression in such cell lines. We would also like to clarify which transcription factors are involved in the upregulation of IER5 mRNA in cancer.
A novel mechanism of HSF1 activation by IER5
The activation and extensive phosphorylation of HSF1 following heat stress has been well characterized6. Phosphorylation occurs on multiple residues, including Ser230, Ser320 and Ser326 and these enhance the transcriptional activity of HSF120,21,22. In this study, we showed that IER5 expression activates HSF1, but this activation involves HSF1 hypo-phosphorylation at multiple residues including Ser121, Ser307, Ser314, Thr323 and Thr367. In addition to the previously known HSF1 phosphorylation sites that are involved in HSF1 inactivation (Ser121 and Ser30723,24,25,26), Ser314, Thr323 and Thr367 are novel sites that were identified in this study. We observed that IER5 forms a ternary complex with HSF1 and PP2A to dephosphorylate and activate HSF1. PP2A is a ubiquitously expressed protein that dephosphorylates many different intracellular targets29,30. PP2A incorporates 15 unique B-regulatory subunits and it has been demonstrated that these B subunits control the substrate specificity and localization of the PP2A holoenzyme. Since it has been reported that IER5 associates with one of the B subunits of PP2A, PPP2R2B, we speculate that induction of IER5 may alter the substrate specificity of PP2A via interaction with one or more of its subunits and thereby promote the dephosphorylation and consequent activation of HSF127.
Very recently, Ishikawa et al. reported that there is a functional connection between HSF1 and IER5 under heat stress conditions34,35. They have identified IER5 as a target gene of HSF1 upon heat stress and reported that IER5 forms a complex with HSF1 and the PP2A regulatory subunit B5534,35. Their data is similar to our results shown in Fig. 5J,I and our data also confirms the importance of IER5 in the response to heat shock stress. In this manuscript, we have further shown that the IER5-mediated HSF1 activation can also occur in the absence of heat stress and is highly important for proliferation of cancer cells. Our results provide several novel insights into HSF1 function, showing IER5 facilitates the generation of a novel hypo-phosphorylated form of HSF1 that is active, yet is distinct from the hyper-phosphorylated form induced by heat shock and involves distinct sites of phosphorylation. While the mechanisms of HSF1 activation in response to heat stress have been well studied, HSF1 activation in response to other signals has not been well understood to date. Thus, our study provides a novel example of HSF1 activation by a signal other than heat stress.
HSF1 activation in cancer requires IER5
Recent findings have highlighted the potential importance of HSF1 activation in cancer. It has been suggested that growth-stimulating signals such as those transduced via the MAPK pathway can activate HSF136, although the precise mechanism by which this might occur is unknown. Because HSF1 activity is normally regulated by post-translational mechanisms, upregulation of HSF1 mRNA or protein expression alone would not be expected to lead to enhanced HSF1 activity or tumorigenicity. Indeed, it has been reported that overexpression of HSF1 does not transform MEFs9. Conversely however, MEFs lacking HSF1 are refractory to transformation, indicating that HSF1 is critical for tumorigenesis9. We have shown that IER5 expression is induced by growth stimuli, is overexpressed in various cancers, that the IER5 gene is associated with super-enhancers in cancer cells and that high IER5 expression is related to poorer prognosis of cancer patients. We also have shown that mRNA expression of IER5 and HSF1 target genes show high correlation in several cancers. In addition, in cancer cells that exhibit enhanced IER5 expression, suppression of IER5 by siRNA results in the downregulation of HSF1 activity and cell proliferation. These results collectively indicate that IER5 is responsible for HSF1 activation in cancers and supports cancer cell proliferation.
The p53-IER5-HSF1 axis in normal and cancer cells
In this report, we have mainly focused on the tumor-promoting functions of IER5. However, HSF1 and the HSP family of genes are well known to regulate cellular homeostasis and promote the survival of normal cells37. Therefore the function of IER5 downstream of p53 in normal cells is presumably to elicit a recovery and survival response under conditions of stress by activating HSF1 and upregulating the HSP family of genes downstream of HSF1 (Fig. 7M).
It is well known that approximately half of all human cancers carry a p53 mutation, illustrating the importance of p53 function in tumor suppression. However, it is of note that the remaining half retains wild-type p53. Indeed, we imagine that some cancer cells may positively select for the retention of wild-type p53. As we have shown, the IER5 gene is associated with super enhancers in cancers, indicating that it is a key gene involved in tumor pathogenesis. Therefore, some cancer cells may select for wild-type p53 because induction of IER5 (and downstream activation of HSF1) by p53 can protect the cells under conditions of stress resulting from oncogenic transformation, such as lack of anchorage or DNA damage. The role of the p53-IER5-HSF1 axis in the maintenance of cellular homeostasis in normal cells and its contribution to tumorigenesis in vivo, particularly, the function of IER5 in tumors containing wild-type p53, is an important future issue, which we would like to address using IER5-deficient mice.
Methods
Northern blotting analysis and microarray expression analysis
RNA was prepared using an RNeasy Mini kit (QIAGEN). Northern blotting was performed as described10. Methylene blue staining of 28S ribosomal RNA confirmed equal loading of RNA in each lane. Probes were prepared using a BcaBEST labeling kit (TaKaRa) and purified by serial purification using a Probe Quant G-50 MicroColumn (Amersham) and NICK Column (Amersham). The full open reading frames of genes obtained from EST clones, purchased from Open Biosystems, were used for probe preparation. The EST clones were IER5 (IMAGE ID 3457186), HSP70 (IMAGE ID 3345864), HSPA6 (IMAGE ID 5723718) and DNAJB1 (IMAGE ID 2821008). Microarray expression analysis was performed as described10.
Western blotting analysis
Cells were lysed in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 1% NP40, 250 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1 mM protease inhibitor (PMSF, aprotinin, leupeptin) and 1 mM DTT. Whole cell lysates were subjected to protein quantification and analyzed by Western blotting. Antibodies used in this study: anti-IER5 rabbit polyclonal antibody (HPA029894), anti-Flag M2 mouse monoclonal antibody (F1804), anti-β-actin mouse monoclonal antibody (A2228) from SIGMA, anti-p53 goat polyclonal antibody (sc-6243-G), anti-p21 rabbit polyclonal antibody (C-19), anti-HSF1 rabbit polyclonal antibody (sc-9144), anti-phospho-HSF1 (Ser230) rabbit polyclonal antibody (sc-30443-R), anti-PP2A-B55 (sc-18330) from Santa Cruz Biotechnology, anti-phospho-HSF1 (Ser320) rabbit monoclonal antibody (#2446-1) and anti-phospho-HSF1 (Ser326) rabbit monoclonal antibody (#2092-1) from Epitomics, anti-phospho HSF1 (Ser121) rabbit polyclonal antibody from Assay bio Tech, anti-phospho-HSF1 (Ser303/Ser307) rabbit monoclonal antibody (ab81281) from abcam, anti-HSPB1 mouse monoclonal antibody (ADI-SPA-800), anti-DNAJB1 rabbit polyclonal antibody (ADI-SPA-400), anti-HSPA6 mouse monoclonal antibody (ADI-SPA-754), anti-HSPA1A/1B mouse monoclonal antibody (ADI-SPA-810) from Enzo Life Sciences, anti-HA mouse monoclonal antibody (12CA5) from Roche and anti-PP2A C subunit (clone 1D6) from Merck Millipore.
Immunoprecipitation
Lysis buffer used for each experiment is shown below.
Figures 4D and 6I–L: 50 mM HEPES (pH 7.0), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% NP-40, 0.1 mM Na3VO4, 1 mM NaF, 0.1 mM DTT and a protease inhibitor cocktail.
Figures 5C–E and 6E: 50 mM Tris-HCl (pH 8.0), 1% NP40, 250 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1 mM protease inhibitor (PMSF, aprotinin, leupeptin) and 1 mM DTT.
For immunoprecipitation of Flag-tagged and HA-tagged proteins, cell lysates were immunoprecipitated with M2-agarose beads (SIGMA) or EZview Red Anti-HA Affinity Gel (SIGMA). For immunoprecipitation of IER5, anti-IER5 rabbit polyclonal antibody (HPA029894) and Protein A beads (GE Healthcare) were used.
Plasmids
IER5 and HSF1 expression constructs: the IER5 and HSF1 cDNAs were each cloned into the pcDNA3 vector as described10. Constitutively active HSF1 was constructed by deleting amino acids 186–202 in the second leucine zipper of HSF131.
Plasmids carrying one or two copies of wild-type or mutant p53RE (p53RE1, p53RE2, p53RE2 mut): Plasmids were obtained by cloning double-stranded oligonucleotides into the pGL3-promoter vector (Promega), which contains a minimal promoter. Oligonucleotide sequences: p53RE1 (5′-AAACAGGTTGAGACATGTCC-3′), p53RE2 (5′-AGGCATGCCCGGGCATGTCT-3′), p53RE2 mut (5′-AGGAATTCCCGGGAATTTCT-3′).
HSPA1A, HSPA1B, HSPA6 promoter-reporter constructs:
The 1kb region upstream of the transcription start site of each gene was amplified by PCR and cloned into the Xho I/Bgl II site of the pGL3-promoter vector (Promega).
Primers used to amplify promoter regions:
HSPA1A.
5′ primer : 5′-TTAGTCGAGAAAAAAAAAAATTAAAAATAAATAA-3′.
3′ primer : 5′-TTAAGATCTGGCCATCCGGTTCCCTGCTCTCTGT-3′.
HSPA1B.
5′ primer : 5′-TTAGTCGAGCTAAAAACGGTAACAGCCTAGGGGT -3′.
3′ primer : 5′-TTAAGATCTCATCCGGTGCCCTGCTGCTGTGGGC -3′.
HSPA6.
5′ primer : 5′-TTAGTCGAGAACCTTCAGAAGTCTCAGAGAAATG -3′.
3′ primer : 5′-TTAAGATCTCATGGCTGAAGCTTCTTGTCGGATG -3′.
HSPA1A 1–4 promoter-reporter constructs:
The corresponding regions were amplified by PCR from human genomic DNA and cloned into the Xho I/Bgl II site of the pGL3-promoter vector.
Primers used to amplify promoter regions:
Promoter 1.
5′ primer : 5′-TTAGTCGAGCCCCCGCCTCCCCCATTGTGGCTGC -3′.
Promoter 2.
5′ primer : 5′-TTAGTCGAGCCCTGCAAATTTGAGACGGCTCCAA -3′.
Promoter 3.
5′ primer : 5′-TTAGTCGAGACAATTAAAAGCCCAGCGCCGACCC -3′.
Promoter 4.
5′ primer : 5′-TTAGTCGAGCAGGACGGGAGGCGAAAACCCTGGA -3′.
3′ primer (commonly used for all constructs);
5′-TTAAGATCTGGCCATCCGGTTCCCTGCTCTCTGT-3′
ChIP sequence
ChIP assays were performed as described38. For p53 induction, cells were treated with 5-FU (0.375 mM for 9 hrs). Antibodies against p53 (FL393, Santa Cruz), H3K27ac, H3K4me1, H3K4me3 (07–473, Millipore) or phospho-RNAP II were used to precipitate immune complexes. Antibodies against H3K27ac, H3K4me1 and phospho-RNAP II were kindly provided by Dr. Hiroshi Kimura, Tokyo Institute of Technology. DNA co-precipitating with p53, H3K27ac, H3K4me1, H3K4me3 or phospho-RNAP II (5–10 ng) was purified by SDS–PAGE to obtain 100–300 bp fragments and sequenced on an Illumina 1G sequencer. Size-fractionated DNA was extracted and a single adenosine was added using Klenow exo– (3′ and 5′ exo minus; Illumina). Illumina adaptors were then added and DNA was subjected to 20 cycles of PCR according to manufacturer’s instructions. We then purified the DNA and performed cluster generation and 36 cycles of sequencing on the Illumina cluster station and 1G analyzer following the manufacturer’s instructions. The resulting sequences were mapped to the build #36 reference human genome. ChIP signal values for p53, k4me3 (5-FU 0, 9h), K4me1 (5-FU 0, 9h), K27ac (5-FU 0, 9h) and Pol2 (5-FU 0, 9h) were 26730379, 17020836, 22559531, 8296440, 9319944, 29174994, 27258981, 15964489 and 15142757, respectively. ChIP signal values (ChIP counts/estimated or input counts) were generated as previously reported39. Probability values were generated as all fragments were randomly mapped to the non-repetitive sequences of the Human Genome.
Circularized Chromosome Conformation Capture (4C)
4C-seq assays were performed as previously reported40,41. Chromatin from 10-million cells was cross-linked with 1% formaldehyde for 10 min at room temperature, then cells were treated with lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA, 0.5% IGEPAL CA-630 (Sigma-Aldrich, I8896), 1% Triton X-100). Nuclei were digested with NlaIII (NEB, R0125L) and ligated with T4 DNA ligase (Roche, 799009). A secondary restriction digestion was performed with DpnII (NEB, R0543M), followed again by ligation. Specific 4C primers were designed near a p53 binding site. Illumina adaptor sequence and index sequence were included in the primer sequence. Sixteen-PCR reactions were performed with Expand Long Template PCR System (Roche, 759 060 001), pooled together and purified using Agencourt AMPure XP beads (Beckman, A63882).
PCR primers for 4C-seq
Following primers were used for 4C-seq library preparation.
p53 RE2_Forward;
5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTTTAAATGCCAGTGTCCATG-3′
p53 RE2_Reverse1;
5′-CAAGCAGAAGACGGCATACGAGATATCACGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTCTACAGAGAGCCAGCTT-3′
p53 RE2_Reverse2;
5′-CAAGCAGAAGACGGCATACGAGATTTAGGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTCTACAGAGAGCCAGCTT-3′
p53 RE2_Reverse3;
5′-CAAGCAGAAGACGGCATACGAGATGATCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTCTACAGAGAGCCAGCTT-3′
Reverse transcription and real-time PCR
Reverse transcription was carried out using the SuperScript First-Strand Synthesis System for RT-PCR (Life Technologies) or ReverTra Ace (TOYOBO) following the manufacturer’s instructions. Total RNA (0.2–5 μg) was used for reverse transcription. Reverse-transcribed cDNAs were subjected to real-time PCR, which was performed with a LightCycler 480 Instrument (Roche Diagnostics) or CFX96 Touch Real-Time PCR System (Bio-Rad). For the detection of IER5, HSF1, HSPA1A and GAPDH, a TaqMan probe (IER5; Hs00275419, HSF1; Hs00232134_m1, HSPA1A; Hs00359163_s1, GAPDH; Hs02758991_g1)from Applied Biosystems were used. For the detection of HSPA6 and HSPA1B, a TaqMan probe from Integrated DNA Technologies was used. In some cases, custom-designed TaqMan Dual-Labeled Probes from Sigma-Aldrich were also used for the detection of HSPA1A and GAPDH.
Transfection and luciferase reporter assay
Transfection and luciferase reporter assays were performed as described (10). Transient transfection assays were performed using Lipofectamine Plus reagent (Life Technologies). For the luciferase reporter assay shown in Fig. 1H, Saos2 cells were seeded in 96-well dishes and co-transfected with 60 ng of a firefly luciferase reporter gene and 3 ng of each p53 gene cloned in the pcDNA3 vector together with 15 ng of a Renilla luciferase expression vector (pGL4 [TK-Rluc] vector, Promega) as an internal control for transfection efficiency. For the luciferase reporter assay shown in Fig. 3, H1299 cells were co-transfected with 30 ng of a firefly luciferase reporter gene and 5 ng of each IER5 gene cloned in pcDNA3 vector together with 3 ng of a Renilla luciferase expression vector pGL4 [TK-Rluc] vector. Cells were harvested 48 hrs (Fig. 1H) or 24 hrs (Fig. 3) post-transfection and analyzed using the Dual-Luciferase Reporter Assay System (Promega). All of the luciferase reporter assay data are the mean-fold activation +/− SD of three independent experiments. The siRNAs were introduced using RNAiMAX (Invitrogen, Carlsbad, CA). Control, ON-target plus IER5-targeting, HSF1-targeting, p53-targeting and PPP2CA-targeting siRNAs were purchased from Dharmacon Research (Lafayette, Colorado).
Analysis of HSF1 modification by liquid chromatography/mass spectrometry/mass spectrometry (LC-MS/MS)
Identification of HSF1 modification was performed using LC-MS/MS. 293T cells were transfected with Flag-tagged HSF1 together with IER5 or control empty vector and harvested 27 hrs post-transfection. Cells were lysed with lysis buffer containing 50 mM Tris-HCl (pH 8.0), 1% NP40, 250 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1 mM protease inhibitor (PMSF, aprotinin, leupeptin) and 1 mM DTT. For immunoprecipitation of Flag-tagged HSF1 protein, the lysate was immunoprecipitated with M2-agarose beads (SIGMA). The immunoprecipitated samples were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were stained with SYPRO Ruby, excised and subjected to in-gel tryptic digestion with modified trypsin (Promega, Madison, WI) as described previously42. The tryptic digests were subjected to liquid chromatography (Paradigm MS4 dual solvent delivery system; Michrom BioResources, Auburn, CA) coupled with tandem mass spectrometry (LTQ Orbitrap XL; Thermo Fisher Scientific, San Jose, CA) equipped with a nanoelectrospray ion source (AMR, Tokyo, Japan). Mascot software (Matrix science, London, UK) was used to search for the mass of the peptide ion peaks against the SWISS-PROT database. The following search parameters were used: tolerance of 2 missed trypsin cleavages, variable modification on the methionine residue (oxidation, +16 Da), maximum precursor ion mass tolerance of ±10 ppm and fragment ion mass tolerance of ±0.8 Da. Proteins with a Mascot score of 34 or more were considered as positively identified.
Immunofluorescence
H1299 cells (6 × 104 cells) were transfected with 320 ng of each expression vector plasmid and harvested 24 hrs post-transfection. Cells were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 in PBST for 2 min. Cells were blocked with BSA (5 mg/ml) and 50 mM Glycine in PBST for 1 h. The cells were sequentially incubated with anti-Flag and anti-HSF1 (H-311) antibody for 1 h at room temperature and AlexaFluor 594 and 488-labeled secondary antibody (Molecular Probes, Eugene, OR, USA) and mounted with DAPI (2.5 μg/ml) in DABCO glycerol. Images were obtained by using a Keyence BZ-9000 fluorescence microscope.
HSF1 crosslink
H1299 cells were transfected with an IER5 expression vector, harvested 25 h post-transfection and lysed in lysis buffer A containing 25 mM HEPES (pH 7.4), 0.5% Triton-X, 100 mM NaCl, 5 mM EDTA, 10 mM NaF, 1 mM Na3VO4, 1 mM protease inhibitor (PMSF, aprotinin, leupeptin) and 1 mM DTT. Whole cell lysates (2 × 106 cells/ml) were crosslinked with 0.1, 0.3 mM EGS [ethylene glycol bis] (Thermo SCIENTIFIC) at room temperature for 30 min. After quenching of crosslinking by addition of 50 mM Tris-HCl (pH 7.2), Western blotting was performed.
HSF1 pull-down assay using biotinylated DNA probe
To analyze HSF1 binding to Heat Shock Elements (HSEs), we performed a streptavidin pull down assay. The HSE sequence from the HSPA1A gene was used (nt −132 to −109 of the HSPA1A gene; 5′-AAACCCCTGGAATATTCCCGACC-3′). A 2x repeat of the HSE element was cloned into the Pikka Gene basic vector (Toyo Ink Co.). Preparation of biotinylated DNA probes: HSE and empty vector were PCR amplified using biotinylated primers. The biotinylated probes were bound to streptavidin-coated beads (Magnesphere Paramagnetic Particles, Promega). Preparation of cell extracts: H1299 cells were transfected with an IER5-Flag expression vector, harvested 24 hrs post-transfection and lysed in lysis buffer A. To avoid non-specific binding, poly dI:dC (7 μg/ml) was added. Biotinylated DNA (1.5 μg) was bound to the magnetic beads and mixed with 1.9 mg of cell lysates. Incubation was performed for 7 hrs and beads were washed 5 times with wash buffer containing 20 mM Tris-HCl (pH 7.4), 0.1% Triton-X, 10% Glycerol, 1 mM EDTA, BSA (0.1 mg/ml). Bead-bound HSF1 was denatured in Laemmli buffer and analyzed by Western blotting.
Cell growth assay
Cells were plated in 96 well adherent culture plates (Corning, 3610) or suspension culture plates (SUMILON, MS-8096R) and the indicated siRNAs were introduced into wells. Cell numbers were analyzed using CellTiter-Glo (Promega) in four or six wells and the mean cell numbers ± SD are shown.
Colony formation assay in soft agar
Control or IER5-targeting siRNAs were introduced into cells. After 24 hrs, the cells (1 × 104 cells) were plated in a layer of 0.33% agarose/RPMI over a layer of 0.5% agarose/RPMI in a 3 cm dish and cultured for 9 days. Colonies with an image size larger than 600 square pixels were analyzed by ImageJ software and counted from four dishes and the mean numbers of colonies ±SD are shown.
Measurement of PP2A phosphatase activity
PP2A phosphatase activities were analyzed in 293T cell lysates using a PP2A immunoprecipitation phosphatase assay kit (Millipore). Cell lysates were prepared by suspending the cells in lysis buffer (20 mM imidazole, 50 mM HEPES (pH 7.0), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA and a protease inhibitor cocktail), followed by sonication (3 times, each 30 sec).
Analysis of IER5 and HSPA6 expression using the Gene Logic SCIANTIS database and the PrognoScan database
IER5 expression was analyzed using the Gene Logic SCIANTIS database (http://www.genelogic.com), a comprehensive genome-wide gene expression database annotated with detailed clinical and biological information. IER5 mRNA expression levels were analyzed in kidney (renal cell carcinoma, clear cell type, primary), pancreas (adenocarcinoma, primary), ovary (adenocarcinoma, papillary serous type, primary), stomach (adenocarcinoma, primary), colon (adenoma), endometrium (adenocarcinoma, endometrioid type, primary), urinary bladder (transitional cell carcinoma, primary) cancer and their corresponding normal tissues.
Correlations between the expression of IER5 and HSPA6 and prognosis in cancer patients were analyzed using the PrognoScan database (http://www.abren.net/PrognoScan/), a large collection of publicly available cancer microarray data sets with clinical annotations and a tool for assessing the biological relationships between gene expression and prognosis.
Statistical analysis
Data were calculated and shown as mean ± SD. Significance of differences was determined by Student t test (Figs 2A,F,G,I–L, 3D,E and 7F,I–K), one-way ANOVA (Figs 2D and 7A–D, S4A and S4B) or two-way ANOVA (Figs 4A,B, 5G,I and 6A,B,D,G,H). Correlation data were analyzed using Prism 6 software. Statistical significance was defined as p < 0.05. (*p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.0001).
Additional Information
How to cite this article: Asano, Y. et al. IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumorigenesis. Sci. Rep. 6, 19174; doi: 10.1038/srep19174 (2016).
References
Levine, A. J. p53, the cellular gatekeeper for growth and division. Cell 88, 323–331 (1997).
Aylon, Y. & Oren, M. Living with p53, dying of p53. Cell 130, 597–600 (2007).
Harris, S. L. & Levine, A. J. The p53 pathway: positive and negative feedback loops. Oncogene 24, 2899–2908 (2005).
Vousden, K. H. & Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 137, 413–431 (2009).
Vousden, K. H. & Ryan, K. M. p53 and metabolism. Nat Rev Cancer 9, 691–700 (2009).
Anckar, J. & Sistonen, L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80, 1089–1115 (2011).
Santagata, S. et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA 108, 18378–18383 (2011).
Mendillo, M. L. et al. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150, 549–562 (2012).
Dai, C., Whitesell, L., Rogers, A. B. & Lindquist, S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130, 1005–1018 (2007).
Ohki, R., Kawase, T., Ohta, T., Ichikawa, H. & Taya, Y. Dissecting functional roles of p53 N-terminal transactivation domains by microarray expression analysis. Cancer Sci 98, 189–200 (2007).
Kawase, T. et al. p53 target gene AEN is a nuclear exonuclease required for p53-dependent apoptosis. Oncogene 27, 3797–3810 (2008).
Kawase, T. et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell 136, 535–550 (2009).
Williams, M. et al. Ier5, a novel member of the slow-kinetics immediate-early genes. Genomics 55, 327–334 (1999).
Yoon, J. H. et al. cDNA Microarray Analysis of Gene Expression Profiles Associated with Cervical Cancer. Cancer Research and Treatment 35, 451–459 (2003).
Oda, E. et al. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058 (2000).
Ohki, R. et al. Reprimo, a new candidate mediator of the p53-mediated cell cycle arrest at the G2 phase. J Biol Chem 275, 22627–22630 (2000).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Vihervaara, A. & Sistonen, L. HSF1 at a glance. J Cell Sci 127, 261–266 (2014).
Holmberg, C. I. et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. Embo Journal 20, 3800–3810 (2001).
Chou, S. D., Prince, T., Gong, J. L. & Calderwood, S. K. mTOR Is Essential for the Proteotoxic Stress Response, HSF1 Activation and Heat Shock Protein Synthesis. PLoS ONE 7(6), e39679. 10.1371/journal.pone.0039679 (2012).
Zhang, Y., Murshid, A., Prince, T. & Calderwood, S. K. Protein Kinase A Regulates Molecular Chaperone Transcription and Protein Aggregation. PLoS One 6(12), e28950. 10.1371/journal.pone.0028950 (2011).
Chu, B. Y., Soncin, F., Price, B. D., Stevenson, M. A. & Calderwood, S. K. Sequential phosphorylation by mitogen-activated protein kinase and glycogen synthase kinase 3 represses transcriptional activation by heat shock factor-1. Journal of Biological Chemistry 271, 30847–30857 (1996).
Kline, M. P. & Morimoto, R. I. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Molecular and Cellular Biology 17, 2107–2115 (1997).
Knauf, U., Newton, E. M., Kyriakis, J. & Kingston, R. E. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Gene Dev 10, 2782–2793 (1996).
Wang, X. Z. et al. Phosphorylation of HSF1 by MAPK-activated protein kinase 2 on serine 121, inhibits transcriptional activity and promotes HSP90 binding. Journal of Biological Chemistry 281, 782–791 (2006).
Glatter, T., Wepf, A., Aebersold, R. & Gstaiger, M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol 5, 237 10.1038/msb.2008.75 (2009).
Xing, H., Hong, Y. & Sarge, K. D. Identification of the PP2A-interacting region of heat shock transcription factor 2. Cell Stress Chaperones 12, 192–197 (2007).
Janssens, V. & Goris, J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353, 417–439 (2001).
Eichhorn, P. J., Creyghton, M. P. & Bernards, R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta 1795, 1–15 (2009).
Uchiyama, T. et al. HSF1 and constitutively active HSF1 improve vascular endothelial function (heat shock proteins improve vascular endothelial function). Atherosclerosis 190, 321–329 (2007).
Mizuno, H., Kitada, K., Nakai, K. & Sarai, A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med Genomics 2, 18 10.1186/1755-8794-2-18 (2009).
Melo, C. A. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49, 524–535 (2013).
Ishikawa, Y., Kawabata, S. & Sakurai, H. HSF1 transcriptional activity is modulated by IER5 and PP2A/B55. FEBS Lett 589, 1150–1155 (2015).
Ishikawa, Y. & Sakurai, H. Heat-induced expression of the immediate-early gene IER5 and its involvement in the proliferation of heat-shocked cells. FEBS J 282, 332–340 (2015).
Dai, C. et al. Loss of tumor suppressor NF1 activates HSF1 to promote carcinogenesis. J Clin Invest 122, 3742–3754 (2012).
Pirkkala, L., Nykanen, P. & Sistonen, L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15, 1118–1131 (2001).
Kaneshiro, K., Tsutsumi, S., Tsuji, S., Shirahige, K. & Aburatani, H. An integrated map of p53-binding sites and histone modification in the human ENCODE regions. Genomics 89, 178–188 (2007).
Wakabayashi, K. et al. The peroxisome proliferator-activated receptor gamma/retinoid X receptor alpha heterodimer targets the histone modification enzyme PR-Set7/Setd8 gene and regulates adipogenesis through a positive feedback loop. Mol Cell Biol 29, 3544–3555 (2009).
Splinter, E., de Wit, E., van de Werken, H. J., Klous, P. & de Laat, W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods 58, 221–230 (2012).
van de Werken, H. J. et al. Robust 4C-seq data analysis to screen for regulatory DNA interactions. Nat Methods 9, 969–972 (2012).
Kondo, T. & Hirohashi, S. Application of highly sensitive fluorescent dyes (CyDye DIGE Fluor saturation dyes) to laser microdissection and two-dimensional difference gel electrophoresis (2D-DIGE) for cancer proteomics. Nat Protoc 1, 2940–2956 (2006).
Acknowledgements
We thank Marc Lamphier for critical reading of the manuscript. This study was partly supported by Grant-in-Aid for Scientific Research (C) (#26430133) (R.O.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Applied Research for Innovative Treatment of Cancer from the Ministry of Health, Labour and Welfare (R.O.), Development of Innovative Research on Cancer Therapeutics (P-DIRECT)/Ministry of Education, Culture, Sports, Science and Technology of Japan (R.O.), Takeda Science Foundation (R.O.), Astellas Foundation for Research on Metabolic Disorders (R.O.), Daiichi-Sankyo Foundation of Life Science (to R.O.), Extramural Collaborative Research Grant of Cancer Research Institute, Kanazawa University, Japan (to R.O.), Cooperative Research Program of Institute for Frontier Medical Sciences, Kyoto University, Japan (to R.O.).
Author information
Authors and Affiliations
Contributions
Y.A., T.K. and R.O. designed the experiments. Y.A., T.K., A.O., S.T., H.I., S.T., I.K., T.K. and R.O. performed the experiments. F.T., H.N., K.S., H.A., Y.T, H.N. and R.O. supervised the study. R.O. directed the study. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Asano, Y., Kawase, T., Okabe, A. et al. IER5 generates a novel hypo-phosphorylated active form of HSF1 and contributes to tumorigenesis. Sci Rep 6, 19174 (2016). https://doi.org/10.1038/srep19174
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19174
This article is cited by
-
Novel role of PAF1 in attenuating radiosensitivity in cervical cancer by inhibiting IER5 transcription
Radiation Oncology (2020)
-
Interplay between HSF1 and p53 signaling pathways in cancer initiation and progression: non-oncogene and oncogene addiction
Cellular Oncology (2019)
-
Functional loss of p53 cooperates with the in vivo microenvironment to promote malignant progression of gastric cancers
Scientific Reports (2018)
-
Regulation of heat shock transcription factors and their roles in physiology and disease
Nature Reviews Molecular Cell Biology (2018)
-
Census and evaluation of p53 target genes
Oncogene (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.