Abstract
Species substitution is a form of seafood fraud for the purpose of economic gain. DNA barcoding utilizes species-specific DNA sequence information for specimen identification. Previous work has established the usability of short DNA sequences—mini-barcodes—for identification of specimens harboring degraded DNA. This study aims at establishing a DNA mini-barcoding system for all fish species commonly used in processed fish products in North America. Six mini-barcode primer pairs targeting short (127–314 bp) fragments of the cytochrome c oxidase I (CO1) DNA barcode region were developed by examining over 8,000 DNA barcodes from species in the U.S. Food and Drug Administration (FDA) Seafood List. The mini-barcode primer pairs were then tested against 44 processed fish products representing a range of species and product types. Of the 44 products, 41 (93.2%) could be identified at the species or genus level. The greatest mini-barcoding success rate found with an individual primer pair was 88.6% compared to 20.5% success rate achieved by the full-length DNA barcode primers. Overall, this study presents a mini-barcoding system that can be used to identify a wide range of fish species in commercial products and may be utilized in high throughput DNA sequencing for authentication of heavily processed fish products.
Similar content being viewed by others
Introduction
Food fraud from species substitution is an emerging risk given the increasingly global food supply chain and potential food safety issues. Economic food fraud is committed when food is deliberately placed on the market, for financial gain, with the intention of deceiving the consumer1. As a result of increased demand and the globalization of the seafood supply, more fish species are being encountered in the market2. In fact, the Seafood List from the U.S. Food and Drug Administration (FDA) contains more than 1,700 acceptable market names that can be used to label seafood in interstate commerce in the U.S.3. Subsequently, the need for accurately labelled food products and full disclosure of product composition has become more critical4,5. One difficulty in this is the authentication process of different seafood products through examination of the physical appearance of specimens. In their whole or unprocessed form, these species can generally be identified based on morphological indicators; however, over half of the fresh/frozen finfish imported into North America is processed from its original form into products such as fillets and steaks, blocks and fish sticks3. Moreover, species identification by morphological indicators requires a certain level of expertise to distinguish between closely related species. Unfortunately, most consumers are unable to detect cases of mislabelling or fraud given that recognizable external morphological features are typically removed when the fish is processed4.
To audit and prevent species fraud on the commercial market, a number of molecular methods have been developed, including use of a unique protein or DNA profiles found in different species6. DNA barcoding provides a rapid, cost-effective method for accurate identification at the species-level through comparative analysis of sequence variation in a short, standardized fragment of the genome7. The designated DNA barcode for animal species identification is a ~650-bp fragment of the mitochondrial gene coding for cytochrome c oxidase 1 (COI)5,8. A number of studies have shown the applicability of DNA barcoding for accurate identification of a wide range of fish species9,10. Recently, DNA barcoding has been employed as a species identification tool for food authentication and safety concerns, including incorrect product labelling11,12, ingredient substitutions2 or food contamination6,13, as well as for regulatory use14. DNA-based methods can also be used to monitor illegal trading involving protected or endangered species5,15,16 or to identify the species origin of commercially processed food13,17,18. However, some of the processing and preservation methods used with seafood products are not conducive to DNA barcoding with the full-length target gene region19,20,21. DNA degradation has been recognized as a considerable limitation in DNA-based analyses of these samples and PCR amplification of full-length (i.e., ~ 650 bp) barcodes from moderately or highly processed samples is significantly challenging. In addition, processed seafood products often contain multiple additives, preservatives and flavors that may affect the quantity and quality of DNA extracted from these products21,22,23. Alternatively, a mini-barcoding approach, which focuses the analysis on shorter DNA fragments (e.g., 100–200 bp) within the full-length barcode, has been shown to be effective in obtaining DNA sequence information from specimens containing degraded DNA24,25. The sequencing information generated from a small (≥100 bp) mini-barcode fragment of COI within the full-length DNA barcode region can provide the information required for identification of individual species with more than 90% species resolution21,24,26. However, extensive mini-barcode primer development and testing specifically for use with commercially processed fish species has not been carried out.
Here, we designed and optimized multiple primer sets to amplify mini-barcodes within the COI barcode region. These mini-barcode primer sets were then used to identify species in a variety of commercially processed fish products obtained in the United States.
Materials and Methods
Sample collection
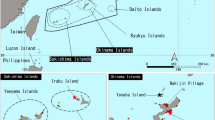
A total of 96 authenticated fish muscle tissue samples were obtained, representing 88 different species. The fish tissue samples were supplied by the FDA-Center for Food Safety and Applied Nutrition (Supplementary Materials-Table S1). These samples are from the FDA’s Reference Standard Sequence Library for Seafood Identification (http://www.fda.gov/Food/FoodScienceResearch/DNASeafoodIdentification/ucm238880.htm) and all are linked to authenticated, vouchered specimens. These samples were used for construction of a DNA barcode library, as described below. Also they were used for optimization of mini-barcode primers designed in this study. For analysis of mini-barcode primers with commercial products, a total of 44 heavily processed seafood products representing a variety of species and product types were purchased in the United States in May 2012 from online retail sources (Fig. 1). Subsamples were collected from each product using sterile forceps and scalpels and stored in 1.5 ml microcentrifuge tubes at −70 °C. These subsamples were shipped to the Biodiversity Institute of Ontario at the University of Guelph for DNA extraction and sequencing.
DNA extraction
For each authenticated or commercial sample, one gram of tissue/product was divided into 10 MP lysing matrix tubes “A” (100 mg each) and homogenized using an MP FastPrep-24 Instrument (MP Biomedicals Inc.) at speed 6 for 40 S. Total DNA of this homogenized slurry was extracted using the Nucleospin tissue kit (Macherey-Nagel Inc.) following the manufacturer’s instructions and eluted in 50 μl of molecular biology grade water.
DNA barcode library construction
The COI standard barcoding region (652 bp) was amplified for each of the 96 authenticated samples using a pair of newly designed degenerate fish primers (Table 1) as well as a primer cocktail previously described27. Each amplification reaction contained 2 μl DNA template, 17.5 μl molecular biology grade water, 2.5 μl 10X reaction buffer, 1 μl MgCl2 (50 μM), 0.5 μl dNTPs mix (10 mM), 0.5 μl forward primer (10 μM), 0.5 μl reverse primer (10 μM) and 0.5 μl Invitrogen’s Platinum Taq polymerase (5 U/μl) in a total volume of 25 μl. The PCR conditions were initiated with a heated lid at 95 °C for 5 min, followed by a total of 35 cycles of 94 °C for 40 S, 51 °C for 1 min and 72 °C for 30 S and a final extension at 72 °C for 5 min and hold at 4 °C. PCR reactions were carried out using Mastercycler ep gradient S (Eppendorf, Mississauga, ON, Canada) thermal cyclers. PCR success was verified by 1.5% agarose gel electrophoresis. A DNA template negative control reaction was included in all experiments to test for contamination. Two microliters of each amplicon were subsequently used directly for bi-directional Sanger sequencing with the M13 primers described in Table 1 using Applied Biosystems’s BigDye Terminator chemistry V3.1 (Foster City, CA, USA). Sequencing reactions were cleaned using EdgeBio’s AutoDTR96 (Gaithersburg, MD, USA) and visualized on an ABI 3730xl sequencer (Applied Biosystems)28,29. Sequence editing and contig assembly were carried out using CodonCode Aligner v 3.7.1.1 (CodonCode Corp., Dedham, MA, USA). Identification of the tested samples was conducted using BLAST in GenBank and a local barcode library for selected taxa with a minimum BLAST cut off of 98% identity for a top match. The accession numbers of the generated sequences are available in the Supplementary Materials-Table S1.
PCR primer design and in silico testing
A total of 8845 fish COI barcodes were downloaded from GenBank (n = 1894) and the Barcode of Life Database (BOLD; n = 6951) using the FDA Seafood List (http://www.accessdata.fda.gov/scripts/fdcc/?set=seafoodlist) as a guide for the target species. All sequences were aligned and multiple copies of identical sequences were removed. Degenerate nucleotides and inosine were used to manually design a fish COI primer set to amplify 652 bp—the standard barcoding region—within a wide range of fish species (Table 1).
The newly designed COI primer set was used to amplify the full-length DNA barcode in the 96 authenticated samples from the FDA. For comparison, a previously designed primer cocktail was also used to amplify these samples27. The COI sequences generated from the authenticated samples, along with the unique COI sequences downloaded from GenBank and BOLD, were then used to design multiple mini-barcode primer sets to amplify partial fragments within the standard COI barcoding region (Fig. 2). The primers were picked according to the availability of highly-conserved priming sites in a wide range of species with consideration of the primer stability in PCR reactions as well as the physical and structural properties of oligos (e.g., annealing temperature, G+C percentage, hairpin formation and self- and hetero-dimer formation). In silico analysis was also carried out using UCLUST30 and MEGA V5.2.231 on the newly designed mini-barcode primers to assess the potential for the amplification targets to differentiate fish species at the 98% and 100% levels (Table 2 and Table S2). The analysis included full-length DNA barcodes representing 200 species and 124 genera obtained from the FDA’s Reference Standard Sequence Library for Seafood Identification. M13 forward and reverse tails were attached to the forward and reverse barcoding primers, respectively, to facilitate high-throughput sequencing. The Integrated DNA Technologies (IDT) analysis tool was used to evaluate all the mentioned parameters32. Six mini-barcode primer sets were selected (Table 1) for further testing with commercial samples.
Mini-barcoding PCR Optimization Strategy
The amplification conditions for all the primer sets were tested using a gradient PCR approach at a wide range of annealing temperatures (43–60 °C). The composition of the amplification reactions, the PCR amplification conditions (except the annealing temperature) and the sequencing conditions were exactly the same as those used previously for amplification and sequencing of the full-length barcode. The optimal annealing temperature of each primer set was determined based on the results of gel electrophoresis of temperature gradient PCR products and is listed in Table 1. The mini-barcode amplification and sequencing steps were carried out on DNA from the 44 commercial fish products using each of the designed six sets of mini-barcode primers. Reagent blanks and a non-template PCR control were included in all PCR and sequencing experiments. Sequence editing and contig assembly of the generated barcodes were carried out as described for the full-length barcodes using CodonCode Aligner v 3.7.1.1 (CodonCode Corp., Dedham, MA, USA). Species identification for each sample was conducted using BLAST against GenBank and a local barcode library for selected taxa with a minimum BLAST cut off of 98% identity for a top match. These results were verified by neighbour-joining analysis33 and subsequent evaluation of the grouping of specimens tested as compared to database sequences5.
Results and Discussion
Full-length DNA barcodes (652 bp) could be recovered using the newly-designed Fish primers (Fish UnivF and Fish UnivR) in 86 out of 88 of fish species (93 out of 96 specimens) within the authenticated fish muscle tissue sample collection obtained from the FDA. Both peak intensities and sequencing qualities of the generated barcodes were compared to the sequences generated with the previously used primer cocktail27. The new full-length barcode fish primer set showed slightly higher success rate (97.7%) among the wide variety of the tested species compared to a success rate of 95.5% for those species sequenced with the previously developed primer cocktail.
Regarding the commercial fish products, the tested products included a wide range of processed products packed as cans, tins, retort pouches, jars, or tubes (Fig. 1). These samples were all shelf-stable, preserved products that had experienced different levels of processing, for instance, smoking, salting, etc. and they also contained multiple additives, preservatives and flavors (Table 3). These traits may negatively impact the quantity and quality of DNA extracted from these samples, which can decrease subsequent DNA barcode recovery.
The standard COI barcoding of the 44 tested fish processed products was achieved in only 9 products (20.5%) using both the newly designed universal fish primer set (Table 1) and the previously used fish primer cocktail27. These full-length barcodes were generated from a variety of samples with different levels of processing and with a variety of additives (Table 3). The major cause of full-barcode failure was most likely due to the degradation of the DNA extracted from these samples as a result of different levels of processing and the presence of multiple additives25. Samples showed low amplicon yields in gel electrophoresis and poor quality sequences with co-amplification of multiple non-targets (results are not shown).
Previous work has shown the applicability of a mini-barcoding approach in different groups of organisms21,34,35. Furthermore, it has been shown that the sequencing information of any 100 bases or more within the standard COI barcoding region can distinguish up to 91–94% of species in different taxonomic groups24,25,36. Here, we developed primers to amplify 6 mini-barcodes for commercial fish species, based on species described in the FDA Seafood List. The target fragment size ranges between 127 bp and 314 bp (Fig. 2). When compared in silico using DNA barcodes from authenticated FDA fish specimens representing 124 genera and 200 species, the mini-barcode amplification targets showed high levels of differentiation at both the species and genus levels (Table 4 and Table S2). Overall, primer sets SH-B, SH-D, SH-E and SH-F showed the greatest ability to resolve sequences at the genus and species levels. All four of these primer sets showed high potential for use in fish species identification, with resolution at the species level for 98–100% of the species analyzed at the 100% sequence identity level and 98–99% of the species analyzed at the 99% sequence identity level. Figure 2 demonstrates the amplification regions of the designed mini-barcode primers within the full-length COI DNA barcode. These mini-barcodes target 5′ (SH-A, SH-C, SH-E) and 3′ (SH-B, SH-F) regions of the standard DNA barcode as well as the middle region (SH-E, SH-D, SH-F). Hence, their combination can maximize recovery of sequence information from across the full-length DNA barcode and should provide sufficient sequence information for species identification24. In support of this, when the results of the in silico taxonomic analyses for all six mini-barcode primer sets were combined, species-level resolution was possible in 100% of sequences analyzed (Table S2). However, it is important to note that this analysis was restricted to sequences from authenticated specimens representing 200 species of commercial fish. Incorporation of sequences from a wider number of fish species may lead to less definitive results and may require slight primer modifications.
Out of the 44 processed fish products tested with the mini-barcode primer sets, 41 products (93.2%) could be mini-barcoded with at least one primer set. Three samples (RB-1_94, RB-1_104 and RB-2_114) were negative in both standard barcoding and mini-barcoding with all primer sets (Table 3). These samples were all labelled as tuna products (2 retort pouches of light tuna and 1 can of albacore) and contained a variety of additives. Although they showed some amplification success with the mini-barcoding primers, all generated amplicons failed at the sequencing stage. Two of these samples were marinated with either lemon or sweet and spicy marinades, which may either interfere with PCR amplification or result in low amplicon yield which cannot be successfully sequenced. Alternatively, DNA barcode failure in these products may be due to the presence of more than one species, which can co-amplify and produce a mixed electrophorogram37,38. For the samples which have multiple closely related species, they generated overlapping traces at few specific sites in the electrophorogram which were called as ambiguous bases. Indeed, the light tuna products may very well have contained multiple species, as FDA regulations allow for multiple species to be used in the production of canned light tuna as long as the color of the tuna meat is not darker than Munsell value 5.339 (FDA, 2013a).
As for the remaining samples (41 products), the 6 mini-barcode primer sets showed different success rates with each tested product group (Table 4). Overall, primer sets SH-B, SH-C, SH-D and SH-E had significantly higher proportions of sequencing success compared to the full barcode (Z-test, two-sided, P values all <0.05). Primer set SH-E (226 bp) showed the highest success rate with 39 samples barcoded (88.6%). The two additional samples that failed with this primer set (RB-1_100, smoked sprat in oil and RB-1_101, chunk light tuna in water) were amplified and sequenced with other sets. Sample RB-1_101 was successfully sequenced only with primer set SH-C, which amplifies 130 bp, indicating the high degradation level in the DNA extracted from this sample. On the other hand, sample RB-1_100 was a can of smoked sprat that was successfully sequenced using primer sets SH-B, SH-D and SH-F (208–314 bp). These results indicate either a lack of specificity at the SH-A/SH-C/SH-E primer binding sites for this sample or degradation of DNA towards the 5′ end of the standard barcoding region, as only the mini-barcodes closer to the 3′ end were amplified (Fig. 2). After SH-E, the primer set with the next-highest success rate was SH-D (208 bp), which achieved 63.6% sequencing success among the 41 samples, followed by primer set SH-B (227 bp) with 54.5% sequencing success. Among individual product groups, the primer sets showed a range of amplification and sequencing success rates (Table 4). For example, for the 13 tuna samples tested, primer set SH-E showed the greatest sequencing success (n = 9) followed by primer set SH-C (n = 8), while the remaining primer sets were only able to obtain sequences for 0–1 of the tuna samples. Interestingly, primer set SH-F showed the greatest PCR amplification success with the tuna samples (n = 10), but none of these amplicons were successfully sequenced. This is most likely due to co-amplification of non-target DNA along with the target DNA. On the other hand, within the eight sardine samples, the highest performing primer set was again SH-E, with amplification and sequencing success for all samples. Primer sets SH-F and SH-D were successful in amplification and sequencing of 7 out of the 8 sardine samples, while SH-C was only able to amplify and sequence one of the products (RB-3_132). Interestingly, this product was only successfully sequenced by one other primer set (SH-E). The only species group where SH-E showed a reduced sequencing success rate compared to the other primer sets was for sprat products, in which case primer sets SH-F, SH-D and SH-B showed the highest success rate (3 out of 3 products), whereas SH-E and SH-C showed success with 2 out of 3 products. Based on the set of commercial products tested here, these results show primer set SH-E to be the most favorable for use in mini-barcoding. However, in instances where this primer set fails to amplify a target sequence, primer sets SH-D or SH-C may be reasonable alternatives (Table 4).
All barcoded products could be identified as the species listed on the product label except in three cases involving tuna products (see below) and in two cases where species substitution was detected (Table 3). Species substitution is a form of seafood fraud in which seafood is mislabelled on imported or exported products. In one case of species substitution detected with the mini-barcodes, a sample labelled as “Wild Alaskan salmon” (RB-1_102) was found to be mislabelled. This sample was expected to be a species of Pacific salmon (genus Oncorhynchus), but it was identified by mini-barcoding as Atlantic salmon (Salmo salar). Atlantic salmon is not commercially harvested in North America, but rather it is a farm-raised fish. Furthermore, farming of Atlantic salmon is not permitted in the state of Alaska (http://www.legis.state.ak.us/basis/statutes.asp?title=16#16.40.100). While some species of Pacific salmon are actually sold at a lower price than Atlantic salmon, wild-caught salmon has certain marketing advantages over farm-raised salmon37, which may be a driving incentive for this form of substitution. Indeed, substitution of farm-raised salmon for wild-caught salmon is one of the examples of known species substitution given by the FDA40 (FDA, 2013b). On the other hand, another sample labelled as “Wild Alaskan sockeye salmon” (RB-1_92) was found to correctly contain the Pacific salmon species stated on the label - Oncorhynchus nerka. In another instance of mislabelling, sample RB-3_131 was labelled as “Mackerel in tomato sauce”, but DNA mini-barcoding identified this sample as Decapterus russelli. While mackerel is an acceptable market name for a number of species, including Scomber scombrus, Gasterochisma melampus and Grammatorcynus bicarinatus, it is not an acceptable market name for D. russelli according to the FDA Seafood List41 (FDA, 2013c). One of the vernacular names associated with this scientific name is mackerel scad, but the only acceptable market name for D. russelli in the U.S. is scad or Indian scad. Consistent with these findings, a previous study reported that Decapterus spp. are known to be substituted for other species of higher value in processed foods and that they share organoleptic properties with species of the genus Scomber, making them difficult to differentiate without laboratory analyses42.
As discussed above, the set of mini-barcode primers developed here were able to identify tuna at the genus or species level for 10 out of the 13 processed tuna products. The three un-sequenced tuna samples could be amplified with at least one COI mini-barcode primer set, but failed in the Sanger sequencing step possibly due to the presence of multiple species in each sample, co-amplification of non-target, or due to DNA degradation. All the tested tuna samples were identified as belonging to the genus Thunnus except sample RB-3_122 which was identified as Katsuwonus pelamis (skipjack tuna). However, it is important to note that it was challenging to discriminate between closely related tuna species and these three products showed multiple Thunnus species matches at the ≥99% level. As a result, the species identifications did not match what was stated on the label for three of the products (RB-1_97, RB-3_125 and RB-3_127). Although the in silico analysis showed high levels of species resolution (Table 2 and Table S2), the group of sequences tested only included two Thunnus species (T. alalunga and T. albacares). Based on these results, COI mini-barcoding may be used for the identification of tuna at the genus level but it is not recommended for the reliable differentiation of species within the Thunnus genus. Previous studies have also reported difficulties in differentiating closely related Thunnus species using DNA barcoding of the COI marker only43,44. To overcome this challenge, we recommend using additional genetic markers for further authentication of tuna samples at the species level23,38.
Besides detecting instances of species mislabelling, DNA mini-barcoding can also be used to clarify the identity of species in products with nonspecific labels. For example, a sample labelled as “Smoked garlic pepper salmon” (RB-2_106), with no species names listed, was found to contain Atlantic salmon by DNA mini-barcoding. Additionally, for the two samples of fish balls (RB-1_91 and RB-3_129), the ingredients list on the label simply stated “fish meat” (61%) and claimed the presence of one or more of the following: Gadus morhua, Melanogrammus aeglefinus, Pollachius virens, or Merluccius merluccius. These two samples were successfully mini-barcoded and identified with at least 3 primer sets as Melanogrammus aeglefinus (Haddock) only.
Although regulations for the safety and quality of commercial seafood exist in North America, the enforcement of proper species labelling has proven to be particularly difficult in heavily processed seafood products. This study sets the stage for the use of DNA information to identify a wide range of fish species in heavily processed products using one or more mini-barcode primer pairs. Basing the mini-barcode primer design on sequences of the full-length DNA barcode has allowed us to build upon the extensive research that has been carried out in this field9,12, including a database that contains DNA barcodes for over 10,000 fish species (i.e., www.fishbol.org).
Conclusion
This study presents a DNA mini-barcoding system for species identification applicable to heavily processed fish products. Six mini-barcode primer sets were developed, with one primer set in particular showing a high rate of success for identification of heavily processed products at the species or genus level. The additional primer sets developed showed promise as supplemental tools to be used in cases where the initial primer set fails. All mini-barcode primer sets showed increased performance for species identification in heavily processed products as compared to full-length DNA barcode primers. Additionally, the mini-barcoding system provides a new avenue for the utility of next-generation DNA sequencing for authentication of mixed products that may contain multiple species and experienced different levels of DNA un-friendly commercial procedures. Overall, the mini-barcode system developed here provides a means to identify species in heavily processed products and may be used for the detection and enforcement of species substitution on the commercial market.
Additional Information
How to cite this article: Shokralla, S. et al. A DNA Mini-Barcoding System for Authentication of Processed Fish Products. Sci. Rep. 5, 15894; doi: 10.1038/srep15894 (2015).
References
Woolfe, M. & Primrose, S. Food forensics: using DNA technology to combat misdescription and fraud. Trends Biotechnol 22, 222–226 (2004).
Marko, P. B. et al. Fisheries: mislabelling of a depleted reef fish. Nature 430, 309–310 (2004).
Hellberg, R. S. & Morrissey, M. T. Advances in DNA-based techniques for the detection of seafood species substitution on the commercial market. J Lab Autom 16, 308–321 (2011).
Wong, E. & Hanner, R. DNA barcoding detects market substitution in North American seafood. Food Res Int 41, 828–837 (2008).
Wallace, L. et al. DNA barcodes for everyday life: routine authentication of Natural Health Products. Food Res Int 49, 446–452 (2012).
Teletchea, F. Molecular identification methods of fish species: reassessment and possible applications. Rev Fish Biol Fisher 19, 265e293 (2009).
Hebert, P. D., Cywinska, A., Ball, S. L. & deWaard, J. R. Biological identifications through DNA barcodes. Proc R Soc Lond B 270, 313–321 (2003).
Hajibabaei, M., Singer, G., Hebert, P. D. & Hickey, D. A. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet 23, 167–172 (2007).
Ward, R. D., Zemlak, T. S., Innes, B. H., Last, P. R. & Hebert, P. D. DNA barcoding Australia’s fish species. Phil Trans R Soc B 360, 1847–1857 (2005).
Ward, R. D., Hanner, R. & Hebert, P. D. The campaign to DNA barcode all fishes, FISH-BOL. J Fish Biol 74, 329–356 (2009).
Hubert, N. et al. Identifying Canadian freshwater fishes through DNA barcodes. PLOS One 3(6), e2490 (2008).
Steinke, D., Zemlak, T. S., Gavin, H. & Hebert, P. D. DNA barcoding of Pacific Canada’s fishes. Mar Biol 156, 2641–2647 (2009).
Nicolè, S. et al. DNA barcoding as a reliable method for the authentication of commercial seafood products. Food Technol Biotech 50, 387–398 (2012).
Handy, S. M. et al. A single laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance. J AOAC Int 94, 201–210 (2011).
Baker, C. S., Lento, G. M., Cipriano, F. & Palumbi, S. R. Predicted decline of protected whales based on molecular genetic monitoring of Japanese and Korean markets. Proc R Soc Lond B 267, 1191–1199 (2000).
Shivji, M. et al. Genetic identification of pelagic shark body parts for conservation and trade monitoring. Conserv Biol 16, 1036–1047 (2002).
Dawnay, N., Ogden, R., McEwing, R., Carvalho, G. R. & Thorpe, R. S. Validation of the barcoding gene COI for use in forensic genetic species identification. Forensic Sci Int 173, 1–6 (2007).
Tanabe, S. et al. PCR method of detecting pork in foods for verifying allergen labeling and for identifying hidden pork ingredients in processed foods. Biosci Biotechnol Biochem 71, 1663–1667 (2007).
Chapela, M. J. et al. Comparison of DNA extraction methods from muscle of canned tuna for species identification. Food Control 18, 1211–5 (2007).
Shokralla, S., Singer, G. A. & Hajibabaei, M. Direct PCR amplification and sequencing of specimens’ DNA from preservative ethanol. Biotechniques 48, 233–234 (2010).
Shokralla, S. et al. Pyrosequencing for mini-barcoding of fresh and old museum specimens. PLOS One 6(7), e21252 (2011).
Civera, T. Species identification and safety of fish products. Vet Res Commun 27 Suppl 1, 481–489 (2003).
Rasmussen-Hellberg, R. S. et al. Interlaboratory evaluation of a real-time multiplex polymerase chain reaction method for identification of salmon and trout species in commercial products. J Agric Food Chem 59, 876–884 (2011).
Hajibabaei, M. et al. A minimalist barcode can identify a specimen whose DNA is degraded. Mol Ecol Notes 6, 959–964 (2006).
Meusnier, I. et al. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9, 214 (2008).
Rasmussen, R. S., Morrissey, M. T. & Hebert, P. D. DNA barcoding of commercially important salmon and trout species (Oncorhynchus and Salmo) from North America. J Agric Food Chem 57, 8379–8385 (2009).
Ivanova, N., Zemlak, T. S., Hanner, R. H. & Hebert, P. D. Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes 7, 544–548 (2007).
Hajibabaei, M. et al. Critical factors for assembling a high volume of DNA barcodes. Phil Trans R Soc B 360, 1959–1967 (2005).
Ratnasingham, S. & Hebert, P. D. BOLD: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes 7, 355–364 (2007).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 (2011).
Owczarzy, R. et al. IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res 36, W163–169 (2008).
Kimura, M. A simple method for estimating evolutionary rates of base substitution through comparative studies of nucleotide sequences. J Mol Evol 16, 111–120 (1980).
Virgilio, M., Backeljau, T., Nevado, B. & De Meyer, M. Comparative performances of DNA barcoding across insect orders. BMC Bioinformatics 11, 206 (2010).
Nagy, Z. T., Sonet, G., Glaw, F. & Vences, M. First large-scale DNA barcoding assessment of reptiles in the biodiversity hotspot of Madagascar, based on newly designed COI primers. PLOS One 7(3), e34506 (2012).
Van Houdt, J. K., Breman, F. C., Virgilio, M. & De Meyer, M. Recovering full DNA barcodes from natural history collections of Tephritid fruitflies (Tephritidae, Diptera) using mini barcodes. Mol Ecol Resour 10, 459–465 (2010).
Gray, S. F. & Evans, R. Dose-response in an outbreak of non-bacterial food poisoning traced to a mixed seafood cocktail. Epidemiol Infect 110, 583–590 (1993).
Shokralla, S., Spall, J., Gibson, J. & Hajibabaei, M. Next-generation sequencing technologies for environmental DNA research. Mol Ecol 21, 1794–1805 (2012).
FDA 2013a. Requirements for specific standardized fish and shellfish:, canned tuna. Code of Federal Regulations, 21CFR161.190.Accessed on 12/01/2015. Available at (http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/cfrsearch.cfm?fr=161.190)
FDA 2013b. Regulatory Fish Encyclopedia: Seafood Species Substitution and Economic Fraud. Accessed on 12/01/2015. Available at: (http://www.fda.gov/Food/FoodScienceResearch/RFE/ucm071528.htm).
FDA 2013c. The Seafood List. Accessed on 12/01/2015. Available at: (http://www.fda.gov/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/Seafood/ucm113260.htm).
Espiñeira, M., Vieites, J. M. & Santaclara, F. J. Development of a genetic method for the identification of salmon, trout and bream in seafood products by means of PCR-RFLP and FINS methodologies. Eur Food Res Technol 229, 785e793 (2009).
Lowenstein, J. H., Amato, G. & Kolokotronis, S. O. The real maccoyii: identifying tuna sushi with DNA barcodes–contrasting characteristic attributes and genetic distances. PLOS One 4(11), e7866 (2009).
Rasmussen, R. S. & Morrissey, M. T. DNA-based methods to identify fish and seafood substitution on the commercial market. Compr Rev Food Sci F 8, 118–154 (2009).
Acknowledgements
Thanks to Michael Kawalek and Khanh Van at the FDA Pacific Regional Laboratory Southwest for help with sample processing and to Jonathan Deeds at the FDA Center for Food Safety and Applied Nutrition for critical reads of the manuscript. This research was funded through grants from the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-050) and by a Discovery Grant from Natural Sciences and Engineering Research Council of Canada to M.H.
Author information
Authors and Affiliations
Contributions
S.S. and R.H. conceived and designed the experiments. R.H. and S.H. collected all samples examined. S.S. designed the mini-barcoding primers and performed molecular analysis. S.S., R.H., S.H., I.K. and M.H. analyzed sequence data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Shokralla, S., Hellberg, R., Handy, S. et al. A DNA Mini-Barcoding System for Authentication of Processed Fish Products. Sci Rep 5, 15894 (2015). https://doi.org/10.1038/srep15894
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep15894
This article is cited by
-
A Fast and Simple DNA Mini-barcoding and RPA Assay Coupled with Lateral Flow Assay for Fresh and Canned Mackerel Authentication
Food Analytical Methods (2023)
-
The gut bacterial microbiome of Nile tilapia (Oreochromis niloticus) from lakes across an altitudinal gradient
BMC Microbiology (2022)
-
Application of DNA barcoding for ensuring food safety and quality
Food Science and Biotechnology (2022)
-
DNA barcoding to assess species identification in museum samples of Amphiliidae and natural samples of Cichlidae from Southern Mozambique
Rendiconti Lincei. Scienze Fisiche e Naturali (2022)
-
Life barcoded by DNA barcodes
Conservation Genetics Resources (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.