Abstract
The identification of microRNA precursors (pre-miRNAs) helps in understanding regulator in biological processes. The performance of computational predictors depends on their training sets, in which the negative sets play an important role. In this regard, we investigated the influence of benchmark datasets on the predictive performance of computational predictors in the field of miRNA identification and found that the negative samples have significant impact on the predictive results of various methods. We constructed a new benchmark set with different data distributions of negative samples. Trained with this high quality benchmark dataset, a new computational predictor called iMiRNA-SSF was proposed, which employed various features extracted from RNA sequences. Experimental results showed that iMiRNA-SSF outperforms three state-of-the-art computational methods. For practical applications, a web-server of iMiRNA-SSF was established at the website http://bioinformatics.hitsz.edu.cn/iMiRNA-SSF/.
Similar content being viewed by others
Introduction
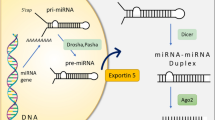
MicroRNAs (miRNAs) are a class of evolutionally conserved, single-stranded, small (approximately 19–23 nucleotides), endogenously expressed and non-protein-coding RNAs that act as post-transcriptional regulators of gene expression in a broad range of animals, plants and viruses1,2,3,4. MiRNAs play an important role as a regulator in biological process5. The aberrant expressions have been observed in many cancers6,7,8,9 and several miRNAs have been convincingly proved to play important roles in carcinogenesis10. The protein architecture in different programmed cell death (PCD) subroutines has been explored, but the global network organization of the noncoding RNA (ncRNA)-mediated cell death system is limited and ambiguous11,12. Thus, the discovery of human miRNAs regulation is an important task.
As traditional experimental methods for miRNA identification are time and money consuming, recently more attention has been paid to the development of computational approaches. Because miRNAs are short, the traditional feature engineering approaches13,14,15 are usually failed to extract features based on their sequences and structures and therefore, computational approaches usually identify the precursors of miRNAs (pre-miRNAs) instead of miRNA. A variety of software tools for this purpose have been proposed. As shown in previous studies, extracting useful features are important for constructing a computational predictor16. Various features and machine learning techniques have been proposed to predict miRNAs. Triplet-SVM17 incorporated a local contiguous sequence-structure composition feature and utilized SVM to construct the predictor. MiPred18 identified the human pre-miRNAs by using an RF classifier with a combined feature set, including local contiguous sequence-structure (Triplet-SS), minimum of free energy feature (MFE) and P-value of randomization test feature (P-value). Compared with Triplet-SVM, MiPred improved the performance by nearly 10% in terms of accuracy. MiRanalyzer19 employed an RF classifier trained with a variety of features associated with nucleotide sequence, structure and energy. Wei, L. et al.20 proposed a SVM-based method called miRNApre using the local contiguous structure-sequence composition feature, primary sequence composition feature and MFE. Recently some predictors have been proposed based on the predicted secondary structure of RNA sequences, such as iMiRNA-PseDPC21, iMcRNA-PseSSC22, miRNA-dis23, deKmer24, etc. These methods using different features and classifiers treat the pre-miRNA identification problem as a binary classification problem. Currently, the widely used classification algorithms include Support Vector Machine (SVM)17,25, Hidden Markov Model (HMM)26, Random Forest (RF)18 and Naive Bayes (NB)27. The widely used features of characterizing pre-miRNAs include stem-loop hairpin structures28,29, MFE of the pre-miRNAs and P-value of randomization test18,19,20,30. Because the importance of the features for constructing a predictor, recently, some web-servers or stand-alone tools were proposed to extract the features from RNA sequences, such as Pse-in-One31 and repRNA32. MiPred18 identified the human pre-miRNAs by combining Triplet-SS, MFE and P-value, in which MFE and P-value were the top 2 most important features. MiRanalyzer19 was trained with a variety of features, in which MFE was the secondary most important feature. miRNApre was built based on Triplet-SS, primary sequence composition feature and MFE. However, based on the feature analysis of miRNApre, MFE feature cannot improve the performance. Therefore, it is interesting to explore the reasons for the different discriminative power of the same feature in different predictors. Furthermore, there are several other challenging problems should be solved in this filed:
-
1
Many features have been proposed to characterize the pre-miRNAs, but their discriminative power is not investigated. Some features showed strong discriminative power in some predictors, while in other predictors, they only showed limited discriminative power, for example MFE played an important role in Triplet-SVM, but it almost had no contribution to the discriminative power of miRNApre. Therefore, the most discriminative features and their combinations for miRNA identification should be investigated.
-
2
The existing benchmark datasets are too small to reflect the statistical profile. Most of these datasets only contain several hundreds of real pre-miRNA samples and pseudo pre-miRNA samples. It is necessary to construct an updated benchmark dataset to fairly evaluate the performance of different methods.
-
3
Most of these methods performed well in cross validation test, but they showed much lower performance on independent testing sets. This is because the samples in the training set are not representative enough, especially for the pseudo pre-miRNA samples (negative samples). There is no golden standard to select or construct the negative samples33,34.
To solve these problems, we investigated the distributions of various benchmark datasets and found that they had large variance, especially for the distributions of negative samples. A series of controlled experiments were conducted to find out how the performance were impacted on different distributions of negative samples. The results showed the negative samples were not representative enough. Therefore, the key to improve predictive performance was to construct an high quality benchmark dataset for miRNA identification. In this regard, a new benchmark dataset was constructed, in which the positive samples were extracted from the miRBase35,36,37 and the negative samples were selected from existing datasets with different data distributions. Finally, we proposed a new computational method for pre-miRNA identification, called iMiRNA-SSF, which employed the sequence and structure features trained with the updated benchmark dataset. The web-server of iMiRNA-SSF can be accessed at http://bioinformatics.hitsz.edu.cn/iMiRNA-SSF/.
Results
Negative samples have significant impact on the discriminative power of features
As reported in literatures, MFE and P-value were the top 2 most important features in MiPred18, but they were not so important in miRNApre20 (out of top 10 features). The main difference between these two methods was their negative samples in benchmark datasets. The negative samples of MiPred were collected from the protein coding regions with parameter filtering method, while the negative samples of miRNApre were collected by multi-level process. For more details, please refer to17,20.
Our hypothesis was that the different discriminative power of the same method was caused by the negative samples. In order to validate this hypothesis, two datasets  and
and  were constructed with the same positive set and the different negative sets:
were constructed with the same positive set and the different negative sets:


where the  ,
,  and
and  are the same as the subsets in Equation (4). The dataset
are the same as the subsets in Equation (4). The dataset  is union of
is union of  and
and  ;
;  is union of
is union of  and
and  .
.
We investigated the discriminative power of all features mentioned in Method section on datasets  and
and  by assessing their information gain related to the classes. The higher information gain value38 means the related feature is more powerful. The top 20 most important features on the two datasets were shown in Table 1(A,B), respectively.
by assessing their information gain related to the classes. The higher information gain value38 means the related feature is more powerful. The top 20 most important features on the two datasets were shown in Table 1(A,B), respectively.
MFE and P-value were the top 2 most important features on  . However, P-value was only ranked at 20th on
. However, P-value was only ranked at 20th on  and MFE was ranked out of top 20. We also found that 14 of the top 20 most important features on
and MFE was ranked out of top 20. We also found that 14 of the top 20 most important features on  belonged to local triplet sequence-structure features (Triplet-SS) category, but only 4 features belonged to primary sequence features (3-gram) category. In contrast, for
belonged to local triplet sequence-structure features (Triplet-SS) category, but only 4 features belonged to primary sequence features (3-gram) category. In contrast, for  , only 5 of the top 20 most important features belonged to Triplet-SS category and 14 features belonged to 3-gram category. The structure features are more powerful than sequence features on
, only 5 of the top 20 most important features belonged to Triplet-SS category and 14 features belonged to 3-gram category. The structure features are more powerful than sequence features on  database, but it is not the case on
database, but it is not the case on  database. The results showed that the negative samples have significant impact on the discriminative power of features.
database. The results showed that the negative samples have significant impact on the discriminative power of features.
We took MFE and P-value as examples to analyse the reasons. Their distributions on positive and negative samples of  and
and  were calculated and the results were shown in Fig. 1. The distributions of MFE and P-value are very similar between
were calculated and the results were shown in Fig. 1. The distributions of MFE and P-value are very similar between  and
and  , but they are different between
, but they are different between  and
and  . A feature has more discriminative power if its distribution has variance on positive and negative sets. This is why MFE and P-value show powerful discriminability on
. A feature has more discriminative power if its distribution has variance on positive and negative sets. This is why MFE and P-value show powerful discriminability on  database, but it is not the case on
database, but it is not the case on  database.
database.
Importance of negative samples for training a classifier
The different distributions of negative samples have significant impact on the performance of a trained classifier. However, how does it come into being and how to avoid this problem?
We conducted the controlled experiments, employing all features (Triplet-SS, MFE, P-value and N-gram). The training sets and testing sets were constructed:

where  and
and  are disjoint subsets of
are disjoint subsets of  , in which respectively contain 1312 and 300 human pre-miRNAs;
, in which respectively contain 1312 and 300 human pre-miRNAs;  and
and  are disjoint subsets of
are disjoint subsets of  , in which respectively contain 1312 and 300 Xue pseudo pre-miRNAs;
, in which respectively contain 1312 and 300 Xue pseudo pre-miRNAs;  and
and  are disjoint subsets of
are disjoint subsets of  , in which respectively contain 1142 and 300 Zou pseudo pre-miRNAs. The numbers of samples in each dataset were carefully chosen to avoid bias.
, in which respectively contain 1142 and 300 Zou pseudo pre-miRNAs. The numbers of samples in each dataset were carefully chosen to avoid bias.
The prediction results were listed in Table 2. The cross validation results were achieved by leave-one-out strategy on  and
and  , whereas the independent testing results were achieved by testing on
, whereas the independent testing results were achieved by testing on  and
and  . In term of Table 2, both two predictors performed well in cross validation test, achieved 87.69% and 98.57% accuracies, respectively. But they showed much lower performance on the independent testing dataset, especially the performance of the classifier trained on
. In term of Table 2, both two predictors performed well in cross validation test, achieved 87.69% and 98.57% accuracies, respectively. But they showed much lower performance on the independent testing dataset, especially the performance of the classifier trained on  and tested on
and tested on  dropped to 51.17% from 98.57% in term of accuracy.
dropped to 51.17% from 98.57% in term of accuracy.
For a SVM-based method, it generates a decision boundary that separates the positive samples from the negative ones. The generated decision boundaries based on different datasets are significant difference. As shown in Fig. 2(A,B), the two generated decision boundaries built on two datasets with different distributions are different. When using a decision boundary to classify samples in another dataset, the majority of samples can’t fall on their own categories. As shown in Fig. 2(C), if samples in  as test samples, the decision boundary Bxue performs badly to classify them into two classes. If samples in
as test samples, the decision boundary Bxue performs badly to classify them into two classes. If samples in  as test sample, the same is to Bzou. But if we merge
as test sample, the same is to Bzou. But if we merge  and
and  into one dataset, the generated new decision boundary BNew based on the new dataset can improve the predictive performance significantly. As shown in Fig. 2(D), the new decision boundary BNew can separate all samples correctly. It indicates that new decision boundary BNew is more general and outperforms Bxue and Bzou.
into one dataset, the generated new decision boundary BNew based on the new dataset can improve the predictive performance significantly. As shown in Fig. 2(D), the new decision boundary BNew can separate all samples correctly. It indicates that new decision boundary BNew is more general and outperforms Bxue and Bzou.
A new predictor built on updated benchmark dataset
We constructed a new benchmark set with different data distributions of negative samples, including real human pre-miRNAs as positive set, Xue pseudo pre-miRNAs  and Zou pseudo pre-miRNAs
and Zou pseudo pre-miRNAs  as negative sets. Trained with this high quality benchmark dataset, a new computational predictor called iMiRNA-SSF was proposed. Four kinds of features were employed to investigate that if they could be combined to improve performance of iMiRNA-SSF, including Triplet-SS, MFE, P-value and N-gram. The performance was obtained by using LibSVM algorithm with leave-one-out crossing validation on updated benchmark dataset. As shown in Table 3, the best performance (ACC = 90.42%, MCC = 0.79) was achieved with the combination of the four kinds of features. Triplet-SS is a local triplet sequence-structure-based feature; MFE and P-value are features based on the on minimum of free energy of the secondary structure; N-gram is a sequence-based feature considering the local sequence composition information. These features describe the characteristics of pre-miRNA from different aspects. Therefore the predictive performance of iMiRNA-SS can be further enhanced by combining all of features.
as negative sets. Trained with this high quality benchmark dataset, a new computational predictor called iMiRNA-SSF was proposed. Four kinds of features were employed to investigate that if they could be combined to improve performance of iMiRNA-SSF, including Triplet-SS, MFE, P-value and N-gram. The performance was obtained by using LibSVM algorithm with leave-one-out crossing validation on updated benchmark dataset. As shown in Table 3, the best performance (ACC = 90.42%, MCC = 0.79) was achieved with the combination of the four kinds of features. Triplet-SS is a local triplet sequence-structure-based feature; MFE and P-value are features based on the on minimum of free energy of the secondary structure; N-gram is a sequence-based feature considering the local sequence composition information. These features describe the characteristics of pre-miRNA from different aspects. Therefore the predictive performance of iMiRNA-SS can be further enhanced by combining all of features.
 with different features combination.
with different features combination.Furthermore, the importance of all features was also investigated. P-value and MFE features are the most discriminative, followed by the local triplet sequence-structure features and the primary sequence based features. The results were shown in Table 4.
Comparison with other methods
Three state-of-the-art methods Triplet-SVM17, MiPred18 and miRNApre20 were selected to compare with the proposed iMiRNA-SSF. MiPred is a classifier using Random Forest algorithm combined with Triplet-SS, MFE and P-value features. miRNApre employed the SVM algorithm with Triplet-SS, N-gram, MFE features. As mentioned in the introduction section, the reported accuracy of these methods were based on small datasets containing only several hundreds of samples without removing redundant sequences, thus, their performance might be overestimated. In order to make a fair comparison among these methods, all these methods were evaluated on the same updated benchmark dataset via leave-one-out crossing validation. Their predictive results were shown in Table 5.
To further illustrate the comparison, receiver operating characteristic (ROC) scores of different methods were provided in Fig. 3. The ROC scores of Triplet-SVM, MiPred, miRNApre and iMiRNA-SSF are 0.90, 0.92, 0.94 and 0.96, respectively. iMiRNA-SSF outperforms the other three state-of-the-art methods.
Web-server description
For the convenience of the vast majority of experimental scientists, we provided a simple guide on how to use the iMiRNA-SSF web-server. It is available at http://bioinformatics.hitsz.edu.cn/iMiRNA-SSF/.
Step 1: The homepage was shown in Fig. 4. The users can input their test data through two ways. One way is to copy pre-miRNA sequences in FASTA format into text area. The other way is to upload test file. Example sequences can be found by clicking on the Example link.
Step 2: Click on the prediction button to submit. iMiRNA-SSF will decide whether the test sequences are real human pre-miRNA sequences or not. Note that the computational cost of P-value feature is expensive, because for each query sequence we need to predict the secondary structures of its random shuffled sequences for 1000 times via running Vienna RNA software.
Step 3: An output example was shown in Fig. 5. If the classification is predicted to Real pre-miRNA, it indicates the query most probably is a pre-miRNA. Besides the predictive classification, we output other useful information, including the secondary structure, MFE and P-value.
Discussion
By exploring two datasets that were constructed with the same positive set and different negative sets, we found that negative samples have significant impact on the predictive results of various methods. Therefore, we constructed an updated benchmark set with different data distributions of negative samples. A new predictor called iMiRNA-SSF was proposed, which was trained with this high quality benchmark dataset. Experimental results showed that iMiRNA-SSF achieved an accuracy of 90.42%, an MCC of 0.79 and an ROC score of 0.96, outperforming three state-of-the-art computational methods, including Triplet-SVM, MiPred and miRNApre. Furthermore, the discriminative power of employed features was investigated on an updated benchmark. The results showed that structure features are more discriminative than the sequence features for pre-miRNA identification.
As shown in this study, the quality of the training samples is very important for improving the predictive performance of a computational predictor. The proposed framework of combining samples with different distributions can be applied to other important tasks in the field of bioinformatics, such as DNA binding protein identification39,40, protein remote homology detection41,42, enhancers and their strength prediction43, etc. Therefore, in our future studies, we will focus on applying the proposed framework to improve the performance of these problems.
Method
Datasets
Our benchmark dataset for pre-miRNA identification (see the Supplementary information) consists of real human pre-miRNAs as positive set and two pseudo pre-miRNAs subsets as negative set. The pre-miRNAs sharing sequence similarity more than 80% were removed using the CD-HIT software44 to get rid of redundancy and avoid bias. The benchmark dataset can be formulated as:


where the positive samples set  contains 1612 human miRNA precursors, which were selected from the 1872 reported Homo sapiens pre-miRNA entries downloaded from the miRBase36,37; the negative samples set
contains 1612 human miRNA precursors, which were selected from the 1872 reported Homo sapiens pre-miRNA entries downloaded from the miRBase36,37; the negative samples set  is the union of
is the union of  and
and  ; the
; the  contains 1612 Xue pseudo miRNAs, which were selected from the 8494 pre-miRNA-like hairpins17; the
contains 1612 Xue pseudo miRNAs, which were selected from the 8494 pre-miRNA-like hairpins17; the  contains 1442 Zou pseudo miRNAs20. As miRNAs locate in the untranslated regions or intragenic regions, both
contains 1442 Zou pseudo miRNAs20. As miRNAs locate in the untranslated regions or intragenic regions, both  and
and  were collected from the protein coding regions. The main difference between them is that they were constructed based on different techniques. The
were collected from the protein coding regions. The main difference between them is that they were constructed based on different techniques. The  was collected by the widely accepted characteristics and the
was collected by the widely accepted characteristics and the  was collected by a multi-level negative sample selection technique. For more information, please refer to17,20.
was collected by a multi-level negative sample selection technique. For more information, please refer to17,20.
Features for characterizing microRNA precursors
Various sequence-based features were used in this study, including primary sequence features, minimum free energy feature, P-value randomization test feature and local triplet sequence-structure features, which were described as followings:
Primary sequence features (N-gram)
For a given RNA sequence R:

where Si {Adenine (A), Cytosine (C), Guanine (G), Uracil (U)}; S1 denotes the nucleic acid residue at sequence position 1, S2 denotes the nucleic acid residue at position 2 and so on. The sequence pattern Si + 1Si + 2Si + 3…Si + N is called N-gram. N-grams refer to all the possible sub-sequences. The different kinds of N-grams are 4n (n is the length of the N-gram). Following previous studies17, we set n as 3 and the number of different 3-grams is 64 (43).
{Adenine (A), Cytosine (C), Guanine (G), Uracil (U)}; S1 denotes the nucleic acid residue at sequence position 1, S2 denotes the nucleic acid residue at position 2 and so on. The sequence pattern Si + 1Si + 2Si + 3…Si + N is called N-gram. N-grams refer to all the possible sub-sequences. The different kinds of N-grams are 4n (n is the length of the N-gram). Following previous studies17, we set n as 3 and the number of different 3-grams is 64 (43).
Minimum of free energy feature (MFE)
The MFE describes the stability of a RNA secondary structure. Some evidences showed that miRNAs have lower folding free energies than random sequences45. The MFE of the secondary structure was predicted by the Vienna RNA software package (released 2.1.6)46 with default parameters.
P-value of randomization test feature (P-value)
In order to determine if the MFE value is significantly different from that of random sequences, a Monte Carlo randomization test was used47. The process can be summarized as follow:
-
1
Infer MFE value of the original sequence.
-
2
Randomize the order of the nucleotides of the original sequence while keeping the dinucleotide distribution (or frequencies) constant48. Then infer the MFE value of the shuffled sequence.
-
3
Repeat step 2 for 999 times to build the distribution of random sequence MFE values.
-
4
Denote Num as the number of shuffled sequences that their MFE value is not greater than the original sequence MFE value, then P-value can be computed based on:

Local triplet sequence-structure features (Triplet-SS)
In the predicted secondary structure, there are only two statuses for each nucleotide, paired or unpaired, represented as brackets “(” or “)” and dots “.”, respectively. The left bracket “(” means that the paired nucleotide is located near the 5′-end and the right bracket “)” means one nucleotide can be paired with another at the 3′-end. When the sequences were represented as vectors, we didn’t distinguish these two situations and used “(” for both situations. For any 3 adjacent nucleotides, there are 8 (23) possible structure compositions: “(((”, “((.”, “(..”, “(.(”, “.((”, “.(.”, “..(” and “…”. Considering the middle nucleotide among the three adjacent nucleotides, there are 32 (4 × 8) possible sequence-structure combinations, which they can be denoted as “U(((”, “A((.”, etc.. The occurrence frequencies of all 32 possible triplet elements were counted along the stem portions of a hairpin segment. Details of the 32 sequence-structure features can be found in17.
Support Vector Machine
Support Vector Machine (SVM) is a supervised machine learning technique based on statistical theory for classification task49. Given a set of fixed length vectors with positive or negative labels, SVM can learn an optimal hyper plane to discriminate the two classes. New test samples can be classified based on the learned classification rule. SVM has exhibited excellent performance in practice and has a strong theoretical foundation of statistical learning.
In this study, the LibSVM algorithm was employed, which is an integrated software tool for SVM classification and regression. The kernel function was set as Radial Basis Function (RBF). The two parameters  and
and  were set as 11 and −9 respectively, which were optimized by using the grid tool in LibSVM package49.
were set as 11 and −9 respectively, which were optimized by using the grid tool in LibSVM package49.
Leave one out cross validation
Three test validation methods, including independent dataset test, sub-sampling (or K-fold cross-validation) test and leave-one-out test, are often used to evaluate the performance of a predictor. Among these three methods, the leave-one-out test is deemed the least arbitrary and most objective as elucidated in49,50,51. It has been widely recognized and adopted by investigators to examine the quality of various predictors. In the leave-one-out test, each sequence in the benchmark dataset is in turn singled out as an independent test sample and all the rule-parameters are calculated with the whole benchmark dataset.
Measurement
For a prediction problem, a classifier can predict an individual instance into the following four categories: false positive (FP), true positive (TP), false negative (FN) and true negative (TN). As shown in previous studies52,53, the total prediction accuracy (ACC), Specificity (Sp), Sensitivity (Sn) and Mathew’s correlation coefficient (MCC) for assessment of the prediction system are given by:




The receiver operating characteristic (ROC) score54 was also employed to evaluate the performance of different methods. Because it can evaluate the trade-off between specificity and sensitivity. An ROC score is the normalized area under a curve that is plotted with true positives as a function of false positives for varying classification thresholds. An ROC score of 1 indicates a perfect separation of positive samples from negative samples, whereas an ROC score of 0.5 denotes that random separation.
Additional Information
How to cite this article: Chen, J. et al. iMiRNA-SSF: Improving the Identification of MicroRNA Precursors by Combining Negative Sets with Different Distributions. Sci. Rep. 6, 19062; doi: 10.1038/srep19062 (2016).
References
Bartel, D. P. MicroRNAs: genomics, biogenesis, mechanism and function. cell 116, 281–297 (2004).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5, 522–531 (2004).
Li, Y. et al. ViRBase:a resource for virus-host ncRNA-associated interactions. Nucleic Acids Res 43, D578–D582 (2015).
Zhang, X. et al. RAID: a comprehensive resource for human RNA-associated (RNA-RNA/RNA-protein) interaction. RNA 20, 989–993 (2014).
Li, Y. et al. Connect the dots: a systems level approach for analyzing the miRNA-mediated cell death network. Autophagy 9, 436–439 (2013).
Shi, H., Wu, Y., Zeng, Z. & Zou, Q. A Discussion of MicroRNAs in Cancers. Curr Bioinform 9, 453–462 (2014).
Zou, Q. et al. Prediction of microRNA-disease associations based on social network analysis methods. Biomed Res Int 2015, 810514 (2015).
Wang, Q. et al. Briefing in family characteristics of microRNAs and their applications in cancer research. BBA-Proteins Proteom 1844, 191–197 (2014).
Zou, Q., Li, J., Song, L., Zeng, X. & Wang, G. Similarity computation strategies in the microRNA-disease network: A Survey. Brief Funct Genomics 10.1093/bfgp/elv024 (2015).
Wang, Y. et al. Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis 4, e765 (2013).
Wu, D. et al. ncRDeathDB: A comprehensive bioinformatics resource for deciphering network organization of the ncRNA-mediated cell death system. Autophagy 11, 1917–1926 (2015).
Cai, R. C., Zhang, Z. J. & Hao, Z. F. Causal gene identification using combinatorial V-structure search. Neural Networks 43, 63–71 (2013).
Cai, R. C., Hao, Z. F., Yang, X. W. & Wen, W. An efficient gene selection algorithm based on mutual information. Neurocomputing 72, 991–999 (2009).
Cai, R. C., Tung, A. K. H., Zhang, Z. J. & Hao, Z. F. What is Unequal among the Equals? Ranking Equivalent Rules from Gene Expression Data. IEEE T Knowl Data En 23, 1735–1747 (2011).
Cai, R. C., Zhang, Z. J. & Hao, Z. F. BASSUM: A Bayesian semi-supervised method for classification feature selection. Pattern Recogn 44, 811–820 (2011).
Liu, B., Liu, F., Fang, L., Wang, X. & Chou, K.-C. repDNA: a Python package to generate various modes of feature vectors for DNA sequences by incorporating user-defined physicochemical properties and sequence-order effects. Bioinformatics 31, 1307–1309 (2015).
Xue, C. et al. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics 6, 310 (2005).
Jiang, P. et al. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res 35, W339–W344 (2007).
Hackenberg, M., Sturm, M., Langenberger, D., Falcon-Perez, J. M. & Aransay, A. M. miRanalyzer: a microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res 37, W68–W76 (2009).
Wei, L. et al. Improved and promising identification of human microRNAs by incorporating a high-quality negative set. IEEE ACM T Comput Bi 11, 192–201 (2014).
Liu, B., Fang, L., Liu, F., Wang, X. & Chou, K.-C. iMiRNA-PseDPC: microRNA precursor identification with a pseudo distance-pair composition approach. J Biomol Struct Dyn 10.1080/07391102.2015.1014422 (2015).
Liu, B. et al. Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS ONE 10, e0121501 (2015).
Liu, B., Fang, L., Jie, C., Liu, F. & Wang, X. miRNA-dis: microRNA precursor identification based on distance structure status pairs. Mol BioSyst 11, 1194–1204 (2015).
Liu, B. et al. Identification of microRNA precursor with the degenerate K-tuple or Kmer strategy. J Theor Biol 385, 153–159 (2015).
Lin, C. et al. LibD3C: Ensemble Classifiers with a Clustering and Dynamic Selection Strategy. Neurocomputing 123, 424–435 (2014).
Nam, J.-W., Kim, J., Kim, S.-K. & Zhang, B.-T. ProMiR II: a web server for the probabilistic prediction of clustered, nonclustered, conserved and nonconserved microRNAs. Nucleic Acids Res 34, W455–W458 (2006).
Yousef, M., Showe, L. & Showe, M. A study of microRNAs in silico and in vivo: bioinformatics approaches to microRNA discovery and target identification. FEBS J 276, 2150–2156 (2009).
Lim, L. P., Glasner, M. E., Yekta, S., Burge, C. B. & Bartel, D. P. Vertebrate microRNA genes. Science 299, 1540–1540 (2003).
Wang, X. et al. MicroRNA identification based on sequence and structure alignment. Bioinformatics 21, 3610–3614 (2005).
Liu, X., He, S., Skogerbo, G., Gong, F. & Chen, R. Integrated sequence-structure motifs suffice to identify microRNA precursors. PloS ONE 7, e32797 (2012).
Liu, B. et al. Pse-in-One: a web server for generating various modes of pseudo components of DNA, RNA and protein sequences. Nucleic Acids Res W1, W65–W71 (2015).
Liu, B., Liu, F., Fang, L., Wang, X. & Chou, K.-C. repRNA: a web server for generating various feature vectors of RNA sequences. Mol Genet Genomics, 1–9 (2015).
Song, L. et al. nDNA-prot: Identification of DNA-binding Proteins Based on Unbalanced Classification. BMC Bioinformatics 15, 298 (2014).
Zou, Q. et al. Survey of MapReduce Frame Operation in Bioinformatics. Brief Bioinform 15, 637–647 (2014).
Ambros, V. et al. A uniform system for microRNA annotation. RNA 9, 277–279 (2003).
Kozomara, A. & Griffiths-Jones, S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 10.1093/nar/gkq1027 (2010).
Kozomara, A. & Griffiths-Jones, S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 10.1093/nar/gkt1181 (2013).
Uğuz, H. A two-stage feature selection method for text categorization by using information gain, principal component analysis and genetic algorithm. Knowl-Based Syst 24, 1024–1032 (2011).
Liu, B. et al. PseDNA-Pro: DNA-Binding Protein Identification by Combining Chou’s PseAAC and Physicochemical Distance Transformation. Mol Inform 34, 8–17 (2015).
Liu, B., Wang, S. & Wang, X. DNA binding protein identification by combining pseudo amino acid composition and profile-based protein representation. Sci Rep 5, 15479 (2015).
Liu, B., Chen, J. & Wang, X. Application of Learning to Rank to protein remote homology detection. Bioinformatics 31, 3492–3498 (2015).
Liu, B. et al. Combining evolutionary information extracted from frequency profiles with sequence-based kernels for protein remote homology detection. Bioinformatics 30, 472–479 (2014).
Liu, B., Fang, L., Long, R., Lan, X. & Chou, K.-C. iEnhancer-2L: a two-layer predictor for identifying enhancers and their strength by pseudo k-tuple nucleotide composition. Bioinformaitcs 10.1093/bioinformatics/btv604 (2015).
Sætrom, P. et al. Conserved microRNA characteristics in mammals. Oligonucleotides 16, 115–144 (2006).
Zhang, B. H., Pan, X. P., Cox, S. B., Cobb, G. P. & Anderson, T. A. Evidence that miRNAs are different from other RNAs. Cell Mol Life Sci 63, 246–254 (2006).
Hofacker, I. L. Vienna RNA secondary structure server. Nucleic Acids Res 31, 3429–3431 (2003).
Bonnet, E., Wuyts, J., Rouzé, P. & Van de Peer, Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20, 2911–2917 (2004).
Workman, C. & Krogh, A. No evidence that mRNAs have lower folding free energies than random sequences with the same dinucleotide distribution. Nucleic Acids Res 27, 4816–4822 (1999).
Chang, C.-C. & Lin, C.-J. LIBSVM: a library for support vector machines. ACM T Intel Syst Tec 2, 27 (2011).
Liu, B., Chen, J. & Wang, X. Protein remote homology detection by combining Chou’s distance-pair pseudo amino acid composition and principal component analysis. Mol Genet Genomics 290, 1919–1931 (2015).
Liu, B. et al. iDNA-Prot|dis: Identifying DNA-Binding Proteins by Incorporating Amino Acid Distance-Pairs and Reduced Alphabet Profile into the General Pseudo Amino Acid Composition. PLoS ONE 9, e106691 (2014).
Zhao, X., Zou, Q., Liu, B. & Liu., X. Exploratory predicting protein folding model with random forest and hybrid features. Curr Proteomics 11, 289–299 (2014).
Liu, B., Liu, B., Liu, F. & Wang, X. Protein binding site prediction by combining Hidden Markov Support Vector Machine and Profile-based Propensities. Sci World J 2014, 464093 (2014).
Fawcett, T. An introduction to ROC analysis. Pattern Recog Lett 27, 861–874 (2006).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 61300112, 61573118 and 61272383), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, the Natural Science Foundation of Guangdong Province (2014A030313695), Shenzhen Foundational Research Funding (Grant No. JCYJ20150626110425228) and Development Program of China (863 Program) [2015AA015405].
Author information
Authors and Affiliations
Contributions
B.L. conceived of the study and designed the experiments, participated in designing the study, drafting the manuscript and performing the statistical analysis. J.J.C. participated in coding the experiments and drafting the manuscript. X.L.W. participated in performing the statistical analysis. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Chen, J., Wang, X. & Liu, B. iMiRNA-SSF: Improving the Identification of MicroRNA Precursors by Combining Negative Sets with Different Distributions. Sci Rep 6, 19062 (2016). https://doi.org/10.1038/srep19062
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19062
This article is cited by
-
Distinguishing mirtrons from canonical miRNAs with data exploration and machine learning methods
Scientific Reports (2018)
-
SkipCPP-Pred: an improved and promising sequence-based predictor for predicting cell-penetrating peptides
BMC Genomics (2017)
-
Categorization of species based on their microRNAs employing sequence motifs, information-theoretic sequence feature extraction, and k-mers
EURASIP Journal on Advances in Signal Processing (2017)
-
On the performance of pre-microRNA detection algorithms
Nature Communications (2017)
-
An improved method for identification of small non-coding RNAs in bacteria using support vector machine
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.



 ,
,  and
and  , respectively. (D, E) and (F) are the comparison of distributions of P-value on
, respectively. (D, E) and (F) are the comparison of distributions of P-value on  ,
,  and
and  , respectively.
, respectively.
 and
and  ; Bzou is the generated decision boundary based on
; Bzou is the generated decision boundary based on  and
and  ; BNewis the generated decision boundary based on
; BNewis the generated decision boundary based on  ,
,  and
and  .
.

