Abstract
Tomato yellow leaf curl virus (TYLCV) is one of the most well-known tomato-infecting begomoviruses and transmitted by Bemisia tabaci. Seed transmission has previously been reported for some RNA viruses, but TYLCV has not previously been described as a seed-borne virus. In 2013 and 2014, without whitefly-mediated transmission, TYLCV was detected in young tomato plants germinated from fallen fruits produced from TYLCV-infected tomato plants in the previous cultivation season. In addition, TYLCV-Israel (TYLCV-IL) was also detected in seeds and their seedlings of TYLCV-infected tomato plants that were infected by both viruliferous whitefly-mediated transmission and agro-inoculation. The seed infectivity was 20–100%, respectively and the average transmission rate to seedlings was also 84.62% and 80.77%, respectively. TYLCV-tolerant tomatoes also produced TYLCV-infected seeds, but the amount of viral genome was less than seen in TYLCV-susceptible tomato plants. When tomato plants germinated from TYLCV-infected seeds, non-viruliferous whiteflies and healthy tomato plants were placed in an insect cage together, TYLCV was detected from whiteflies as well as receiver tomato plants six weeks later. Taken together, TYLCV-IL can be transmitted via seeds and tomato plants germinated from TYLCV-infected seeds can be an inoculum source of TYLCV. This is the first report about TYLCV seed transmission in tomato.
Similar content being viewed by others
Introduction
Tomato yellow leaf curl virus (TYLCV) is a tomato (Solanum lycopersicum)-infecting plant virus transmitted by whitefly Bemisia tabaci1,2. It belongs to the genus Begomovirus of the family Geminiviridae and has a single-stranded circular DNA genome of about 2.8 kb encapsidated in a twinned icosahedral virion3. TYLCV-infected tomato plants show severe symptoms such as stunting, leaf curling and yellowing, which cause critical production loss in tomato cultivation4. In addition to tomato, other cultivated plants including pepper (Capsicum species), common bean (Phaseolus vulgaris), cucurbit (Cucumis species) and eustoma (Eustoma grandiflora) have been reported to be TYLCV hosts3,5,6,7,8. After the first report from the Middle East in 1931, TYLCV has occurred uninterruptedly in tropical and subtropical areas1. In Korea, TYLCV has been reported continuously across the country since the first outbreak in 20089,10.
The management of TYLCV in tomato is difficult and expensive both in cultivation under a structure and open field production11. Many different approaches for controlling TYLCV disease such as removing whiteflies, killing intermediate weeds and changing cultivation season have been applied to decrease losses due to TYLCV because a single approach is not frequently effective and certain other approaches cannot be used in different agricultural environments and locations3,12. Therefore a combination of chemical and biological control techniques for integrated pest management should be employed to reduce the population and migration of the whitefly vector and minimize or eliminate inoculum sources of TYLCV.
The transmission of most plant viruses depends on vectors such as parasitic fungi, root nematodes, mites and insects13,14. Some viruses can also be mediated by the sap of virus-infected plants (mechanical transmission), vegetative propagation and grafting15. However, these are restricted to horizontal transmission among neighboring plants16. On the other hand, seed transmission can play a critical role in survival from season to season and from parent to offspring16,17. According to previous studies, there are three major methods of vertical transmission of plant viruses via seed contamination18. A few stable viruses, such as tobamoviruses, remain only in the seed coat without embryo contamination and then are transmitted to the seedling after germination19. Other methods of seed contamination correspond to virus invasion of the embryo, which is usually protected against virus invasion, either from infected maternal tissues or via infected pollen18. Approximately 231 plant virus and viroid diseases have been reported to be seed-transmissible20. From tomato plants, Arabis mosaic virus, Pepino mosaic virus, Tomato apical stunt viroid (TASVd), Tomato black ring virus, Tomato chlorotic dwarf viroid (TCDVd), Tomato mosaic virus and Tomato streak virus (TSV) were confirmed as transmittable by seeds21,22,23,24,25,26. Among them, TCDVd exhibited 85.5–94.4% transmission though seeds in tomato and high transmission yields were also identified from TASVd (80%) and TSV (66%)21,24.
There have been no previous reports addressing the potential of TYLCV to be a seed-borne virus on any host plant. Begomoviruses including TYLCV have been believed to be inoculated by only whitefly-mediated transmission and artificial inoculation with an infectious clone27. In some previous papers, Abutilon mosaic virus and Beet curly top virus affiliated with the family Geminiviridae were listed as seed-borne viruses, but they were considered to be erroneous reports with nomenclature problems20,28. Therefore, geminiviruses were thought to spread only via insect vectors in nature. Recently, however, Sweet potato leaf curl virus (SPLCV) belonging to the genus Begomovirus was first reported as a seed-transmissible virus in sweet potatoes29. More than 70% of the seeds harvested from SPLCV-infected sweet potato plants were infected by SPLCV. The transmission rate of SPLCV from seeds to seedlings was up to 15%. Southern blot hybridization showing SPLCV-specific single- and double-stranded DNAs in seedlings germinated from SPLCV-infected seeds confirmed SPLCV seed transmission. Therefore, the possibility of TYLCV seed transmission has been reinvestigated based on the seed transmission results of SPLCV as another begomovirus in the family Geminiviridae.
Seed-transmissible viruses can be an initial source of inoculum for vector-mediated transmission and viruses can be disseminated over a long range and perpetuated in seeds30. Therefore, understanding the possibility of TYLCV seed transmission is important for plant protection against infection and the spread of TYLCV. The careful investigation of TYLCV transmission through seeds can provide answers to unresolved problems about sporadic TYLCV occurrence in cultivation under structures or in bionurseries without outbreak of whiteflies. In this study, TYLCV detection was performed in floral tissues, seeds and their young seedlings harvested from TYLCV-infected plants inoculated by whitefly-mediated infection and agro-inoculation to evaluate the possibility of TYLCV seed transmission in tomato plants. In addition, we also investigated whether seedlings germinated from TYLCV-infected seeds could act as TYLCV reservoirs for whitefly transmission.
Results
TYLCV was detected from newly grown tomato plants under no-whitefly conditions
At the tomato-cultivation farm in Jeju Island located in the southern part of Korea, small tomato plants germinated from seeds of fallen tomato fruits from TYLCV-infected tomato plants in the previous cultivation season, were observed and harvested for further TYLCV detection analysis (Fig. 1B). These tomato plants had no visible TYLCV symptoms, but PCR results using the TYLCV-specific primer set showed that 17 plants from 20 randomly selected newly grown small tomato plants were TYLCV infected (Fig. 1C). A similar experiment was performed using tomato plants from just outside of the greenhouse in another tomato farm located in Goheung, Korea and TYLCV was also detected from 14 of 20 tomato plants (Fig. 1F). No whiteflies were observed on either of these two farms.
TYLCV detection from seedlings.
(A) and (D) Symptomatic tomato infected by TYLCV. (B) and (E) Seedlings naturally germinated from fallen fruits of TYLCV-infected tomato plants. (C) and (F) PCR results using a TYLCV-specific primer set. Lane N, no template control; lane P, positive control with TYLCV-infected tomato genomic DNA; and lanes 1-20, genomic DNA from collected tomato seedlings.
Floral tissues, seeds of TYLCV-infected tomato plants and their seedlings tested positive to TYLCV
All five bulked floral tissues (petal, stamen and pistil respectively) and fruit flesh harvested from TYLCV-infected tomatoes tested positive for TYLCV on PCR analysis. Whole dried seeds, surface-sterilized seeds and embryos with endosperm tissues were also infected with TYLCV (Fig. 2). The TYLCV detection rates of these tissues were determined to be 20–100%, representing the theoretically minimum to maximum values, as described in the Materials and Methods (5/5), respectively (Fig. 2C, Table 1). TYLCV was also detected in all cotyledons (22/26) and true leaves (41/45) of their young tomato plants germinated from seeds, which were collected from whitefly-transmitted TYLCV-infected tomatoes (Fig. 2C, Table 1). In addition, similar results were observed from samples of tomato plants which were agro-inoculated with TYLCV-infectious clones (Fig. 2C, Table 1). All samples, including floral tissues, dried seeds, embryos with endosperms, cotyledons and young seedlings, were infected by TYLCV (Fig. 2C). Analyzed sequences from all tested organs, including floral tissue, seeds and seedlings, exhibited 100% identity with a sequence of the TYLCV-infectious clone (NCBI GenBank accession number JN680149) (data not shown). To analyze the systemic infection of TYLCV in young seedlings, TYLCV was also tested in leaf, stem and root samples of young tomato seedlings and all samples showed positive TYLCV amplicons in the PCR reaction (Fig. 2D).
PCR analysis for TYLCV from reproductive tissues and seeds of whitefly-mediated infected and agro-inoculated tomato plants.
(A) Schematic diagram of preparation of TYLCV-infected tomato plants by whitefly-mediated (WMT) and agro-inoculation (AI) methods. Fruits were harvested from each prepared tomato plant and seeds were collected from tomato fruits. (B) Tomato embryos of seeds collected from tomato fruits and cotyledons and true leaves from seeds. (C) PCR analysis with symptomatic leaves, floral tissues (petal, stamen and pistil), fruits, whole seeds, sterilized seeds, endosperm and embryos of TYLCV-infected tomato plants and cotyledons and the true leaves of their offspring. Lane N, no template control; and lane P, positive control with TYLCV-infected tomato genomic DNA. (D) PCR analysis with leaf, stem and root samples from tomato plants germinated from seeds which were collected from TYLCV-infected tomatoes. Lane N, no template control; lane P, positive control with TYLCV-infected tomato genomic DNA; lane L, leaf; lane S, stem; and lane R, root samples from tomato plants germinated from TYLCV-infected seeds.
TYLCV DNA accumulated more in seeds and their seedlings harvested from the TYLCV-susceptible tomato cultivar than those of the TYLCV-resistant cultivar after TYLCV infection
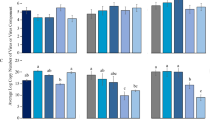
To investigate whether or not TYLCV-tolerant tomato plants also transmit TYLCV into seeds and seedlings, TYLCV was agro-inoculated in tomato plants of both a susceptible cultivar (Seogwang cv.) and a resistant cultivar (Bacchus cv.), which were all commercially purchased. TYLCV-tolerant cultivar plants did not show any symptoms and they grew just like the mock-inoculated healthy tomato plants (Fig. 3A). TYLCV-tolerant cultivar (Bacchus cv.) plants were confirmed as having Ty-1 and Ty-3a genes by PCR-RFLP analysis; however, TYLCV-susceptible cultivar plants did not have the TYLCV-tolerant gene (Fig. 3B). Figure 3C showed that TYLCV could infect both tomato cultivar plants and it could also be transmitted to the seeds of two cultivar plants, respectively, even though TYLCV DNA accumulation in TYLCV-tolerant cultivar plants was more reduced than that of TYLCV-susceptible cultivar plants. These different TYLCV DNA accumulation patterns amplified in the seeds of TYLCV-tolerant and TYLCV-susceptible cultivars were similar to the TYLCV DNA accumulation shown in leaf tissues. To analyze whether viral DNA accumulation patterns in seedlings germinated from TYLCV-infected seeds were different between TYLCV-susceptible and -tolerant cultivar plants, a real-time amplification test was conducted with genomic DNA from the seedlings of two different tomato cultivars. Relative amounts of TYLCV DNA in TYLCV-susceptible Seogwang tomato were about 66–164 times more than those in TYLCV-tolerant Bacchus tomato (Fig. 3D). Taken together, all TYLCV DNA accumulation patterns shown in leaves, seeds and seedlings showed strong correlations based on TYLCV susceptibility. The high replication efficiency and high viral DNA accumulation that occur in TYLCV-susceptible plants may provide a greater opportunity for transmission of viral DNA to seeds or seedlings than occurs in TYLCV-resistant plants. These data suggested that the TYLCV tolerance gene may not be involved in the seed transmission process.
Relationship between different TYLCV DNA amounts and TYLCV susceptibility.
(A) Symptom development in TYLCV-susceptible Seogwang cultivar tomato showing typical symptoms such as yellowing and curling of leaves and severe stunting (left) and TYLCV-tolerant Bacchus tomato showing no symptoms (right). (B) Identification of Ty-1, 2 and 3 loci. Two different bands were shown in Bacchus cultivar tomato (left). The Ty-2 locus was not confirmed (middle) and the Ty-3a/ty-3 loci was identified from the Bacchus cultivar tomato (right). Lane M, iVDye 100-bp DNA ladder (GenDEPOT, Barker, TX, USA); lane S, TYLCV-susceptible Seogwang cultivar tomato; lane B, TYLCV-tolerant Bacchus cultivar tomato; and lane N, no template control. (C) PCR analysis with leaf and seed samples from TYLCV-inoculated TYLCV-susceptible and -tolerant tomato plants. Lane N, no template control; lane P, positive control with TYLCV-infected tomato genomic DNA; lane L, leaf; lane S1-5, genomic DNA form leaves or seeds of TYLCV-susceptible tomato plants; and lane T1-5, genomic DNA from leaves or seeds of TYLCV-tolerant tomato plants. (D) Relative DNA amounts of TYLCV in TYLCV-susceptible and -tolerant tomato plants germinated from seeds collected from TYLCV-infected tomatoes. Five individual samples for each cultivar were used for analysis. Relative TYLCV DNA amounts were normalized against T1 tomato. Data analyses were conducted using the 2−ΔΔCt method.
Seed-borne TYLCV was transmitted to healthy tomato plants by whiteflies
To investigate the possibility of TYLCV transmission from seedlings germinated from TYLCV-infected seeds to new healthy plants via non-viruliferous whiteflies, a transmission assay was performed by placing a donor tomato plant germinated from TYLCV-infected seeds, three healthy receiver tomato plants and around 100 non-viruliferous whiteflies together in an insect-rearing tent (Fig. 4A). After 8 weeks of co-cultivation in an insect-rearing tent, three healthy tomato plants were all infected by TYLCV (3/3) via new viruliferous whiteflies, which obtained TYLCV from a TYLCV-infected donor plant. Three receiver plants produced typical TYLCV disease symptoms on newly developed leaves (Fig. 4E) and these symptoms were confirmed by both PCR and Southern blot hybridization (Fig. 5A,B). In both donor and receiver plants 8 weeks after co-cultivation, Southern blot showed that TYLCV replicated viral DNA in both plants by showing TYLCV-specific ssDNA and dsDNA (Fig. 5B). In the case of new viruliferous whitefly, PCR and Southern hybridization data supported that non-viruliferous whiteflies were TYLCV-viruliferous (Fig. 5A,B). Sequence analysis also supported that all examined samples of the donor plant, receiver plants and whiteflies were infected by the same TYLCV originating from donor tomato plants via whiteflies (Fig. 5C). Taken together, non-virulferous whiteflies transmitted TYLCV from a donor tomato plant, which were germinated from seeds harvested from TYLCV-infected plants, to healthy receiver tomato plants (Fig. 6).
Whitefly-mediated transmission of seed-borne TYLCV from infected to healthy tomato plants.
(A) Experimental scheme of whitefly-mediated transmission analysis. TYLCV-infected plants germinated from seeds collected from TYLCV-infected tomatoes (donor) and healthy tomato plants (recipients) were put in an insect tent with non-viruliferous whiteflies. During the first 4 weeks, whiteflies became TYLCV viruliferous and seed-borne TYLCV was transmitted to tomato plants. (B) and (C) TYLCV-infected donor tomato plants. (D) and (E) Recipient tomato plants.
PCR analysis confirming whitefly-mediated transmission of seed-borne TYLCV from infected to healthy tomato plants.
(A) PCR analysis using a TYLCV-specific primer set and (B) Southern hybridization using a TYLCV-specific probe at 0 and 8 weeks after whitefly release in tents. Lane N, negative control; lane P, positive control with TYLCV-infected tomato genomic DNA; lane D, donor plant; lane V, vector (whitefly); and lane R, recipient plant genomic DNA. OC, open-circular double-stranded DNA; SC, supercoiled double-stranded DNA; SS, single-stranded DNA. (C) Linearized diagram of TYLCV DNA. Red and green arrows indicate coding sequences and the gray dotted box indicates the region used for sequence analysis containing the intergenic region of TYLCV. (D) Multiple sequence alignments of the TYLCV partial genome from donor and recipient plants and a vector with the control sequence (JN680149).
Discussion
Tomato is one of the most valuable vegetable crops globally, cultivated in many countries31. In Korea, tomato has been widely cultivated in almost every area and consumption of tomato has gradually been increased. However, tomato-infecting pathogens were continuously flowing into Korea and numerous economic losses have been reported9,32. Among them, TYLCV did a great deal of damage to tomato cultivation and has been reported consistently throughout the country after the first occurrence in 20089,10. Based on sequence analyses, Korean TYLCV isolates were suggested to originate from two Japanese TYLCV isolates9. As a hypothesis introduced in the previous report, TYLCV-infected seedlings or viruliferous whitefly can serve as an initial inoculum of TYLCV in Korea, where TYLCV was spread rapidly by non-viruliferous whiteflies observed in many greenhouses since 20059,20. In the occurrence or outbreak of some viruses, seeds can be considered the initial viral source (Sastry, 2013). However, in the case of TYLCV as a member of the genus Begomovirus and family Geminiviridae, the possibility of TYLCV inflow or transmission through seeds was completely excluded because TYLCV was known to be transmitted not by seeds or mechanical inoculation, but only by the insect vector Bemisia tabaci27,28.
Even though TYLCV transmission in nature has been thought to be restricted to whitefly-mediated methods, farmers in Korea periodically raised concerns regarding the potential for seed transmission of TYLCV. By chance, in this study, TYLCV-infected tomato seedlings were identified from whitefly-free conditions in two geographically different regions, Jeju, Jeju Island and Goheung, Chunnam Province (Fig. 1). With the previous documentation of geminivirus seed transmission of SPLCV in sweet potato plants29, we were concerned about the vertical transmission of TYLCV from infected tomato plants to offspring via seeds. Like TYLCV, SPLCV, which belongs to the Begomovirus genus and Geminiviridae family, is the first geminivirus documented to be a seed-transmissible virus. To investigate whether TYLCV could infect floral tissues and seeds from vegetative tissues, floral tissues (petal, stamens and pistils) and seeds (dried seeds, surface-sterilized seeds and embryos) harvested from whitefly transmitted TYLCV-infected tomato plants were tested. In addition, cotyledons and true leaves of young plants were also analyzed. In all tissues, TYLCV was detected with high infection rates (73–91%) (Fig. 2C, Table 1). Sequence analysis of all PCR products showed that TYLCV sequences were identical among the samples and this result indicated that TYLCV of all samples originated from the initial inoculum (data not shown). Systemic infection of TYLCV in young plants germinated from TYLCV-infected seeds was also confirmed from organ-specific PCR results (Fig. 2D). This result indicated that seed-transmitted TYLCV can infect whole plants, replicate viral DNA and move systemically just like TYLCV-inoculated via agrobacteria or whiteflies. Seed transmission rates vary from 0% to 100% depending on interactions between the virus and host plant33. Seed transmission rates could also vary according to the detection methods used for virus detection such as serological methods or molecular biological methods. New detection methods such as loop-mediated isothermal amplification can yield higher sensitivity and specificity34. High seed transmission rates were not a universal feature of plant viruses, but some tomato-infecting viruses and viroids reached high infection rates such as TCDVd (85.5–94.4%), TASVd (80%) and TSV (66%)21,24. Whitefly-transmitted TYLCV-infected tomato plants showed 20–100% floral infection rates and 20–100% seed transmission rates. Like whitefly-transmitted TYLCV-infected tomato plants, agro-inoculated TYLCV-infected tomato plants also showed similarly high infection rates on different floral tissues (20–100%) and seeds (20–100%) (Fig. 2C, Table 1). Therefore, these inoculation studies indicated that TYLCV seed transmission on tomato plants was confirmed with both inoculation systems, whitefly-mediated infection and agro-inoculation.
The results of different viral genome accumulation rates in young plants germinated from TYLCV-infected seeds between TYLCV-susceptible Seogwang and TYLCV-tolerant Bacchus tomatoes were also confirmed by PCR analysis as well as Southern hybridization (Fig. 3C,D). These TYLCV DNA accumulation patterns in seeds and their seedlings are similar to those in TYLCV-infected tomatoes of these two cultivars (Fig. 3A). Previous studies showed that symptom severity by TYLCV infection was correlated to viral genome methylation, which represented one of the important anti-viral mechanisms of plants called transcriptional gene silencing35,36. Low TYLCV DNA amounts shown in TYLCV-tolerant tomato are thought to be the result of siRNA production and DNA methylation targeted to viral sequences35. Moreover, Fig. 3 showed that TYLCV of a TYLCV-tolerant tomato plant was transmitted via the seeds to seedlings even though viral DNA accumulation was much lower than a TYLCV-susceptible tomato plant. This result may indicate that DNA methylation by the Ty gene(s) in a TYLCV-tolerant plant is not a main factor for TYLCV seed transmission because TYLCV of a TYLCV-tolerant plant was able to be transmitted to the seedlings via the seeds. However, in this study, we have not investigated the correlation between the degree of TYLCV DNA methylation in the DNA of the seeds or their seedlings and seed transmission; however, this should be investigated in the near future to understand the different amounts of TYLCV in the seeds of TYLCV-tolerant and TYLCV-susceptible cultivars.
Finally, the transmission of seed-borne TYLCV to other healthy tomato plants by whitefly was tested (Figs 4 and 5) in order to investigate whether TYLCV-infected seeds or their seedlings can be a virus reservoir for further virus spread. Even though TYLCV was identified as a seed-transmissible virus in tomato plants, if TYLCV cannot be transmitted from donor tomato plants germinated from TYLCV-infected seeds to healthy receiver tomato plants, the TYLCV-infected tomato plants may be a dead-end host. Figures 4 and 5 show that TYLCV can be transmitted from a donor plant to a receiver plant via whitefly. Southern data supported that TYLCV in donor and receiver plants can replicate its DNA and move systemically. In addition, in terms of insect transmission, TYLCV in seedlings can be normally acquired by whiteflies and delivered to new healthy plants. This means that TYLCV is a seed-transmissible geminivirus.
TYLCV has caused a great deal of economic damage around the world such that many TYLCV management protocols have been developed3,12. To prevent TYLCV spread or management, the following approach is recommended: (1) virus- and whitefly-free transplants should be planted, (2) insecticides or insect repellents for whiteflies should be used to reduce whitefly feeding and virus transmission, (3) TYLCV-infected and TYLCV-infected-looking tomatoes should be eliminated from fields and placed in plastic bags immediately at the beginning of the season, (4) plantings of tomatoes should be separated according to time and space from plantings of other TYLCV crop hosts or weeds that are good sources of whiteflies and (5) TYLCV-resistant tomato cultivars should be used if available in a given production area12,37. In previous reports and plant pathology fact sheets, the warning about the possibility of TYLCV seed transmission has not been introduced. If TYLCV is a seed-transmissible virus, the summarized TYLCV management protocol is not good enough to prevent TYLCV outbreak in fields. Based on our results, we suggest that two more TYLCV management protocols should be added in succession to the previous protocol: (6) the possibility of TYLCV seed infection should be tested before display on the market and (7) more strict investigation and drastic monitoring of TYLCV infection or whitefly occurrence on tomato seed gathering fields should be performed.
This is the first report of TYLCV seed transmission in tomato plants. TYLCV can be transmitted by two different transmission cycles: (1) whitefly-mediated transmission and (2) seed-mediated transmission (Fig. 6). Since TYLCV is a notorious virus in tomato plants, in many countries, enormous efforts have been carried out to prevent the infection or spread of TYLCV concerning transmission by whitefly. However, this study showed that TYLCV-infected seeds can be a more critical source for TYLCV spread than TYLCV-viruliferous whiteflies within a country and between countries. Therefore, due to this new observation of TYLCV seed transmission, a new strategy for TYLCV control is needed.
Methods
Collecting naturally germinated seedlings from TYLCV-infected tomato fruits
In the spring of 2013 and 2014, tomato seedlings from tomato-cultivating farms on Jeju and Goheung in the Republic of Korea were observed (Fig. 1) and these seedlings were evaluated for TYLCV infection. These small seedlings were not artificially planted, but germinated from the seeds of fallen tomato fruits where TYLCV was present during the last cultivation season. No whitefly was detected in or near the greenhouses and no egg or larva was present on the abaxial side of leaves. No disease symptoms were identified among the newly planted tomato plants. To prevent the occurrence of TYLCV, cultivated tomato plants and weeds were eliminated and insecticide was sprayed by farmers. For the analysis of the inoculation status, 20 tomato seedlings were randomly gathered from Jeju and 20 seedlings were collected from Goheung.
DNA extraction and PCR analysis for TYLCV detection from seedlings
Viral DNA was isolated from the leaves of each tomato seedling using the Viral Gene-spinTM Viral DNA/RNA Extraction Kit (iNtRON Biotechnology, Seongnam, Korea) following the manufacturer’s instructions. PCR was performed using a T100TM Thermal Cycler (Bio-Rad, Hercules, CA, USA) with a final reaction volume of 20 μl containing 20 ng of isolated DNA, 1× AccuPower® PCR Master Mix (Bioneer, Daejeon, Korea) and the TYLCV-specific primer set (Table 2). The PCR conditions were as follows: an initial denaturation at 94 °C for 3 min followed by 35 cycles (denaturation at 94 °C for 30 s, annealing at 70 °C for 30 s and an extension at 72 °C for 30 s) and a final extension at 72 °C for 10 min. Amplified DNA fragments were analyzed and electrophoresed on 1% agarose gels. Each reaction was performed three times.
Sample collection, DNA isolation and PCR analysis using whitefly- and agrobacterium-mediated inoculated tomato samples
Tomatoes cvs. Seogwang (TYLCV susceptible) and Bacchus (TYLCV tolerant) (Seminis, Anseong, Korea) were planted in a walk-in growth chamber in Sungkyunkwan University, Korea and fifteen 4-week-old plants were inoculated with agrobacteria containing TYLCV-Isreal (TYLCV-IL)-infectious clones prepared in a previous study10, or placed with TYLCV-IL-viruliferous whitefly (B. tabaci, Q biotype) in an insect-free BugDorm-2120 insect rearing tent (MegaView Science Education Services Co., Taipei, Taiwan) for five days (Fig. 2A). Before planting tomato plants on soil, seeds were tested for TYLCV contamination or infection in advance. They were transplanted in a greenhouse covered with tightly woven mesh located in Yesan and Iksan to prevent the effects of insects on the plants. Whitefly-mediated TYLCV-inoculated tomato plants were treated with insecticide three times to ensure whitefly removal before transplanting. Four weeks after agrobacterium- and whitefly-mediated inoculation, the leaves, flowers and fruits were collected from symptomatic tomato plants. Seeds were harvested from ripe tomato fruits and washed with distilled water several times to remove the tomato fruit flesh. Washed seeds were surface-sterilized with 70% ethanol for 10 min and then 10% Clorox for 20 min38. Embryos were separated carefully from the seed coat of each seed using a sterile scalpel30 (Fig. 2B). The petal, stamen and pistil from the floral tissues were also prepared from mature flowers. Seeds harvested from TYLCV-infected tomato fruits were planted in pots and placed in a BugDorm-2120 insect rearing tent to maintain whitefly-free conditions. Cotyledons were harvested 10 days after planting and true leaves were collected 30 days after planting. Stem and root samples were also collected from the same plants. Twenty-five sampled tissues for each organ were bulked into five pools (five sampled tissues for one pool) and each pool was used for DNA extraction with the Viral Gene-spinTM Viral DNA/RNA Extraction Kit (iNtRON Biotechnology). DNA was isolated from each cotyledon and true leaf individually. PCR was performed as described above. The detection rate range for seeds and the detection rate for embryos varied from the minimum and maximum numbers because the infection rates were calculated based on the number of bulks that were confirmed to be TYLCV infected by PCR.
Identification of the TYLCV-tolerant loci using sequence-characterized amplified region/cleaved amplified polymorphic sequence (SCAR/CAPS) markers
To identify tolerant loci (Ty-1, 2 and 3) in the TYLCV-tolerant Bacchus cultivar tomato, sequence-characterized amplified region/cleaved amplified polymorphic sequence (SCAR/CAPS) markers of Ty-1, Ty-2 and Ty-3 introduced in previous studies39,40,41 were detected. PCR was performed with specific primers (Table 3). PCR products for the Ty-2 and Ty-3 markers were directly loaded on an agarose gel and amplicons for the Ty-1 marker were eluted and digested with TaqI restriction enzyme at 65 °C for 1 hr. TaqI-treated products were also confirmed through agarose gel electrophoresis.
Real-time quantitative PCR
To analyze the relationship between TYLCV DNA amounts and TYLCV susceptibility, amounts of genomic DNA of TYLCV were tested by real-time quantitative PCR from the leaves of four-week-old seed-borne TYLCV-infected plants of two different cultivars, TYLCV-susceptible cv. Seogwang and TYLCV-tolerant cv. Bacchus. Reactions were performed using the SYBR premix Ex Taq (Tli RNase H Plus, TaKaRa, Shiga, Japan) with specific primer sets (Table. 1). Cycling of PCR consisted of pre-denaturation at 95 °C for 5 min followed by 40 cycles of a denaturation step at 95 °C for 10 min, an annealing step at 60 °C for 15 sec and an extension step at 72 °C for 20 sec using a Rotor Gene Q thermocycler (QIAGEN, Hilden, Germany). The annealing temperature was modified following the melting temperature of each primer and each reaction was repeated at least three times. Relative TYLCV DNA amounts were normalized against T1 tomato. Data analyses were conducted by the 2−ΔΔCt method42.
Transmissibility test of seed-borne TYLCV to healthy tomato by whitefly
The transmissibility of seed-borne TYLCV in tomato to other healthy tomato plants was analyzed using sweet potato whiteflies. Thirty non-viruliferous Bemisia tabaci (Q biotype) were reared with one TYLCV-donor plant (TYLCV-infected tomato plants germinated from TYLCV-infected seeds, Seogwang cv.) and three TYLCV-receiver plants (TYLCV-free tomato plants, Seogwang cv.) in an insect rearing tent together at 25 °C with a day length of 12 hrs. Another insect tent rearing non-viruliferous whiteflies with healthy plants of tomato cv. Seogwang were prepared as a negative control group. After 8 weeks, each plant and whiteflies were harvested and analyzed for TYLCV infection by PCR, Southern hybridization and sequence analysis. This experiment was performed three times independently.
Southern blot hybridization analysis
Southern hybridization analysis was conducted to confirm the viral replication of TYLCV in tomato plants using the modified method from Southern et al.43. Total DNA isolated from tomato tissues (15 μg) was loaded on 1% agarose gel. After the depurination, denaturation and neutralization steps, DNA loaded on the gel was transferred to a positively charged nylon membrane (Hybond-N+ membrane, GE Healthcare Life Sciences, Waukesha, WI, USA) using the capillary transfer method for up to 16 h and the transferred DNA was linked covalently to the nylon membrane using an ultraviolet crosslinker (UVC 500 crosslinker, GE Healthcare Life Sciences). The TYLCV DNA fragment, amplified with the detection primer set (Table 2), was gel purified and labeled with [α-32P] dCTP using the Rediprime II Random Primer Labeling System (GE Healthcare Life Sciences). Hybridization was conducted at 65 °C for 16 h. After washing, the membrane was then exposed to X-ray film (Kodak, Rochester, NY, USA) for approximately 48 h in a −70 °C freezer.
Sequence analysis
PCR was performed in order to amplify the full-length DNA genome of TYLCV (Table 2). PCR products were ligated into a pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced (Macrogen, Seoul, Korea). The sequences were analyzed using a multiple alignment program MultAlin (http://multalin.toulouse.inra.fr/multalin/)44 and basic local alignment search tool (BLAST, http://blast.ncbi.nlm.nih.gov/Blast.cgi)45.
Additional Information
How to cite this article: Kil, E.-J. et al. Tomato yellow leaf curl virus (TYLCV-IL): a seed-transmissible geminivirus in tomatoes. Sci. Rep. 6, 19013; doi: 10.1038/srep19013 (2016).
References
Czosnek, H. & Laterrot, H. A worldwide survey of tomato yellow leaf curl viruses. Archives of Virology 142, 1391–1406 (1997).
Ghanim, M., Morin, S., Zeidan, M. & Czosnek, H. Evidence for Transovarial Transmission of Tomato Yellow Leaf Curl Virus by Its Vector, the Whitefly Bemisia tabaci. Virology 240, 295–303 (1998).
Moriones, E. & Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Research 71, 123–134 (2000).
Papayiannis, L., Katis, N., Idris, A. & Brown, J. Identification of weed hosts of Tomato yellow leaf curl virus in Cyprus. Plant Disease 95, 120–125 (2011).
Cohen, J. et al. Lisianthus leaf curl a new disease of lisianthus caused by tomato yellow leaf curl virus. Plant Disease 79, 416–420 (1995).
Navas-Castillo, J., Sánchez-Campos, S., Díaz, J. A., Sáez-Alonso, E. & Moriones, E. Tomato yellow leaf curl virus-Is causes a novel disease of common bean and severe epidemics in tomato in Spain. Plant Disease 83, 29–32 (1999).
Reina, J., Morilla, G., Bejarano, E., Rodríguez, M. & Janssen, D. First report of Capsicum annuum plants infected by tomato yellow leaf curl virus. Plant Disease 83, 1176–1176 (1999).
Anfoka, G., Haj Ahmad, F., Abhary, M. & Hussein, A. Detection and molecular characterization of viruses associated with tomato yellow leaf curl disease in cucurbit crops in Jordan. Plant pathology 58, 754–762 (2009).
Lee, H. et al. Phylogenetic analysis and inflow route of Tomato yellow leaf curl virus (TYLCV) and Bemisia tabaci in Korea. Molecules and Cells 30, 467–476 (2010).
Kil, E.-J. et al. Sweet pepper confirmed as a reservoir host for tomato yellow leaf curl virus by both agro-inoculation and whitefly-mediated inoculation. Archives of Virology 159, 2387–2395 (2014).
Lapidot, M. & Polston, J. E. In Natural Resistance Mechanisms of Plants to Viruses 503–520 (Springer, 2006).
Polston, J. E. & Lapidot, M. In Tomato Yellow Leaf Curl Virus Disease 251–262 (Springer, 2007).
Ziegler-Graff, V. & Brault, V. In Plant Virology Protocols 81–96 (Springer, 2008).
Blanc, S., Uzest, M. & Drucker, M. New research horizons in vector-transmission of plant viruses. Current Opinion in Microbiology 14, 483–491 (2011).
Tomlinson, J. Epidemiology and control of virus diseases of vegetables. Annals of Applied Biology 110, 661–681 (1987).
Simmons, H., Holmes, E., Gildow, F., Bothe-Goralczyk, M. & Stephenson, A. Experimental verification of seed transmission of Zucchini yellow mosaic virus. Plant Disease 95, 751–754 (2011).
Johansen, E., Edwards, M. C. & Hampton, R. O. Seed transmission of viruses: current perspectives. Annual Review of Phytopathology 32, 363–386 (1994).
Fabre, F. et al. Narrow bottlenecks affect pea seedborne mosaic virus populations during vertical seed transmission but not during leaf colonization. PLoS pathogens 10, e1003833 (2014).
Broadbent, L. The epidemiology of tomato mosaic. Annals of Applied Biology 56, 177–205 (1965).
Sastry, K. S. Seed-borne plant virus diseases. (Springer, 2013).
Berkeley, G. & Madden, G. Transmission of streak and mosaic diseases of tomato through seed. Sci Agric 13, 194–197 (1932).
Lister, R. & Murant, A. Seed‐transmission of nematode‐borne viruses. Annals of Applied Biology 59, 49–62 (1967).
Hadas, R. et al. Indexing system forTomato mosaic virus (ToMV) in commercial tomato seed lots. Phytoparasitica 32, 421–424 (2004).
Antignus, Y., Lachman, O. & Pearlsman, M. Spread of Tomato apical stunt viroid (TASVd) in greenhouse tomato crops is associated with seed transmission and bumble bee activity. Plant disease 91, 47–50 (2007).
Córdoba-Sellés, M. d. C., García-Rández, A., Alfaro-Fernández, A. & Jordá-Gutiérrez, C. Seed transmission of Pepino mosaic virus and efficacy of tomato seed disinfection treatments. Plant Disease 91, 1250–1254 (2007).
Singh, R. P. & Dilworth, A. D. Tomato chlorotic dwarf viroid in the ornamental plant Vinca minor and its transmission through tomato seed. European Journal of Plant Pathology 123, 111–116 (2009).
Stanley, J., Boulton, M. I. & Davies, J. W. Geminiviridae. eLS, doi: 10.1038/npg.els.0000750 (2001).
Mink, G. Pollen and seed-transmitted viruses and viroids. Annual Review of Phytopathology 31, 375–402 (1993).
Kim, J. et al. Seed transmission of Sweet potato leaf curl virus in sweet potato (Ipomoea batatas). Plant Pathology, doi: 10.1111/ppa.12366 (2015).
Ali, A. & Kobayashi, M. Seed transmission of Cucumber mosaic virus in pepper. Journal of virological methods 163, 234–237 (2010).
Lapidot, M. et al. Pelargonium zonate spot virus is transmitted vertically via seed and pollen in tomato. Phytopathology 100, 798–804 (2010).
Kil, E.-J. et al. Identification of natural weed hosts of Tomato chlorosis virus in Korea by RT-PCR with root tissues. European Journal of Plant Pathology 142, 419–426 (2015).
Sastry, K. S. In Seed-borne plant virus diseases 55–66 (Springer, 2013).
Notomi, T. et al. Loop-mediated isothermal amplification of DNA. Nucleic acids research 28, e63–e63 (2000).
Yang, X. et al. Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS pathogens 7, e1002329 (2011).
Butterbach, P. et al. Tomato yellow leaf curl virus resistance by Ty-1 involves increased cytosine methylation of viral genomes and is compromised by cucumber mosaic virus infection. Proceedings of the National Academy of Sciences 111, 12942–12947 (2014).
Momol, T., Olson, S., Funderburk, J. & Sprenkel, R. Management of Tomato Yellow Leaf Curl Virus (TYLCV) in Tomato in North Florida. Fact Sheet PP- 184 (2001).
Trolinder, N. L. & Goodin, J. Somatic embryogenesis in cotton (Gossypium) I. Effects of source of explant and hormone regime. Plant cell, tissue and organ culture 12, 31–42 (1988).
Garcia, B. E. et al. Co-dominant SCAR marker for detection of the begomovirus-resistance Ty-2 locus derived from Solanum habrochaites in tomato germplasm. Rep. Tomato Genet. Coop 57, 21–24 (2007).
Ji, Y., Schuster, D. J. & Scott, J. W. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Molecular Breeding 20, 271–284 (2007).
Ji, Y., Scott, J. W., Hanson, P., Graham, E. & Maxwell, D. P. In Tomato Yellow Leaf Curl Virus Disease 343–362 (Springer, 2007).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods 25, 402–408 (2001).
Southern, E. Southern blotting. Nature Protocols 1, 518–525 (2006).
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Research 16, 10881–10890 (1988).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. Journal of molecular biology 215, 403–410 (1990).
Acknowledgements
This research was supported by a grant (311058-05-4-HD140) from Agricultural Biotechnology Development Program, Ministry of Agriculture, Food and Rural Affairs of Republic of Korea.
Author information
Authors and Affiliations
Contributions
S.L. and H.-S.C. conceived and designed experiments, E.-J.K., S.K., Y.-J.L., H.-S.B., J.-H.P., H.S. and J.-K.S. conducted experiments, E.-J.K., C.-S.K. and S.L. analyzed the results, E.-J.K., S.K. and S.L. wrote the manuscript and prepared figures, J.-H.L., J.-K.K., K.-Y.L., H.-S.C. and S.L. revised and approved the article. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kil, EJ., Kim, S., Lee, YJ. et al. Tomato yellow leaf curl virus (TYLCV-IL): a seed-transmissible geminivirus in tomatoes. Sci Rep 6, 19013 (2016). https://doi.org/10.1038/srep19013
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19013
This article is cited by
-
A virus drives its vector to virus-susceptible plants at the cost of vector fitness
Journal of Pest Science (2024)
-
Study on mosaic disease of sponge gourd (Luffa cylindrica L.) caused by tomato leaf curl New Delhi virus
Journal of Plant Pathology (2023)
-
Occurrence, distribution, and management of tomato yellow leaf curl virus in China
Phytopathology Research (2022)
-
Metagenomic analyses and genetic diversity of Tomato leaf curl Arusha virus affecting tomato plants in Kenya
Virology Journal (2021)
-
Development of a real-time RT-PCR method for the detection of Citrus tristeza virus (CTV) and its implication in studying virus distribution in planta
3 Biotech (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.