Abstract
The two-spotted spider mite, Tetranychus urticae Koch is a major pest that feeds on >1,100 plant species. Many perennial crops including hop (Humulus lupulus) are routinely plagued by T. urticae infestations. Hop is a specialty crop in Pacific Northwest states, where 99% of all U.S. hops are produced. To suppress T. urticae, growers often apply various acaricides. Unfortunately T. urticae has been documented to quickly develop resistance to these acaricides which directly cause control failures. Here, we investigated resistance ratios and distribution of multiple resistance-associated mutations in field collected T. urticae samples compared with a susceptible population. Our research revealed that a mutation in the cytochrome b gene (G126S) in 35% tested T. urticae populations and a mutation in the voltage-gated sodium channel gene (F1538I) in 66.7% populations may contribute resistance to bifenazate and bifenthrin, respectively. No mutations were detected in Glutamate-gated chloride channel subunits tested, suggesting target site insensitivity may not be important in our hop T. urticae resistance to abamectin. However, P450-mediated detoxification was observed and is a putative mechanism for abamectin resistance. Molecular mechanisms of T. urticae chemical adaptation in hopyards is imperative new information that will help growers develop effective and sustainable management strategies.
Similar content being viewed by others
Introduction
As a flavoring and stability ingredient in beer, hop (Humulus lupulus) is an economically important crop in the Pacific Northwest (PNW) of the United States. The U.S. hop industry is concentrated in the three PNW states, Washington, Oregon and Idaho, which represent over 99% of the nation’s1 and 30% of the world’s2 hop acreage in 2013. The preliminary production of the U.S. hops crop was valued at $249 million in 20131. Hop is a dioeciously perennial specialty crop that is planted in female monoculture3. Hops bloom in the PNW initiated by long days and un-pollinated flowers develop into cones that ripen between mid-August to mid-September. The commercial products from hops are resin and hop oil extracted from the lupulin gland of the hop cone4. In the State of Washington hops are only grown commercially in the Yakima Valley including three distinct growing areas: the Moxee Valley, the Yakama Indian Reservation and the Lower Yakima Valley. Each of these areas are within a 50-mile radius (80 kilometers) of each other in shrub-steppe habitats characterized by low annual winter precipitation and hot dry summers. Climate, experienced growers, established infrastructure and modern drip irrigation techniques enhance optimal hop production in the Yakima Valley and make it among the most productive hop growing regions in the world5. In 2013, Washington growers produced 79% of the U.S. hops crop1.
Integrated pest management strategies have been developed to optimize production of high-quality hops6. To date, several plant pathogens and arthropods have been reported as pests of hops in the PNW6. Among these, Tetranychus urticae is the most serious and prevalent arthropod pest in warmer dry climates6,7 and efficient control of this pest is a priority for the hop industry. In most situations, hops growers apply various acaricides to suppress T. urticae populations. Grower records indicate that up to nine pesticides were used over the course of the hop growing seasons each year in 2012 and 20138. These pesticides include several classes of acaricides with different modes of action. Unfortunately T. urticae has been documented to quickly develop tolerance and resistance to these acaricides, which has been linked to control failures6. The accelerated development of resistance in T. urticae is not only due to the extensive exposure to acaricides, but is also exacerbated by the biology of T. urticae, including an extremely short life span with relatively high fecundity and arrhenotokous reproduction9,10. As its name implies, T. urticae is able to produce webs from silk glands located at each palp11. The webbings made by T. urticae may work as a shelter to avoid pesticide exposure and protect it from other life-threatening conditions including wind, rain and natural enemies12. Moreover, T. urticae undergoes diapause in soil, tree bark, ground cover and dried leaves when decreasing temperature, photoperiod and decline in the quality of food supply occur due to plant senescence13. Diapause facilitates T. urticae adaptation to the agroecosystem and offers a refuge to escape pesticide exposure. Indeed, the two-spotted spider mite has been reported to be the world’s most resistant arthropod; this species has been found to be resistant to 94 unique insecticide/acaricide active ingredients in 468 documented cases worldwide14,15.
The mechanisms of pesticide resistance exhibited by arthropods typically evolve along several trajectories, including behavioral avoidance16, decreased cuticular penetration17, enhanced sequestration or metabolic detoxification18 and target site insensitivity19,20,21. Among these, target site insensitivity to acaricides in T. urticae have been investigated extensively22,23. For example, several mutations in the acetylcholinesterase (AChE) gene have been documented in organophosphate resistant T. urticae strains from Europe and Korea24,25. Two mutations were identified in the Glutamate-gated chloride channel (GluCl) genes that are correlated with abamectin resistance in T. urticae populations23,26. Studies reported that resistance to bifenazate commonly used for T. urticae control was tightly linked to multiple mutations at the Quinol oxidation (Qo) site of mitochondrial cytochrome b (cytb)27,28. Pyrethroid resistance in T. urticae has been associated with several amino acid substitutions in the voltage-gated sodium channel (VGSC) gene29,30. Additionally, recent studies revealed that a mutation on the chitin synthase gene may contribute to resistance to etoxazole31, hexythiazox and clofentezine32 in T. urticae.
In order to design the most effective and sustainable T. urticae management strategy, our long-term goals include revealing the mechanisms underlying the chemical adaptation of T. urticae in the field. We initially calculated the baseline concentration response curves of T. urticae population susceptible to three acaricides: abamectin, bifenazate and bifenthrin. We chose these three compounds because they are currently the most commonly used acaricides for T. urticae control in hopyards according to the spray records we investigated (Fig. 1). Recently, field control failures with these acaricides have been observed in the Yakima Valley of Washington State8. We collected 31 T. urticae field populations from hopyards in the Yakima Valley during summer 2013 (Fig. 2) and evaluated the acaricide resistance levels in most of these populations compared with a susceptible strain. We also investigated the distribution pattern of resistance-associated target site mutations in these field collected T. urticae populations. Finally, the relative expressions of several detoxification-related P450 genes in field T. urticae populations were compared with that of the susceptible population.
The acaricide spray model at hopyards during hop season in 2013.
Several acaricides with different mode of actions were applied to control T. urticae. Among them, abamectin, bifenazate and bifenthrin were commercially important acaricides used in hopyards10.
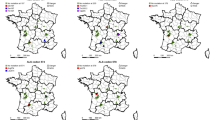
The geographic distribution of 31 T. urticae populations collected from 5 major locations.
The map of Washington counties was modified from a public domain picture (https://commons.wikimedia.org/wiki/File%3AMap_of_Washington_counties%2C_blank.svg).
Results
Baseline toxicity of the lab susceptible population
To establish baseline levels of susceptibility and discriminating concentrations for three acaricides, their toxicity was first evaluated in the susceptible T. urticae population. All acaricides tested caused 100% mortality of spider mites at concentrations equivalent to the field rates of 23 mg a.i./L (abamectin), 899 mg a.i./L (bifenazate) and 120 mg a.i./L (bifenthrin). Probit analysis showed that the dose responses of susceptible T. urticae to these three acaricides are significantly lower than field rates (Table 1).
Acaricide resistance levels in field populations
The toxicities of abamectin and bifenazate were assessed for T. urticae populations collected from 13 and 12 hopyards, respectively (Tables 2 and 3). In the bioassays with abamectin, the LC50s ranged from 1.36 to 26.05 mg a.i./L and the resistant ratios (RRs) compared with the susceptible strain varied from 5.96 to 114.25 (Table 2). Low resistance levels (RR < 10) were observed in 10.5% of the surveyed populations, 10.5% had high resistance (RR > 100) and the majority of the surveyed populations (79%) exhibited moderate resistance (RR = 10–100) to abamectin (Fig. 3A). The RR of the T. urticae population in the organic hopyard (Granger 2) compared with the susceptible population was 11.23, which is the 3rd lowest resistance among surveyed populations and the highest level of mortality (100%) at the field rate. Samples collected from the Granger 4 hopyard showed the lowest resistance ratio (RR = 5.96) compared with the susceptible population. There were three 1st year (baby) hopyards (Prosser 3, 4 and 5) surveyed in 2013. The RRs of samples collected from these baby hopyards ranged from 21.80 to 114.25, exhibiting a moderate to high degree of resistance (Table 2). There were multiple collections from certain hopyards (Prosser 2, 3 and 4) during the course of summer 2013. Specifically, six collections were taken from the Prosser 2 hopyard starting from middle of June till just prior to harvest in late August during which abamectin was applied twice10. The RR increased 6-fold from the middle of July to mid-August (Table 2). The RRs in samples collected from Prosser 3 and 4 increased 1.7-fold and 2.3-fold in four and five weeks, respectively. The highest resistance level to abamectin was recorded at the Prosser 4 (RR = 114.25) (Table 2).
In the bioassays with bifenazate, the LC50s ranged from 3.93 to 78.97 mg a.i./L and the RRs varied from 4.79 to 96.30 (Table 3). Populations exhibiting low resistance levels (RR < 10) accounted for 37.5% of the populations surveyed and 62.5% of the populations exhibited moderate resistance (RR = 10–100) to bifenazate (Fig. 3B). The lowest RR to bifenazate, 4.79, was recorded from the samples collected from the organic hopyard (Granger 2). The RRs of samples collected from the 1st year hopyards showed low to moderate level of resistance (Table 3). The highest RR to bifenazate was observed in the sample collected from Granger 3 (RR = 96.30) (Table 3). Due to the limited number of collected T. urticae individuals in four populations, only the discriminating dose of bifenazate was evaluated (Table 3).
Evaluation of target site mutations
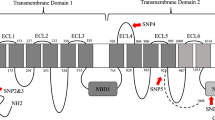
The occurrence of 16 mutations in four target genes, GluCl1 and GluCl3 (target of abamectin; Fig. S1), cytb (target of bifenazate; Fig. S2) and VGSC (target of bifenthrin; Fig. S3), was examined in T. urticae field populations by direct sequencing of PCR products. By visual examination of sequencing chromatographs at the mutation sites, we could identify samples that contained wild-type, resistant, or both alleles. The combination of mutations in field T. urticae populations collected from PNW hopyards exhibited a unique pattern (Table 4). Only two mutations, G126S and F1538I, in cytb and domain III of VGSC, respectively, were identified (Table 4). There were no mutations observed in GluCl1, GluCl3 and other region of cytb and VGSC.
No mutations observed in Glutamate-gated chloride channel genes
Inhibitory Glutamate-gated chloride channels (GluCls), members of the cys-loop ligand-gated ion channel (cysLGIC) superfamily, are extrajunctional or postsynaptic receptors found in muscle or neural ganglion of most protostome phyla including Chelicerates such as T. urticae23,33. The genome of T. urticae contains six orthologous GluCl genes23. Previous studies revealed that two mutations in two different GluCl channel subunits, GluCl1 and GluCl3, were related to abamectin resistance in T. urticae23,26. Thus we designed primers to sequence the fragments containing these two mutations (Fig. S1) from susceptible and all hop field populations of T. urticae. Surprisingly, there were no mutations identified from the samples tested (Table 4), suggesting target site insensitivity-mediated resistance is not the mechanism leading to the abamectin resistance that we observed in T. urticae field populations.
Identification of mutations in the cytb gene
Recent studies suggested that bifenazate resistance was closely correlated with mutation(s) in the mitochondrial cytb27. A combination of at least two cd1 helix mutations in the Qo pocket (G126S and I136T or G126S and S141F) and one mutation in the ef helix of Qo pocket (P262T) were linked with a high level of bifenazate resistance in T. urticae. We sequenced an 828 bp fragment of the T. urticae cytb gene, which included the G126, I136, S141, D161 and P262 sites (Fig. S2) that have been demonstrated to confer bifenazate resistance in T. urticae27. One amino acid substitution, G126S, was detected in T. urticae field populations. 35% of field samples field samples contained only the resistant allele, 20% contained both alleles (G/S) and 15% only the susceptible allele (G) (Table 4; Fig. 4A). Since the G126S mutation alone only causes low to moderate bifenazate resistance27, this result is consistent with the bifenazate resistance phenotype observed (Table 3).
Identification of mutations in the voltage-gated sodium channel gene
The voltage-gated sodium channel (VGSC) is an integral transmembrane protein that is responsible for the rapidly rising phase of action potentials on the neuronal membranes. Due to its essential role in electrical signaling, VGSC is the target of several neurotoxins, including pyrethroids and DDT34. Many amino acid substitutions associated with pyrethroid resistance in arthropods are located in transmembrane segments 4–6 of domain II (IIS4-IIS6) including M918 (super kdr), L925, T929, L932, V1010, L1014 (kdr) and L102430,34,35,36. One mutation within the intracellular inter linker connecting domains II and III (A1215D) and one mutation in domain III (F1538I) were detected in a highly bifenthrin resistant T. urticae strain from Greece29. Thus we amplified three fragments of the VGSC from the domain II, II-III inter linker and domain III regions (Fig. S3). We identified only one amino acid substitution, F1538I. It was observed in 16 out of 24 field samples tested (66.7%), 12 of which contained both alleles (F/I) and 4 of which were only contained the isoleucine substitution (I) (Table 4; Fig. 4B).
Cytochrome P450-mediated metabolic detoxification
Besides target site insensitivity, cytochrome P450-mediated detoxification had been shown to be one of the most important mechanisms in acaricide resistance of T. urticae37,38,39. The genome of T. urticae contains 86 P450 genes. We examined the relative expression of three P450s, CYP385C4, CYP389A1 and CYP392D8, belonging to the CYP3, CYP4 and CYP2 clans, respectively. We chose these three P450s because they have been shown to exhibit more than two-fold up regulation after switching host plants and their expression patterns have been linked to acaricide resistance in T. urticae9. The expressions of these three P450s in five field populations from five major locations were compared with their expressions in the susceptible population. As shown in Fig. 5, CYP385C4 had significantly higher expression in all five field populations. However, this increase in expression was not large (less than two-fold). CYP389A1 only showed significantly higher expression in the Prosser 2 population. The expression of CYP392D8 was strikingly higher in all five field populations, exhibiting levels 5 to 40-fold higher than the susceptible strain. It indicates that CYP392D8 may play an important role in acaricide resistance of T. urticae populations in hopyards.
Relative expression of CYP385C4, CYP389A1 and CYP392D8 in field T. urticae populations compared with that of the susceptible strain.
The mRNA levels were quantified by qRT-PCR and normalized with reference genes Actin and RP49. The data shown are mean + SEM (n = 3). Statistical significance of the gene expression between two samples was calculated using Student’s t test (two-tailed distribution). *p-value < 0.05, **p-value < 0.01.
Discussion
Due to a very short residual effectiveness, abamectin has become the predominant acaricide applied to control T. urticae outbreaks in August as the hops near harvest. Annually, approximately 98% of the hop acreage in Washington is treated with abamectin at least once and 80% is treated at least three times. The widespread use of abamectin on hops raises the distinct possibility of control failure as a result of resistance. From sampling the same hopyard over multiple time points in the same season, we found increasing levels of abamectin resistance, suggesting selection pressure from abamectin applications was driving increasing resistance. For instance, multiple collections in the Prosser 2 hopyard showed that the RR to abamectin increased 6-fold from the middle of July to mid-August (Table 2). The highest LC50s to abamectin were recorded in the Moxee 1 and Prosser 4 T. urticae populations (Table 2). However, Moxee 1 had only two acaricide applications during 20138. The possibility for the reported highest abamectin resistance ratio in Moxee 1 could be the entire application history of abamectin in this field that remains unknown. There may have been high abamectin selective pressure over multiple overwintering populations in this field prior to 2013. The high level of abamectin resistance in the Prosser 4 population was also unexpected because this sample was collected from the 1st year baby hopyard8. Prior to planting hops, the crop planted in Prosser 4 was Concord grapes. A recent study reported infestation of T. urticae in grape yards and high abamectin resistance in 45% T. urticae populations from these grape yards in Brazil40. Nevertheless, our previous investigation suggested that T. urticae is not a pest of grape yards in Washington State41 and thus we do not expect the fields have been extensively sprayed with abamectin. However, Bradenburg and Kennedy42 reported that wind dispersal was a key factor causing the infestations of T. urticae from corn fields to surrounding crops. Thus, the resistant T. urticae populations we detected may have been transported from adjacent crops to the 1st year hopyard through wind dispersal.
GluCls together with gamma-aminobutyric acid (GABA)-gated channels and histamine-gated chloride channels (HisCls) are known targets of the macrocyclic lactones, the avermectins (including abamectin) and ivermectins26,33,43,44. The point mutation G323D in GluCl1 was tightly linked to a moderate abamectin resistance (17.9-fold) in the AbaR strain26. Two point mutations, G323D and G326E, in GluCl1 and GluCl3, respectively, were identified in a > 2,000-fold abamectin resistant strain23. However, there was no mutation on GluCl subunits detected in any hop samples (Table 4), suggesting target site insensitivity is not likely the mechanism involved in resistance to abamectin in T. urticae field populations. Our results are comparable with a study by Khajehali et al.45 which found no GluCl mutations in 15 T. urticae strains collected from rose greenhouses in the Netherlands, although 10 of those strains displayed abamectin resistance. Many recent studies also suggested that abamectin target site mutations are not especially common in T. urticae populations worldwide. For example, the G326E was detected in only seven out of 51 T. urticae populations sampled from 27 countries and five continents46. The G323D mutation was only found in two Greek samples in the same survey46. In another study with 25 Korean T. urticae populations, only one field-collected T. urticae sample contains G323D mutation47.
Previous synergism tests and transcriptomic data indicated that additional mechanisms such as enhanced metabolic detoxification by cytochrome P450s may be implicated in the abamectin resistance phenotype37,48,49. A genome microarray analysis revealed several cytochrome P450 genes were up-regulated in an abamectin resistant strain49. Further evidence confirmed the function of one of these P450s, CYP392A16, in metabolizing abamectin50. Unfortunately, this study was published after the completion of our study and we did not have enough sample material remaining to test for expression of this gene. However, of the three P450s we did examine in our study, one Clan 2 P450, CYP392D8, showed constitutive over-expression in all five field collected samples compared to the susceptible population, indicating its potential function in abamectin resistance (Fig. 5).
Bifenazate is a hydrazine carbazate acaricide that was discovered in 1990 by Uniroyal Chemical and first registered in the state of Washington in 20028,51. Because of the quick knockdown and long residual effects on many economically important phytophagous mite species and low toxicity on predatory mites and beneficial insects, bifenazate is widely used as a selective acaricide to control T. urticae in hopyards. Our bioassay data demonstrated that the majority of field T. urticae populations (62.5%) in hopyards exhibit moderate levels of resistance to bifenazate (Fig. 3B). Our target site mutation screening revealed that a mutation G126S on cytb gene occurs in 35% of T. urticae populations (Fig. 4A). It should be noted that G126S (GGA to AGA) is the same mutation as described in previous studies27,28. G126S is the most common substitution on cytb gene of T. urticae that was identified in several bifenazate resistant populations22,27,28,45,46. Previous studies showed that mutations on the G137 site in Saccharomyces cerevisiae (equivalent to G126 in T. urticae) contributed to respiratory-deficiency through affecting stability of FeS52,53. However, the G126 mutation alone only confers low to moderate level of resistance to bifenazate28. In our results, the G126S mutation was observed in populations of Granger 4, Moxee 1 & 2 and Prosser 1, 4 & 5, which all demonstrated low to moderate level of bifenazate resistance (RR = 8.37–23.02 or mortalities at discriminating dose ranged from 90% to 100%) (Table 3), suggesting the resistance phenotypes of these samples are consistent with their genotypes. Other mutations or mutation combinations on cytb gene such as P262T, G126S with I136T/S141F that are responsible for high bifenazate resistance with RR > 277827,28 were not detected in any of our samples.
Bifenthrin, a pyrethroid, has been introduced for T. urticae control in hopyards since it was registered in 199351. Because of their safety, longevity of residual activity and low cost, pyrethroids are extensively used for pest control, with about a 20% insecticide market share23. Unfortunately, ubiquitous resistance to pyrethroids had been broadly reported in various insect populations19,20,34,36,54. In T. urticae, two mutations, F1538I in domain IIIS6 and A1215D within the intracellular inter linker connecting domains II and III were linked with high bifenthrin resistance in a Greek population29. The function of the F1538I mutation in pyrethroid resistance has been confirmed34,55,56 while the function of A1215D is still unknown. Another substitution, L1024V in domain IIS6 was reported to play an important role in the fenpropathrin resistance of T. urticae from Korea30. Pyrethroids are not used very often in the hopyards in PNW since they are linked with subsequent T. urticae outbreaks. Therefore we omitted the toxicity evaluation of field collected T. urticae samples. However, based on our record of acaricide sprays in hopyards, bifenthrin is still used in August as the hops near harvest8 (Fig. 1). Our DNA diagnostic results demonstrated that a mutation in the VGSC gene (F1538I) was observed in 66.7% T. urticae populations (Fig. 4B). Particularly, F1538I was fixed in four samples collected from Grandview and Prosser 4 & 5, three of which were collected in late August or September (Table 4). Additionally, esterase-mediated metabolic detoxification had also been proposed to confer resistance to pyrethroids in T. urticae57,58,59. This result suggests that developing of pyrethroid resistance in hopyards should be of concern.
In summary, T. urticae populations in hopyards exhibit a low to moderate level of acaricide resistance. The mechanisms of acaricide resistance in T. urticae are likely mediated by a number of different pathways: not only target site insensitivities but also enhanced metabolic detoxification. It is a common phenomenon that multiple genes or mechanisms confer resistance simultaneously to a certain pesticide18,20,60,61,62,63,64. Therefore, we plan a genome-wide investigation to identify a more complete set of candidate resistance genes from T. urticae populations of hopyards. Our data also suggests that acaricide spray history, neighboring plants and time of the season are important factors in correctly diagnosing acaricide resistance in T. urticae. Developing a baseline effective dose for commonly used acaricides and screening local T. urticae populations with resistance-associated molecular markers would be a proactive approach toward T. urticae resistance management. Our study reveals a unique phenotypic and genotypic pattern underpinning the chemical adaptation of T. urticae in hop fields which will be of assistance in developing diagnostic tools for integrated T. urticae management.
Methods
Mite samples
The susceptible acaricide naïve T. urticae strain (SS) was originally collected from weeds in Montana in 1995 and reared under laboratory conditions without exposure to any pesticides8. This population was reared on 2-week-old lima bean plants (Phaseolus lunatus L.) at 28 ± 2 °C, 70 ± 5 RH and a photoperiod of 16:8 (L:D) h in an isolated walk-in growth chamber at the Irrigated Agricultural Research and Extension Center (IAREC) in Prosser, WA. Bean plants were grown from seeds (Buckeye Seed Supply, Canton, OH) with medium grade vermiculite (Therm-o-rock West Inc.) soaking in water in the greenhouse. New, healthy, lima bean plants were provided for T. urticae and plants were replaced every seven days. To prevent mite migration, the colonies were maintained in 27-L plastic tubs filled with soapy water Huffaker moats8.
Thirty-one field T. urticae populations were collected from commercial hopyards located within the Yakima Valley of Washington State from June to September in 2013. There were five major locations: one sample was collected in Grandview, WA (46°15′13″N 119°54′36″W), eight in Granger, WA (46°20′40″N 120°11′29″W), four in Mabton, WA (46°12′42″N 119°59′47″W), two in Moxee, WA (46°33′23″N 120°23′14″W) and 16 in Prosser, WA (46°12′25″N 119°45′56″W) (Fig. 2; Tables 2, 3, 4, 2, 2, 3, 3, 3, 3, 4). The samples collected from the same location at different times were treated as different populations. Mite-infested hop leaves were stored in a plastic bag and transported to the lab in a cooling box within a few hours of collection. Spider mites were identified under a dissecting scope according to morphological characteristics13. Approximately 50–100 adults were stored in 95% ethanol for genomic DNA extraction. About 300 adult T. urticae from each of five major locations listed in Table 4 were also stored in RNAlater® (Sigma-Aldrich, Saint Louis, MO) for RNA extraction. Remaining mites were used for bioassays directly. Because three samples had a low number of mites (Table 4), we reared them on lima bean plants in an isolated walk-in growth chamber for one month to increase population size before sampling them for DNA extraction and bioassays.
Bioassays and data analysis
Leaf disc bioassays were used to estimate the LC50 (lethal concentration required to kill 50% of the individuals in a population) of abamectin and bifenazate for lab susceptible and field spider mite populations. The method followed that of Knight et al.65. Briefly, ten female adult spider mites were placed on the back of a bean leaf disc (2 cm diameter) with a fine brush. Two leaf discs were arranged on water-saturated cotton (4 cm × 4 cm) in a single petri dish (9 cm diameter, 1.5 cm height; Alkali Scientific, Pompano Beach, FL). The water-saturated cotton was pushed up against the perimeter of the leaf disc to prevent mites from walking off the disc65. Two commercially formulated acaricides for leaf disc bioassay are Epi-mek® 0.15 EC (2% a.i. Abamectin, Syngenta Crop Protection) and Acramite® 50WS (50% a.i. Bifenazate, Chemtura Agro Solutions). The recommended field concentrations for these two acaricides are 23 mg a.i./L and 899 mg a.i./L, respectively. The field rate solutions were prepared in the lab using commercial formulated acaricides and distilled water. These solutions were serially diluted in distilled water for 4–7 concentrations ranged from 0.1–67 mg a.i./L and 0.44–889 mg a.i./L for Epi-mek® and Acramite®, respectively.
The sticky tape method was used to estimate the LC50 to bifenthrin for the lab susceptible strain because pyrethroids are shown to have repellent effects on mites66. In this method, ten female adult spider mites were placed dorsal side down on a strip of double-sided sticky Scotch® tape (3cm × 1.2 cm) stuck on a glass slide (7.5 cm × 2.5 cm). The commercially formulated bifenthrin was Bifenture® EC, a pyrethroid provided by United Phosphorus (25.1% a.i. Bifenthrin). These bifenthrin solutions were serially diluted in distilled water for 4–7 concentrations ranged from 6–120 mg a.i./L.
Leaf discs or glass slides were treated topically with 2 ml of acaricide solutions with a Potter spray tower (Burkard Manufacturing, Richmansworth, Herts, UK)67. The tower was calibrated to deliver 1.1 kg/cm2 which allowed 2.0 ± 0.1 mg/cm2 spray fluid. Each bioassay consisted of 4–7 acaricide concentrations with 4–6 replicates for each concentration. The spider mites exposed to distilled water in the Potter spray tower were used as the non-treated control. The treated leaf discs or glass slides were maintained at 25 ± 2 °C and a photoperiod of 16:8 (L:D) h after the initiation of the bioassay. Mortality was evaluated after 24 h. Mortality was assessed by gently touching each individual spider mite with a fine camel hair paint brush under a dissecting stereomicroscope. The individuals with no response were counted as dead. The few moribund individuals that were not able to maintain balance and show uncoordinated twitching were also recorded as dead. The slope, intercept and LC50 (corrected against the untreated control) were evaluated with Abbott’s formula68 calculated by log-dose probit analysis (POLO Probit 2014). The statistical analysis of LC50 values was based on non-overlapping 95% CI. Resistance ratios (RRs) were calculated through dividing LC50 values of field samples by the LC50 value of the lab susceptible population.
Resistance-associated amino acid substitution screening
Genomic DNA was extracted using a DNeasy Blood & Tissue kit (QIAGEN) from 10 adult mites for each population. The DNA was stored at −20 °C till use. The genomic DNA was used as a template for PCR performed in a Peltier-Effect thermal cycler (MJ Research, Inc., Canada). Primers for PCR amplification of regions with resistance-associated point mutations are listed in Table S1. PCR was performed using Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Pittsburgh, PA) under the following cycling parameters: 95 °C for 3 min 50 s, 35 cycles of 94 °C for 35 s, 55 °C for 35 s and 72 °C for 3 min, with final extension for 10 min at 72 °C. PCR products were purified using DNA Clean & Concentrator (Zymo Research, Irvine, CA) following the manufacturer’s protocol. The purified DNA from each individual was directly sequenced using primers described above (Table S1) for PCR amplification. Each individual PCR product was sequenced using ABI BigDye Terminator Version 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) on an ABI 3730 at the Center for Reproductive Biology Molecular Biology and Genomics Core facility at Washington State University. The obtained sequences were analyzed with BioEdit 7.0.1 software (Ibis Biosciences, Carlsbad, CA). The occurrence of mutations was evaluated according to the inspection of sequencing chromatographs, as containing one or both alleles. Each sample was sequenced three times with independently prepared genomic DNAs.
RNA extraction, cDNA synthesis and qRT-PCR
Total RNA from 100 spider mites per population was extracted using TRIZOL reagent (Invitrogen) following manufacturer’s protocol. The quality of total RNA was checked by gel electrophoresis and spectrometry analyses. The total RNA was treated with DNase I (Ambion Inc., Austin, TX) to remove contaminating DNA. DNase I treated total RNA was used as a template for cDNA synthesizes by M-MLV reverse transcriptase (Promega, Madison, WI). qRT-PCR was performed using a CFX96™ Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). Each qRT-PCR reaction (10 μl final volume) contained 5 μl iQ™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), 1.0 μl of cDNA, 3.6 μl ddH2O and 0.4 μl forward and reverse gene specific primers (Table S4, stock 10 μM). An initial incubation of 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s, 55 °C for 60 s settings were used. The qRT-PCR for each sample was conducted with two technique replicates and three biological replicates. The no-template control and internal controls were included in each plate. Actin and rp49 were used as reference genes for internal controls49. Relative expression levels for target genes, in relation to two reference genes, actin and rp49 were calculated by the 2−∆∆CT method69. Both the PCR efficiency and R2 (correlation coefficient) value were taken into consideration in estimating relative quantities. PCR efficiency between 95% and 105% and R2 value >0.99 for each gene were considered as qualified for further analysis.
Additional Information
How to cite this article: Piraneo, T. G. et al. Molecular mechanisms of Tetranychus urticae chemical adaptation in hop fields. Sci. Rep. 5, 17090; doi: 10.1038/srep17090 (2015).
References
USDA National Agriculture Statistics Service (NASS), National Hop Report (2013). Available at: http://www.nass.usda.gov/Statistics_by_State/Washington/Publications/Hops/index.asp (Accessed: 12th Oct 2015).
Barth-Haas Group, The Bath Report-Hops 2013/2014. (2014) Available at: http://www.barthhaasgroup.com/en/news-and-reports/the-barth-report-hops. (Accessed: 12th Oct 2015).
Neve, R. A. In Hops. (ed. Neve, R. A. ) 1–272 (Chapman and Hall, London, 1991).
Clarke, B. J. Hop products. J. Inst. Brew. 92, 123–130 (1986).
USAHOPS.ORG, Hop growing in Washington. (2008) Available at: http://www.usahops.org/ (Accessed: 12th Oct 2015).
Gent, D. H. et al. Field Guide for Integrated Pest Management in Hops. 1st Edition. Oregon State University, University of Idaho, USDA Agricultural Research Service and Washington State University (2009).
Cranham, J. E. & Helle, W. Pesticide resistance in Tetranychidae in Spider mites, their Biology natural enemies and control (eds. Helle, W. & Sabelis, M. W. ) 405–421 (Elsevier Press, Amsterdam, 1985).
Piraneo, T. G. Acaricide resistance of the two-spotted spider mite (Tetranychus urticae) in Pacific Northwest hops. MS thesis, Washington State University (2013).
Grbic, M. et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492 (2011).
Van Leeuwen, T., Dermauw, W., Grbic, M., Tirry, L. & Feyereisen, R. Spider mite control and resistance management: does a genome help? Pest Manag. Sci. 69, 156–159 (2013).
Alberti, G. & Crooker, A. R. Internal anatomy in Spider Mites, Their Biology Natural Enemies and Control (eds. Helle, W. & Sabelis, M. W. ) 29–62 (Elsevier Press, Amsterdam, 1985).
Davis, D. W. Influence on population density on Tetranychus multisetis. J. Econ. Entomol. 45, 652–654 (1952).
Veerman, A. Diapause in Spider mites, their biology natural enemies and control (eds. Helle, W. & Sabelis, M. W. ) 279–316 (Elsevier Press, Amsterdam, 1985).
Van Leeuwen, T., Tirry, L., Yamamoto, A., Nauen, R. & Dermauw, W. The economic importance of acaricides in the control of phytophagous mites and an update on recent acaricide mode of action research. Pesticide Biochem. Physiol. 121, 12–21 (2015).
Whalon, M. E., Mota-Sanchez, R. M. & Hollingworth, R. M. Arthropods resistant to Pesticides Database (ARPD) (2015). Available at: http://www.pesticideresistance.org (Accessed: 12th Oct 2015).
Wada-Katsumata, A., Silverman, J. & Schal, C. Changes in taste neurons support the emergence of an adaptive behavior in cockroaches. Science 340, 972–975 (2013).
Koganemaru, R., Dini, M., Miller, D. M., Zach, N. & Adelman, Z. N. Robust cuticular penetration resistance in the common bed bug (Cimex lectularius L.) correlates with increased steady state transcript levels of CPR-type cuticle protein genes. Pest Biochem. Physiol. 106, 190–197 (2013).
Feyereisen, R. Insect CYP genes and P450 enzymes in Insect molecular biology and Biochemistry (ed. Gilbert, L. I. ) Ch. 8, 236–316 (Elsevier/Academic Press, Amsterdam, 2012).
Liu, N., Xu, Q., Zhu, F. & Zhang, L. Pyrethroid resistance in mosquitoes. Insect Sci. 13, 159–166 (2006).
Zhu, F. et al. Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci. Rep. 3, 1456 (2013).
Zhu, F., Cui, Y., Walsh, D. B. & Lavine, L. C. Application of RNA interference toward Insecticide resistance management in Short views on insect biochemistry and molecular Biology, Vol. 2 (eds. Chandrasekar, R., Tyagi, B. K., Gui, Z. & Reeck, G. R. ) Ch. 27, 595–619 (International Book Mission, Academic Publisher, Manhattan, 2014).
Van Leeuwen, T., Vontas, J., Tsagkarakou, A., Dermauw, W. & Tirry, L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important acari: a review. Insect Biochem. Mol. Biol. 40, 563–572 (2010).
Dermauw, W. et al. The cys-loop ligand-gated ion channel gene family of Tetranychus urticae: implications for acaricide toxicology and a novel mutation associated with abamectin resistance. Insect Biochem. Mol. Biol. 42, 455–465 (2012).
Khajehali, J. et al. Acetylcholinesterase point mutations in European strains of Tetranychus urticae (Acari: Tetranychidae) resistant to organophosphates. Pest Manag. Sci. 66, 220–228 (2010).
Kwon, D. H. et al. Acetylcholinesterase point mutations putatively associated with monocrotophos resistance in the two-spotted spider mite. Pest Biochem. Physiol. 96, 36–42 (2010).
Kwon, D. H., Yoon, K. S., Clark, J. M. & Lee, S. H. A point mutation in a Glutamate-gated chloride channel confers abamectin resistance in the two-spotted spider mite, Tetranychus urticae Koch. Insect Mol. Biol. 19, 583–591 (2010).
Van Leeuwen, T. et al. Mitochondrial heteroplasmy and the evolution of insecticide resistance: non-Mendelian inheritance in action. Proc. Natl. Acad. Sci. USA 105, 5980–5985 (2008).
Van Nieuwenhuyse, P., Van Leeuwen, T., Khajehali, J., Vanholme, B. & Tirry, L. Mutations in the mitochondrial cytochrome b of Tetranychus urticae Koch (Acari: Tetranychidae) confer cross-resistance between bifenazate and acequinocyl. Pest Manag. Sci. 65, 404–412 (2009).
Tsagkarakou, A. et al. Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol. Biol. 18, 583–593 (2009).
Kwon, D. H., Clark, J. M. & Lee, S. H. Cloning of a sodium channel gene and identification of mutations putatively associated with fenpropathrin resistance in Tetranychus urticae. Pest Biochem. Physiol. 97, 93–1000 (2010).
Van Leeuwen, T. et al. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. USA 109, 4407–4412 (2012).
Demaeght, P. et al. High resolution genetic mapping uncovers chitin synthase-1 as the target- site of the structurally diverse mite growth inhibitors clofentezine, hexythiazox and etoxazole in Tetranychus urticae. Insect Biochem. Mol. Biol. 51, 52–61 (2014).
Wolstenholem, A. J. Glutamate-gated chlorid channels. J. Biol. Chem. 287, 40232–40238 (2012).
Dong, K. et al. Molecular biology of insect sodium channels and pyrethroid resistance. Insect Biochem. Mol. Biol. 50, 1–17 (2014).
Soderlund, D. M. & Knipple, D. C. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect Biochem. Mol. Biol. 33, 563–577 (2003).
Wu, M., Gotoh, H., Waters, T., Walsh, D. B. & Lavine, L. C. Identification of an alternative knockdown resistance (kdr)‐like mutation, M918L and a novel mutation, V1010A, in the Thrips tabaci voltage‐gated sodium channel gene. Pest Manag. Sci. 70, 977–981 (2014).
Stumpf, N. & Nauen, R. Biochemical markers linked to abamectin resistance in Tetranychus urticae (Acari: Tetranychidae). Pesticide Biochem. Physiol. 72, 111–121 (2002).
Van Nieuwewnhuyse, P., Van Leeuwen, T., Khajehali, J., Vanholme, B. & Tirry, L. Mutations in the mitochondrial cytochrome b of Tetranychus urticae Koch (Acari: Tetranychidae) confer cross-resistance between bifenazate and acequinocyl. Pest Manag. Sci. 65, 404–412 (2009).
Demaeght, P. et al. Molecular analysis of resistance to acaricidal spirocyclic tetronic acids in Tetranychus urticae: CYP392E10 metabolizes spirodiclofen, but not its corresponding enol. Insect Biochem. Mol. Biol. 43, 544–554 (2013).
Monteiro, V.B., Gondim Jr., M. G. C., Oliveira, J. E. M., Siqueira, H. A. A. & Sousa, J. M. Monitoring Tetranychus urticae Koch (Acari: Tetranychidae) resistance to abamectin in vineyards in the Lower Middle São Francisco Valley. Crop Protect. 69, 90–96 (2015).
Walsh, D. B. Pests of grapes. In Pest management guide for grapes in Washington. Washington State Extension Publications, 15–22 (2013).
Bradenburg, R. L. & Kennedy, G. G. Relationship of Neozygites floridana (Entomophtrorales, Entomorphoraceae) to two-spotted spider mite (Acari: Tetranychidae) populations in field corn. J. Econ. Entomol. 75, 691–694 (1982).
Clark, J. M., Scott, J. G., Campos, F. & Bloomquist, J. R. Resistance to avermectins: extent, mechanisms and management implications. Annu. Rev. Entomol. 40, 1–30 (1995).
McCavera, S., Walsh, T. K. & Wolstenholme, A. L. Nematode ligand-gated chlorid channels: an appraisal of their involvement in macrocyclic lactone resistance and prospects for developing molecular markers. Parasitology 134, 1111–1121 (2007).
Khajehali, J., Van Nieuwenhuyse, P., Demaeght, P., Tirry, L. & Van Leeuwen, T. Acaricide resistance and resistance mechanisms in Tetranychus urticae populations from rose greenhouses in the Netherlands. Pest Manag. Sci. 67, 1424–1433 (2011).
Ilias, A., Vontas, J. & Tsagkarakou, A. Global distribution and origin of target site insecticide resistance mutations in Tetranychus urticae. Insect Biochem. Mol. Biol. 48, 17–28 (2014).
Kwon, D. H., Lee, S. W., Ahn, J. J. & Lee, S. H. Determination of acaricide resistance allele frequenceies in field populations of Tetranychus urticae using quantitative sequencing. J. Asia-Pacific Entomol. 17, 99–103 (2014).
Yorulmaz, S. & Ay, R. Multiple resistance, detoxification enzyme activity and inheritance of abamectin resistance in Tetranychus urticae Koch (Acarina: Tetranychidae). Turk. J. Agr. For. 33, 393–402 (2009).
Dermauw, W. et al. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. USA 110, E113–E122 (2013).
Riga, M. et al. Abamectin is metabolized by CYP392A16, a cytochrome P450 associated with high levels of acaricide resistance in Tetranychus urticae. Insect Biochem. Mol. Biol. 46, 43–53 (2014).
Anonymous, Pesticide Information Center Online Databases. (1993 & 2002) Available at: http://cru66.cahe.wsu.edu/LabelTolerance.html. (Accessed: 7th May 2015).
Giessler, A., Geier, B. M., Derago, J. P., Slonimski, P. P. & Vonjagow, G. Analysis of cytochrome b amino acid residues forming the contact face with the iron-sulfur subunit of ubiquinol:cytochrome c reductase in Saccharomyces cerevisiae. Eur. J. Biochem. 222, 147 154 (1994).
Brasseur, G., Saribas, A. S. & Daldal, F. A. A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc1 complex. Biochim. Biophys. Acta 1275, 61–69 (1996).
Zhu, F. et al. Widespread distribution of knockdown resistance mutations in the bed bug, Cimex lectularius (Hemiptera: Cimicidae), populations in the United States. Arch. Insect Biochem. Physiol. 73, 245–257 (2010).
He, H. Q., Chen, A. C., Davey, R. B., Ivie, G. W. & George, J. E. Identification of a point mutation in the para-type sodium channel gene from a pyrethroid-resistant cattle-tick. Biochem. Biophys. Res. Commun. 261, 558–561 (1999).
Tan, J. et al. Identification of amino acid residues in the insect sodium channel critical for pyrethroid binding. Mol. Pharmacol. 67, 513–522 (2005).
Yang, X., Buschman, L. L., Zhu, K. Y. & Margolies, D. C. Susceptibility and detoxifying enzyme activity in two spider mite species (Acari: Tetranychidae) after selection with three insecticides. J. Econ. Entomol. 95, 399–406 (2002).
Van Leeuwen, T., Van Pottelberge, S. & Tirry, L. Comparative acaricide susceptibility and detoxifying enzyme activities in field-collected resistant and susceptible strains of Tetranychus urticae. Pest Manag. Sci. 61, 499–507 (2005).
Van Leeuwen, T. & Tirry, L. Esterase-mediated bifenthrin resistance in a multiresistant strain of the two-spotted spider mite, Tetranychus urticae. Pest Manag. Sci. 63, 150–156 (2007).
David, J. et al. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genomics 15, 174 (2014).
Zhu, F., Feng, J. N., Zhang, L. & Liu, N. Characterization of two novel cytochrome P450 genes in insecticide-resistant house-flies. Insect Mol. Biol. 17, 27–37 (2008).
Zhu, F., Li, T., Zhang, L. & Liu, N. Co-up-regulation of three P450 genes in response to permethrin exposure in permethrin resistant house flies, Musca domestica. BMC Physiol. 8, 18 (2008).
Zhu, F., Moural, T. W., Shah, K. & Palli, S. R. Integrated analysis of cytochrome P450 gene superfamily in the red flour beetle, Tribolium castaneum. BMC Genomics 14, 174 (2013).
Liu, N., Zhu, F., Xu, Q., Pridgeon, Y. & Gao, X. Behavioral change, physiological modification and metabolic detoxification: mechanisms of insecticide resistance. Acta Entomologica Sinica 48, 672–680 (2006).
Knight, A. L., Beers, E. H., Hoyt, S. C. & Riedl, H. Acaricide bioassay with spider mites (Acari: Tetranychidae) on pome fruits: Evaluation of methods and selection of discriminating concentrations for resistance monitoring. J. Econ. Entomol. 83, 1752–1760 (1990).
Mochizuki, M. Variations in insecticide susceptibility of the predatory mite, Amblyseius womersleyi Schicha (Acarina: Phytoseiidae), in the tea fields of Japan. Appl. Entomol. Zool. 29, 203–209 (1994).
Potter, C. Apparatus for applying direct sprays. Ann. Appl. Biol. 39, 2–20 (1952).
Abbott, W. S. A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 18, 265–267 (1925).
Huggett, J., Dheda, K., Bustin, S. & Zumula, A. Real-time RT-PCR normallisation, strategies and considerations. Genes Immun. 6, 279–284 (2005).
Acknowledgements
We are grateful to Tora Brooks, Bianca Mendoza and Christina Nguyen (Washington State University) for their technical assistance. Special thanks go to Timothy W. Moural and Mark Lavine (Washington State University) for their help on English editing. This work was supported by a USDA/NIFA grant (D.B.W and L.C.L), Hop Research Council, WSU Proposal Development Stimulus Program and WSU Emerging Research Issues Program.
Author information
Authors and Affiliations
Contributions
D.B.W., L.C.L., T.G.P. and F.Z. designed the experiments. T.G.P. collected all field spider mite samples and performed bioassays. F.Z., J.B. and M.M. performed the molecular experiments and analyzed the data. F.Z., T.G.P. and J.B. prepared all figures and tables. F.Z., T.G.P., J.B., M.M., L.C.L. and D.B.W. contributed reagents/materials/analysis tools. F.Z. wrote the manuscript. T.G.P., D.B.W. and L.C.L. revised the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Piraneo, T., Bull, J., Morales, M. et al. Molecular mechanisms of Tetranychus urticae chemical adaptation in hop fields. Sci Rep 5, 17090 (2015). https://doi.org/10.1038/srep17090
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep17090
This article is cited by
-
Evaluation of sublethal effects of abamectin nanoformulation on Tetranychus urticae Koch (Acari: Tetranychidae): nanoliposomal versus nanoniosomal abamectin
International Journal of Tropical Insect Science (2022)
-
The G126S substitution in mitochondrially encoded cytochrome b does not confer bifenazate resistance in the spider mite Tetranychus urticae
Experimental and Applied Acarology (2021)
-
Mechanisms and management of acaricide resistance for Tetranychus urticae in agroecosystems
Journal of Pest Science (2021)
-
Performance of Tetranychus urticae (Acari: Tetranychidae) on three hop cultivars (Humulus lupulus)
Experimental and Applied Acarology (2021)
-
Economic injury level for Tetranychus urticae Koch on parthenocarpic cucumber under protected environment in north-western Indian Himalayas
Phytoparasitica (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.