Abstract
Circulating tumor cells (CTCs) in the blood of cancer patients are recognized as important potential targets for future anticancer therapies. As mediators of metastatic spread, CTCs are also promising to be used as ‘liquid biopsy’ to aid clinical decision-making. Recent work has revealed potentially important genotypic and phenotypic heterogeneity within CTC populations, even within the same patient. MicroRNAs (miRNAs) are key regulators of gene expression and have emerged as potentially important diagnostic markers and targets for anti-cancer therapy. Here, we describe a robust in situ hybridization (ISH) protocol, incorporating the CellSearch® CTC detection system, enabling clinical investigation of important miRNAs, such as miR-10b on a cell by cell basis. We also use this method to demonstrate heterogeneity of such as miR-10b on a cell-by-cell basis. We also use this method to demonstrate heterogeneity of miR-10b in individual CTCs from breast, prostate and colorectal cancer patients.
Similar content being viewed by others
Introduction
Circulating tumor cells (CTCs) have been demonstrated to be an independent prognostic marker of clinical outcome in patients with solid tumors1,2,3. Numerous assays for the enrichment and detection of CTCs have been developed and used, however the semi-automated CellSearch® system (Veridex, Raritan, NJ, USA) remains the only device cleared by the Food and Drug Administration (FDA) for clinical use in metastatic breast, prostate and colon cancer4,5. To date, little is known about phenotypic characteristics of clinically significant CTCs capable of establishing metastasis at secondary sites, or the functional significance of CTC heterogeneity6,7. As a link between the primary tumor and the formation of metastasis throughout the body, CTCs are not only a promising target for future anti-metastasis therapeutic strategies, but can also be analyzed via a ‘liquid biopsy’ that can be obtained by a simple blood draw4,8. Furthermore, CTC analysis enables continuous monitoring of patient condition during the clinical course of their disease allowing early detection of recurrence and monitoring response to treatment5,8. However, the number of CTCs can be very small; as few as one CTC in 109 peripheral blood mononuclear cells (PBMNCs). This rarity makes the enrichment, detection and analysis of CTCs challenging4,5,8. Molecular analysis of cell fractions enriched for CTCs [e.g. after epithelial cell adhesion molecule (EpCAM) enrichment] often face the problem of an over-representation of leukocytes, making data obtained by bulk CTC isolation protocols difficult to interpret8,9. Moreover, small but clinically significant subpopulations are often missed during routine analysis of primary tumor biopsies4,6,10,11 and only a few CTCs are capable of extravasating into surrounding tissue and growing as a metastasis at a secondary site6,7. These cells give important clues as to the genotypic and phenotypic changes that enable metastasis and ultimately patient death. Thus, collective analysis of individual CTCs is the most promising way of overcoming these critical issues and improving the utility of CTC analysis.
MiRNAs function as post-transcriptional regulators of gene expression and mediate a range of pathways linked to disease and development12,13. Several miRNAs, such as miR-10b have been directly linked to the epithelial-mesenchymal transition (EMT)12,13,14 and metastasis in breast and prostate cancer15,16,17,18. Furthermore, increased expression of miR-10b has been demonstrated in glioma, as well as metastatic pancreatic cancers19,20. Notably, increased miR-10b blood serum levels have been described in gastric cancer, non-small lung cancer and metastatic breast cancer18,21,22. However, as there is no association between miR-10b levels in the primary tumor and metastasis the link between miR-10b and metastasis remains controversial. It has been suggested that miR-10b may have a biological effect on a few cells at the leading edge of the tumor, or a small number of cancer cells shed into the blood23.
For analysis of cell-based miRNA expression ISH protocols using locked-nucleic acid (LNA) probes have been developed and applied to cell preparations, fresh frozen tissue and paraffin embedded tissues24,25,26. To enable the detection of miR-10b in distinct cell types present in a complex cellular environment such as CTCs in peripheral blood we have adapted current ISH protocols, combined with antibody based immunofluorescence (IF), to be used with the CellSearch® CTC detection system (Fig. 1). Using this protocol we demonstrate miR-10b heterogeneity in CTCs isolated from the blood of metastatic cancer patients for the first time.
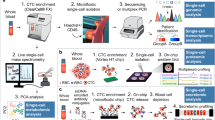
Schematic representation of the combined ISH/IF procedure.
The three day stepwise protocol for analysis of CTCs detected with the CellSearch® system. CTC enrichment and staining initially occurs using the CellSearch®. CTCs are then enumerated using the CellTracks® Analyzer and fixed in the CellSearch® cartridge. The ISH/IHC protocol using LNA probes proceeds in cartridge, followed by manual visualization of miR expression in CTCs on a cell-by-cell basis. DIG labelled probes detected with FITC labelled anti-DIG antibody allowed visualisation and localization of miR expression in cells, which are also re-labelled with cell specific, CTC identifying antibodies from the Circulating Epithelial Cell Kit (Veridex). CTCs can then be labelled as either miR+, CK+, DAPI+ and CD45−; or miR−, CK+, DAPI+ and CD45−, in a semi-quantitative way.
Results
MiR-10b expression in cancer cell lines
To identify suitable cells for method development and evaluation, commercially available breast cancer (MCF-7 and MDA-MB-231) and colorectal cancer (HCT-116 and SW480) cells were analyzed for miR-10b expression by ISH (Fig. 2). MiR-10b expression was detected in all cell lines. Notably, a slightly increased expression of miR-10b could be detected in the highly metastatic MDA-MB-231 cells and SW480 cells compared to less metastatic MCF-7 and HCT-116 cells, respectively.
MiR-10b expression analysis in breast and colorectal cancer cell lines by ISH.
Higher expressions levels of miR-10b in MDA-MB-231 cells compared with MCF-7 breast cancer cells and in SW480 compared with HCT-116 colorectal cancer cells. Cells were stained with miR-10b as well as scrambled=negative and U6-positive control probes. Nuclei were visualized with DAPI (blue). Resolution 40×. Scale bars, 20 μM.
Spike experiment using MCF-7 and MDA-231 cells
To adapt the ISH procedure for analyzing CTCs detected in the blood of cancer patients with the CellSearch® system, spiking experiments using blood of a healthy volunteer donor were performed. MDA-MB-231 and MCF-7 cells were spiked into 7.5 ml of blood from the healthy donor (for miR-10b probe). Additionally, two blood samples were spiked with MCF-7 cells to be used as positive (U6 probe) and negative control (scrambled probe). All samples were processed with the CellSearch® system, screened with the CellTracks® Analyzer to identify CK+ DAPI+ CD45− tumor cells and fixed in the CellSearch® cartridge. The combined ISH/IF protocol was applied using either miR-10b, U6-positive or scrambled-negative probes with IF staining using antibodies from the CellSearch® Epithelial Cell Kit (Veridex). CTC image analysis was performed directly in the CellSearch® cartridge using IF microscopy. MCF-7 cells and MDA-MB-231 cells could be detected and analyzed for miR-10b expression. As observed in previous cell line experiments MDA-MB-231 cells revealed a high miR-10b expression in comparison to the moderate miR-10b expression in MCF-7 cells (Fig. 3). No signal was detected with the scrambled-negative control probe and strong positive signal was observed with the U6-positive probe. Cell loss during re-fixation and ISH procedure was found to be minimal (~1%) for freshly processed samples (within 48 h after fixation) determined by re-enumeration of spiked MCF-7 cells after ISH analysis (134 of 135 cells detected with the CellSearch®).
MiR-10b ISH analysis of breast cancer cell lines captured by the CellSearch® system.
MiR-10b+ CK+ DAPI+ MCF-7 and MDA-MB-231 cells (white arrows) spiked into blood, captured by the CellSearch® system and identified by image analysis. Also shown are miR-10b− CK− DAPI+ (red arrows) and miR-10b+ CK− DAPI+ (yellow arrows) cells detected by IF microscopy. Also shown are the results U6-positive and scrambled-negative control probe analysis, respectively. Nuclei were visualized with DAPI. Resolution 40×. Scale bars, 20 μM.
MiR-10b heterogeneity in CTCs from cancer patients
To investigate miR-10b expression in CTCs in the blood of cancer patients, cells collected from 11 metastatic cancer patients (eight breast cancer, two prostate cancer and one colorectal cancer patient), were analyzed using the combined ISH/IF in cartridge protocol. MiR-10b positive (miR-10b+ ) as well as miR-10b negative (miR-10b−) CTCs could be detected in 10 of 11 patients (Figs 4 and 5a). One breast cancer patient (Breast 2) presented with 19 miR-10b+ CTCs only. The abundance of miR-10b+ CTCs in the analyzed CTC population of these 11 patients was 67% in the colorectal cancer patient, 40% and 20% in the prostate cancer and ranged from 20-94% (mean: 67%) in the breast cancer patients (Fig. 5a).
MiR-10b expression analysis of CTCs from metastatic cancer patients by ISH.
Shown, are miR-10b+ CK+ DAPI+ CD45− CTCs and miR-10b− CK+ DAPI+ CD45− CTCs (white arrows) captured from the blood of metastatic breast, prostate and colorectal cancer patients. Nuclei were visualized with DAPI. Resolution 40×. Scale bars, 20μM.
Heterogeneity of miR-10b expression in CTCs from metastatic cancer patients.
(a) Distribution of miR-10b+ and miR-10b− CTCs in the analyzed CTC populations from eight breast, one colorectal and two prostate cancer patients. (b) Immunoscoring based on miR-10b signal intensity for semi-quantitative analysis of miR-10b expression. Shown are four CTCs from one prostate cancer patient: one miR-10b− (scoring: 0) and one with weak (+1), moderate (+2) and strong (+3) miR-10b expression. Nuclei were visualized with DAPI. Resolution 20×, Scale bars, 10 μM.
For semi-quantitative analysis of miR-10b expression in individual CTCs we established a miR-10b immunoscoring (Fig. 5b). Except for one breast cancer patient (Breast 2) with only miR10b+ CTCs, expression levels among CTC populations varied from ‘negative (0)’ to ‘strong (+3)’ in all patients (Supplementary Table S1).
Cell losses during the fixation and ISH procedure were exemplarily determined by re-enumeration of CTCs from five patients that initially presented with a total number of less than 100 CTCs. In these samples an average cell loss (±SEM) of 24.9 ± 7.6% (range: 0–55.8%), varying with length of storage prior ISH analysis and sample quality, was observed.
The application of our combined ISH/IF protocol for analyzing CTCs proved to be a robust method which revealed a heterogeneous distribution of miR-10b expression among CTCs from the same patient, across three different epithelial tumor types.
Discussion
In this study, we describe a novel, robust combined ISH and IF protocol that enables expression analysis of microRNAs in definite cell types, such as CTCs. Using tumor cell line spiking experiments we adapted the protocol to be used on CTCs after being detected with the CellSearch® CTC detection system. As a proof-of-principle the expression of miR-10b was investigated in 511 CTCs isolated from metastatic cancer patient with different epithelial tumor types (breast, colorectal and prostate cancer) revealing a heterogeneous miR-10b expression pattern in the CTC populations.
For method development we performed ISH on MCF-7 and MDA-MB-231 breast and HCT-116 and SW480 colon cell line cell preparations, detecting moderate but clear miR-10b expression in MCF-7 and HCT-116 and a high expression in MDA-MB-231 and SW480 cells. This expression pattern is consistent with previous miRNA expression studies in cancer cell lines using qPCR and array-based technologies27.
Though various ISH protocols for the analysis of messenger RNA (mRNA) have been described previously28,29,30 the analysis of miRNA expression is less common and requires specified adaptions24,25,26. In particular, the use of locked-nucleic-acid incorporated DNA probes enhances the hybridization efficiency and thus improves the detection of small miRNA molecules31. Several recent methods, have adapted traditional ISH for the analysis of miRNAs24,25,26. These protocols generally use LNA probes in combination with single spectrum (colorimetric or fluorescence) detection technologies, applied to formalin fixed paraffin embedded (FFPE) or fresh frozen tissues25,26 or preparations of cultured cells28. By combining ISH with IF our technique enables the analysis of specific cell subpopulations in heterogeneous tissue or cell preparation (e.g. CTCs surrounded by normal blood cells). Unlike other ISH protocols24,25,29, our protocol includes a short hybridization step only, enabling the procedure to be performed in one day. Moreover, the inherent stability across the steps allows periods of convenient storage and shipping between different sites of analysis.
Based on the heterogeneity of CTCs described previously4,11 and their prominent role in the metastasis, molecular characterisation of individual CTCs is an emerging research field. Variations in the expression of specific proteins, as well as a heterogeneous distribution of gene amplifications/mutations have been observed in several studies. For example, human epidermal growth factor receptor 2 (HER2) and epidermal growth factor receptor (EGFR) expression patterns were analyzed in CTCs detected with the CellSearch® system in metastatic breast and colon cancer patients respectively4,32. Genotypic heterogeneity regarding gene mutations (e.g. in p53, KRAS, PIK3CA) and gene amplifications (e.g. EGFR, HER2, leucine-rich g-protein receptor 5, Aurora A kinase and androgen receptor) could be observed in individual CTCs from colon4,11, breast6 and prostate cancer patients33. Recently, Raemskoeld et al. established a protocol enabling full-length mRNA sequencing of single cells and performed mRNA expression profiling in single CTCs from a melanoma patient34. Our protocol is the first to enable the down-stream analysis of individual CTCs for microRNA expression. The only other study addressing microRNA analysis in CTCs of Sieuwerts et al. was performed on bulk CTCs after being processed using the CellSearch® Profile Kit (Veridex) and procedure9. Unlike the CellSearch® Epithelial Cell Kit this procedure includes the EpCAM enrichment only, resulting in an EpCAM-enriched cell fraction containing all CTCs and numerous remaining leukocytes. Though demonstrating the feasibility of CTC-associated microRNA profiling in a leukocyte-rich background, information regarding the distribution of miRNA expression in individual CTCs and also about the localization of the miRNA expression are not provided.
Using our technique we detected heterogeneous miR-10b expression in CTCs from patients with metastatic breast, prostate and colon cancer, respectively. MiR-10b expression has been described by Ma et al. and others to be linked to EMT and metastasis in breast cancer15,16,18. First studies also emphasize a role of miR-10b in colorectal and prostate cancer malignancy17,23,35,36. However, work from Gee et al. could not confirm the correlation of miR-10b expression in human breast tumors with metastasis and progression23, suggesting that only small highly malignant subpopulations of the primary tumor to be miR-10b positive. In this context, the detection of miR-10b+ CTCs, as putative founders of metastatic outgrowth, is an important finding that emphasizes the importance of miR-10b in cancer malignancy and provides avenues for further research in this field.
Conclusion
Our protocol incorporates ISH with fluorescent antibody detection that allows miRNA analysis of well-defined cell populations, such as CTCs in blood of cancer patients. This has the potential to be used for various other applications such as miRNA-profiling of endothelial or immune response cells in tissue sections. The protocol was adapted for microRNA analysis of individual CTCs detected from the blood of cancer patients with the CellSearch® CTC detection system. Detecting CTCs with varying miR-10b expression and the crucial role of miR-10b in metastasis and disease progression points out the potential of our ISH protocol to be useful for identifying and studying clinically relevant CTC subpopulations.
Material and Methods
Cell lines
The human mammary carcinoma cell lines MCF-7 and MDA-MB-231 as well as colorectal cancer cells HCT-116 and SW480 were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). MCF-7 and MDA-MB-231 cells were maintained in DMEM, HCT-116 and SW480 cells in RPMI medium (5% CO2, 37 °C). Cell medium was supplemented with 4.5 g/L glucose, 10% fetal bovine serum (FBS), 2 mM L-glutamine (Lonza, Basel, Switzerland), as well as 10 U/mL penicillin and 10 μg/mL streptomycin (Life Technologies, Carlsbad, CA, USA). For ISH analysis cells were either grown on chamber slides (Thermo Fisher Scientific, Waltham, MA, USA) or spiked into 7.5 mL of blood from a healthy donor that was subjected to the CellSearch® system (Veridex, Raritan, NJ, USA).
Patient samples
Blood from patients diagnosed with either metastatic breast, prostate cancer or colorectal cancer was collected at the Princess Alexandra Hospital, QLD, Australia, Royal Perth Hospital, WA, Australia, St John of God Subiaco Hospital, WA, Australia, or the University Hospital Heidelberg, Germany under protocols reviewed and approved by the respective Human Research Ethics Committees. For spiking experiments blood from a healthy volunteer was used. Written informed consent was obtained from all patients and the healthy volunteer. Patients were chosen based on their clinical diagnosis of late stage metastatic disease.
For CellSearch® analysis at least 7.5 mL of blood were drawn into a CellSave® tube (Veridex) and stored at ambient temperature until processing (within 96 h after blood sampling).
Locked nucleic acid (LNA) probes
ISH was performed using 5′ and double digoxigenin (DIG) conjugated miCURY LNA detection probes from Exiqon (Vedbæk, Denmark): (i) a U6-small nuclear RNA (positive) probe: CACGAATTTGCGTGTCATCCTT; melting temperature (Tm): 84 °C; (ii) a scrambled (negative) probe: GTGTAACACGTCTATACGCCCA; Tm:87 °C; and (iii) a specific anti-miR-10b probe: CACAAATTCGGTTCTACAGGGTA; Tm:83 °C. For hybridization the following final concentrations of probe were used: U6-positive (10 nM), scrambled-negative (40 nM) and miR-10b (40 nM).
ISH analysis of cancer cell lines
ISH analysis of cancer cell line cells was conducted to validate miR-10b expression levels. Cells grown on chamber slides were prepared for analysis by washing: once with 0.1% Triton X-100, PBS (10 min) and twice with PBS (10 min). They were then fixed (4% PFA, 5 min) and washed twice (PBS, 10 min) before blocking in hybridization buffer (50% formamide, 5 × SSC, 1 × Denhardt’s solution, 0.5 mg/mL yeast tRNA, 0.5 mg/mL salmon sperm DNA) at 50 °C (10 min). Following blocking the buffer was replaced with hybridization buffer containing DIG labeled LNA probe and the slides left to incubate (50 °C, 45 min). The slides were then subjected to three stringency wash steps: (i) twice with 0.1 × SSC (52 °C, 10 min); (ii) once with 2 × SSC (5 min); and (iii) three times with TN Buffer (0.1 M Tris-HCl pH7.5; 0.15 M NaCl) (3 min). Prior to antibody staining, the slides were incubated in antibody blocking buffer (1% blocking reagent, Roche Diagnostics, 10% FCS, 0.15 M NaCl, 0.1 M Tris-HCL pH 7.5) for 30 min.
For probe detection cells were incubated in 1:400 diluted (in blocking buffer) anti-DIG-fluorescein (FITC)-Fab fragments (Roche Diagnostics, Basel, Switzerland) for 3 h. The slides were then washed: once with TNT buffer (1% Triton X-100 in TN-buffer, 5 min) and three times with TN buffer (3 min each). Cell nuclei were visualized using DAPI staining solution (1 ng/μL DAPI, PBS, 5 min). Cells were then washed: (i) once with 0.1% Triton X-100/PBS (5 min); (ii) twice with PBS (10 min); and (iii) twice PBS (20 min). The slides were mounted in Prolong® Gold Antifade (Life Technologies, Carlsbad, California, USA). In this way slides can be stored at 4 °C for 2–4 days prior to microscopic analysis. The entire procedure was implemented in an RNAse-free working environment, with RNAse-free reagents. Unless otherwise stated steps were conducted at room temperature.
CTC detection using the CellSearch® system
CTC enrichment and detection with the semi-automated CellSearch® system using the Circulating Epithelial Kit (Veridex) was performed as described previously1,4,37. Blood (7.5 mL) from cancer patients, or blood from the healthy volunteer spiked with cancer cell line cells (MCF-7 and MDA-MB-231) was mixed with dilution buffer (6.5 mL) and centrifuged at 800 g (10 min) prior to loading the sample into the CellSearch® Autoprep system (Veridex). CTCs were immunomagnetically enriched using a ferrofluid-conjugated anti-EpCAM antibody (Veridex); and stained with 4′,6-Diamidin-2-phenylindol (DAPI), phycoerythrin (PE) conjugated anti-cytokeratin (CK) antibody and a Allophycocyanin (APC) conjugated anti-CD45 antibody as included in the Circulating Epithelial Cell Kit.
At the end of the run the enriched cell fraction (final sample volume approx. 350 μL) was collected in the CellSearch® cartridge placed in a MagNest (Veridex) to direct EpCAM+ cells into a focal plane for microscopic analysis by the CellTracks® Analyzer (Veridex). CTCs were identified as CK+, DAPI+ and CD45− and by morphology (i.e. round to oval shape, high nucleus-to-cytoplasm ratio). The cartridge was not removed from the MagNest and the assembly was stored at 4 °C until fixation (within 24 h).
Cell fixation in the CellSearch® cartridge
To prepare cells for ISH analysis they were fixed directly in the CellSearch® cartridge38. The buffer in the cartridge was removed and replaced with 250 μL methanol:glacial acetic acid (3:1) solution. This was removed after 2 min and replaced with another 250 μL methanol:glacial acetic acid (3:1) solution. After another 2 min all liquid was carefully removed from the cartridge before it was dried using compressed air (10 min). The cartridge was removed from the MagNest and stored at 4 °C for up to three days or at −20 °C for long-term storage prior to ISH analysis. All pipetting steps were performed with Mμltiflex pipette tips (PEQLAB, Erlangen, Germany).
Combined ISH and IF analysis in CellSearch® cartridge
Combined ISH and IF analysis was conducted directly in CellSearch® cartridges previously subjected to fixation (Fig. 1). Cartridges were filled with 0.1% Triton X-100, PBS. After 10 min it was replaced with PBS and left for another 10 min. Hybridization buffer (350 μL) for blocking was added and the cartridge left to incubate (30 min, 50 °C). Hybridization buffer containing LNA probes was then added and the cartridge left to incubate for a further 45 min (50 °C). After hybridization cartridges were subjected to three stringency washes: (i) twice with 0.1× SSC (10 min, 52 °C); (ii) once with 2× SSC (5 min); and (iii) three times with TN Buffer (3 min each).
For LNA probe detection and re-staining of antigens (CK and CD45), cells were blocked with antibody blocking buffer (30 min) and incubated (3 h) with 1:400 dilution of anti-DIG Fab fragments (Roche Diagnostics), as well as 1:6 dilution of CellSearch® antibody mixture (Cell Search Epithelial Cell Kit, Veridex,). The cartridge was then washed: (i) once with TNT buffer (5 min); and (ii) three times with TN buffer (5 min each). Cell nuclei were visualised with DAPI (1 ng/μL in PBS, 5 min) and cells washed again: (i) once in 0.1% Triton X-100, PBS (10 min); (ii) four times with PBS (10 min). Cartridges were filled with PBS and stored at 4 °C for up to two days prior microscopic analysis. All pipetting steps were performed with Mμltiflex pipette tips (PEQLAB). Unless otherwise stated steps were conducted at room temperature.
Microscopic analysis
Imaging of cells grown on slides and of cells in the CellSearch® cartridges was performed on either a Zeiss LSM5 Live (Carl Zeiss, Jena, Germany) or an Olympus IX71 inverted fluorescence microscope (Olympus, Tokio, Japan). Image analysis was therefore conducted using the AxioVison Rel. 4.8 Imaging software (Carl Zeiss) or OlyVia ver. 2.8. Imaging software (Olympus), respectively.
Additional Information
How to cite this article: Gasch, C. et al. Heterogeneity of miR-10b expression in circulating tumor cells. Sci. Rep. 5, 15980; doi: 10.1038/srep15980 (2015).
References
Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–904 (2004).
Wallwiener et al. Serial enumeration of circulating tumor cells predicts treatment response and prognosis in metastatic breast cancer: a prospective study in 393 patients. BMC Cancer 14, 512 (2014).
Scher et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 10, 233–9 (2009).
Gasch, C. et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin. Chem. 59, 252–60 (2013).
Gorges, T. M. & Pantel, K. Circulating tumor cells as therapy-related biomarkers in cancer patients. Cancer Immunol. Immunother. 62, 931–9 (2013).
Fernandez, S. V. et al. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 16, 445 (2014).
Bacelli et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–44 (2013)
Alix-Panabieres, C. & Pantel K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 14, 623–31 (2014).
Sieuwerts, A. M. et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin. Cancer Res. 17, 3600–18 (2011).
Yang, Y., Swennenhuis, J. F., Rho, H. S., Le Gac, S. & Terstappen, L. W. Parallel single cancer cell whole genome amplification using button-valve assisted mixing in nanoliter chambers. PLoS One 9, e107958 (2014).
Heitzer, E. et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 73, 2965–75 (2013).
Farazi, T. A., Spitzer, J. I., Morozov, P. & Tuschl, T. MiRNAs in human cancer. J. Pathol. 223, 102–15 (2011).
Slack, F. J. & Weidhaas, J. B. MicroRNA in cancer prognosis. New Engl. J. Med. 359, 2720–2 (2008).
Trang, P., Weidhaas, J. B. & Slack, F. J. MicroRNAs as potential cancer therapeutics. Oncogene 27, S52–7 (2008).
Ma, L., Teruya-Feldstein, J. & Weinberg, R. A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449, 682–8 (2007).
Ma, L. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotech. 28, 341–347 (2010).
Prueitt, R. L. et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate 68, 1152–64 (2008).
Roth, C., Rack, B., Müller, V., Janni, W., Pantel, K. & Schwarzenbach, H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 12, R90 (2010).
Sasayama, T., Nishihara, M., Kondoh, T., Hosoda, K. & Kohmura, E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int. J. Cancer 125, 1407–13 (2009).
Nakata, K., et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness and correlates with a poor prognosis. Surgery 150, 916–22 (2011).
Roth, C., Kasimir-Bauer, S., Pantel, K. & Schwarzenbach, H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol. Oncol. 5, 281–91 (2011).
Nishida, N., et al. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann. Surg. Oncol. 19, 3065–71 (2012).
Gee, H. E., et al. MicroRNA-10b and breast cancer metastasis. Nature 455, E8–9 (2008).
Pena, J. T., et al. MiRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods 6, 139–41 (2009).
Thompson, R. C., Deo, M. & Turner, D. L. Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Methods 43, 153–61 (2007).
Jorgensen, S., Baker, A., Moller, S. & Nielsen, B. S. Robust one-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods 52, 375–81 (2012).
Park, S. M., Gaur, A. B., Lengyel, E. & Peter, M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 (2008).
Zimmerman, S. G., Peters, N. C., Altaras, A. E. & Berg, C. A. Optimized RNA ISH, RNA FISH and protein-RNA double labeling (IF/FISH) in Drosophila ovaries. Nat. Protoc. 8, 2158–79 (2013).
Speel, E. J., Herbergs, J., Ramaekers, F. C. & Hopman, A. H. Combined immunocytochemistry and fluorescence in situ hybridization for simultaneous tricolor detection of cell cycle, genomic and phenotypic parameters of tumor cells. J. Histochem. Cytochem. 42, 961–6 (1994).
Van Raamsdonk, C. D. & Tilghman, S. M. Optimizing the detection of nascent transcripts by RNA fluorescence in situ hybridization. Nucleic Acids Res. 29, E42–2 (2001).
Kubota, K., Ohashi, A., Imachi, H. & Harada, H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl. Environ. Microbiol. 72, 5311–7 (2006).
Riethdorf, S. et al. Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant GeparQuattro trial. Clin. Cancer Res. 16, 2634–45 (2010).
Attard, G. et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 69, 2912–8 (2009).
Ramskold, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–82 (2012).
Hur, K. et al. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J. Natl. Cancer Inst. 107 (2015).
Fendler, A. et al. MiRNAs can predict prostate cancer biochemical relapse and are involved in tumor progression. Int. J. Oncol. 39, 1183–92 (2011).
Riethdorf, S. et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin. Cancer Res. 13, 920–8 (2007).
Swennenhuis, J. F., Tibbe, A. G., Levink, R., Sipkema, R. C. & Terstappen, L. W. Characterization of circulating tumor cells by fluorescence in situ hybridization. Cytometry 75, 520–7 (2009).
Acknowledgements
The authors would like to thank Dr Kelly Rogers at the Walter and Elisa Hall institute (WEHI), Melbourne and Dr Sandrine Roy at the Translational Research Institute (TRI), Brisbane, for assistance with imaging and microscopy. We would also like to acknowledge the contribution of Dr Lawrence F Lau, Department of Surgery, Austin Health, University of Melbourne and Dr Handoo Rhee, Department of Urology, Princess Alexandra Hospital, Queensland Health & APCRCQ, for sample collection and assistance in diagnosis; as well as Ms Helen Barry, School of Medicine, Deakin University, Geelong, for Technical Assistance. Funding: This work was supported by the German Research Foundation—Deutsche Forschungsgemeinschaft (DFG; GA2027/1-1) (C.G.), as well as, National Health & Medical Research Council (NHMRC) grants (A.S.M) and the ERC Advanced Investigator Grant “DISSECT” (K.P.). E.W.T was supported in part by the EMPathy National Collaborative Research Program funded by the National Breast Cancer Foundation, Australia and NHMRC Project Grant 1027527. The funders had no role in the design, data collection, analysis, decision to publish, or manuscript preparation.
Author information
Authors and Affiliations
Contributions
C.G., P.N.P. and L.J. performed experimental design, optimization and implementation. C.G., P.N.P. and D.E.W. performed microscopy and data analysis. L.J. and S.R. processed blood samples on the CellSearch system and performed in cartridge fixation. L.M.M., C.M.S., A.S. and M.W. provided the patient samples. C.G., P.N.P. and A.S.M. wrote the manuscript. C.N., K.J.S., E.W.T., K.P. and A.S.M. designed, monitored and directed the work. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Gasch, C., Plummer, P., Jovanovic, L. et al. Heterogeneity of miR-10b expression in circulating tumor cells. Sci Rep 5, 15980 (2015). https://doi.org/10.1038/srep15980
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep15980
This article is cited by
-
The double agents in liquid biopsy: promoter and informant biomarkers of early metastases in breast cancer
Molecular Cancer (2022)
-
A cancer stem cell-like phenotype is associated with miR-10b expression in aggressive squamous cell carcinomas
Cell Communication and Signaling (2020)
-
Tracking cancer progression: from circulating tumor cells to metastasis
Genome Medicine (2020)
-
Liquid biopsy and minimal residual disease — latest advances and implications for cure
Nature Reviews Clinical Oncology (2019)
-
Liquid biopsy - emergence of a new era in personalized cancer care
Applied Cancer Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.