Abstract
Understanding how natural processes affect population genetic structures is an important issue in evolutionary biology. One effective method is to assess the relative importance of environmental and geographical factors in the genetic structure of populations. In this study, we examined the spatial genetic variation of thirteen Myriophyllum spicatum populations from the Qinghai-Tibetan Plateau (QTP) and adjacent highlands (Yunnan-Guizhou Plateau, YGP) by using microsatellite loci and environmental and geographical factors. Bioclim layers, hydrological properties and elevation were considered as environmental variables and reduced by principal component analysis. The genetic isolation by geographic distance (IBD) was tested by Mantel tests and the relative importance of environmental variables on population genetic differentiation was determined by a partial Mantel test and multiple matrix regression with randomization (MMRR). Two genetic clusters corresponding to the QTP and YGP were identified. Both tests and MMRR revealed a significant and strong correlation between genetic divergence and geographic isolation under the influence of environmental heterogeneity at the overall and finer spatial scales. Our findings suggested the dominant role of geography on the evolution of M. spicatum under a steep environmental gradient in the alpine landscape as a result of dispersal limitation and genetic drift.
Similar content being viewed by others
Introduction
Understanding how natural processes affect population genetic structures and gene flows is an important issue in evolutionary biology. Environmental variables can influence the movement of gametes, individuals and groups of individuals among natural populations to affect gene flow patterns1, which may have a profound influence on the spatial and temporal distribution of genetic variation and further, evolutionary progress of natural populations2,3. Recent advances in molecular ecology and landscape genetics make it feasible to dissect the independent roles of landscape features on population genetic differentiation4,5,6. It can also deepen our understanding of how landscape variables affect dispersal and gene flow and shape patterns of genetic variation in nature5,7.
For aquatic plants, given that they live in fragmented islands in terrestrial landscapes, severe genetic differentiation by geographic isolation may be common8,9, representing an increasing genetic differentiation among populations with increasing geographic distance, as a result of reduced gene flow (isolation by distance, IBD10). When ecological characteristics of habitats are dissimilar among natural populations, the probability of successful establishment of immigrants or their offspring may reduce. In this situation, the effective gene flow is reduced as a consequence of local adaptation, resulting in a pattern of isolation by adaptation/environment (IBA or IBE11), or monopolization under the influence of founder effects (IBM4,12). However, relative studies on aquatic plants with incorporating environmental variations in the analysis of population divergence are limited13 (but see Zhao et al.14).
Eurasian watermilfoil (Myriophyllum spicatum L.) is native to Europe, Asia and northern Africa15 and is recognized as a noxious weed in North America due to its ability to rapidly spread in new habitats16. This species occurs in various types of inland water bodies as it is tolerant to a wide range of water and climatic conditions17, which may be partly responsible for its cosmopolitan occurrences. Eurasian watermilfoil is widely distributed in China and previously, we found that both geographic barriers and climate significantly affected population genetic differentiation across the entire country (Wu et al. unpublished). In the waters of the Qinghai-Tibetan Plateau (QTP) and adjacent highlands, M. spicatum is a common submerged macrophyte18. The large range in elevation and the complex landscape of this region offer an ideal place for us to explore the spatial genetic patterns of M. spicatum and the underlying factors.
In this study, thirteen Eurasian watermilfoil populations were collected in the QTP and adjacent highlands (Yunnan-Guizhou Plateau, YGP) for genetic analysis using microsatellite markers. The Bioclim layers (climate) and elevation were recruited as environmental variables along with the hydrological properties, which are important for aquatic plants19,20, for landscape analyses. Our aims were to (1) reveal population genetic structure and gene flow pattern; (2) assess the relative role of geographic isolation and environment heterogeneity in population genetic structure; and (3) find the main force driving genetic divergence of M. spicatum in the alpine environment.
Results
Genetic diversity and population genetic structure
The number of alleles (NA), the number of effective alleles (NEA) and the genotype diversity (D) ranged from 30 to 75, 25.76 to 50.596 and 0.111 to 1, respectively (Table 1). Globally, the mean NA, NEA and D were 47.846, 38.474 and 0.803, respectively. A total of 102 genets were identified and all of the obtained genotypes were population specific.
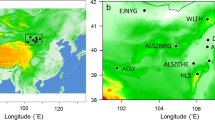
The pairwise Bruvo distance ranged from 0.31 to 0.592, with comparisons involving individuals from AR-DR and DR-DL presenting the greatest and smallest genetic dissimilarity, respectively (Table 2). The STRUCTURE analysis suggested K = 2 as the optimal number of clusters based on the calculation of ∆K (Supplementary Fig. S1). When the studied M. spicatum populations were divided into two clusters, we found that the eight populations from the QTP were assigned to one cluster and the five populations of the adjacent highlands (YGP) were assigned to another cluster (Figs 1 and 2a). In separate STRUCTURE analyses for each cluster, obviously internal structure was revealed in both clusters. The QTP cluster was further divided into three subclusters (Supplementary Fig. S1). One of these was comprised five western populations and the other two subclusters comprised three eastern populations (Fig. 2b). The YGP cluster was further divided into two subclusters (Supplementary Fig. S1). Two western populations were assigned to one subcluser and three eastern populations were assigned to another subcluster (Fig. 2c). DAPC analyses revealed a pattern consistent with STRUCTURE analyses, with the individuals from the QTP and YGP assigned to distinct clusters without genetic admixture (Supplementary Table S1).
Thirteen sampling sites of Myriophyllum spicatum in the QTP and adjacent highlands mapped using ArcGIS.
Pie charts represent the probability of assignment to subclusters when K = 2, as identified by STRUCTURE based on microsatellite data. Population codes are shown on the side. The elevation range and main rivers are visualized.
Isolation by distance
Using Mantel tests, we revealed a significantly positive correlation between geographic and genetic distances (r = 0.578, p = 0.001, Fig. 3a) at the overall scale. Similarly, a significantly positive correlation was revealed in the QTP cluster (QTP: r = 0.559, p = 0.001, Fig. 3b). For the YGP cluster, although the correlation was not significant (r = 0.602, p = 0.063, Fig. 3c), the p value of Mantel test was close to 0.05, suggesting that influence of IBD on genetic differentiation were close at the overall and finer scales.
Landscape genetic analysis
With PCA, we kept the first axis as an environmental variable because it explained 69.3% of the environmental variables, whereas the percentage of inertia explained by other axes of PCA was low (the results of landscape analysis changed little when one or two more axes were included; not shown). For the QTP and YGP clusters, the first two axes explained 69.8% and 80.2% of environmental variables, respectively, which were used to calculate environmental dissimilarity. The relative contributions of environmental variables to the axes are presented in Table S2.
When the influence of the other factor was controlled, the genetic-geographical association (r = 0.330, p = 0.012) was significant, whereas we could not detect significant correlations between genetic differentiation and environmental distance (r = 0.077, p = 0.554) (Table 3). With MMRR analysis, the geographic distance had a higher regression coefficient, whereas the effects of environmental factors were not significant (geographic distance: βD = 0.49, p = 0.010; environment: βE = 0.108, p = 0.559; Table 4). Similarly, in the QTP cluster, both partial Mantel tests and MMRR revealed significant genetic-geographical association (r = 0.556, p = 0.001; βD = 0.619, p = 0.002) and not significant genetic-environmental association (r = 0.044, p = 0.849; βE = 0.041, p = 0.831) (Tables 3 and 4). In the YGP cluster, no significant correlation between genetic distance and geographic distance or environmental distance was found when the influence of the other factor was controlled, but the regression coefficients of genetic-geographical association were about three times higher than those of genetic-environmental association (Tables 3 and 4).
Discussion
In the present study, a genetic structure was revealed where the QTP and YGP populations were assigned to independent genetic clusters (Fig. 1) corresponding to distinct geographic ranges. Further analyses revealed obviously internal phylogeographic structure in both regions (Fig. 2b,c). A strong association between geographic and genetic distances at different spatial scales (Fig. 3) indicated the pattern of IBD in the alpine populations of M. spicatum. The IBD pattern was further confirmed by landscape analyses at the overall and finer spatial scales when the influence of environmental factors was considered (Tables 3 and 4). It suggested that the geographic isolation was the main factor influencing gene flow among populations of M. spicatum in the QTP and adjacent highlands.
The IBD pattern was highly associated with limited dispersal among populations in alpine environments. Suren & Ormerod21 and Lacoul22 suggested that the dispersal of freshwater macrophytes among alpine lakes was limited by the isolation and infrequent visitation of animal vectors. Given few connections by water systems, the inter-population gene flow of M. spicatum should rely only on passive dispersal mediated by animals. Waterfowl play important roles in the seed dispersal of many aquatic plants9,23 and the fact that some waterbirds (e.g., Anas crecca) help to transport the seeds of watermilfoils through digestive track24,25 indicates that the dispersal of Eurasian watermilfoil is likely to be closely linked with waterfowl. However, in China, few waterfowl breed in or migrate across the QTP and YGP regions26,27. High genetic differentiation and limited gene flow were also detected in some species of aquatic plants in the same region28,29. Therefore, the single means of dispersal and lack of mediators might aggravate the isolation of M. spicatum populations in alpine landscapes, which could explain the greater prediction of IBD on genetic differentiation as a result of dispersal limitation and genetic drift.
A series of environmental factors varied along the elevation gradient and among them, climate was a fundamental factor that could cause genetic divergence in the population30,31. Higher-altitude environments contained severe constraints (e.g., short growing seasons and low temperature) on the survival and reproduction of plants and populations located at different altitudes; thus, the divergence might be due to local adaptation30,32. In the present study, no significant correlation existed between genetic differentiation and the environmental dissimilarity that mainly arose from elevational differences when the effects of geographic isolation were considered, suggesting that the genetic adaptation of M. spicatum populations might be limited. Wang et al.33 found that aquatic plants in the QTP could offset the stress induced by low temperatures and a short growing season via nutrient accumulation and an expedited growth rate, which indicated that physiological adaptation might be important for M. spicatum in reacting to alpine environmental stress. In addition, phenological shifts (e.g., flowering period) reduced pollen exchanges between the populations with different elevations and were thus considered to be an important mechanism for creating genetic divergence among populations along the elevational gradient34. However, this mechanism should not greatly contribute to genetic divergence in M. spicatum populations with different elevations because the inter-population gene flow of this submerged macrophyte mainly relies on vegetative propagules for local propagation and on seeds for long-distance propagation rather than on pollen diffusion17.
In conclusion, we highlighted the influence of geographic isolation on the pattern of the gene flow and genetic differentiation of M. spicatum under a steep environmental gradient in an alpine landscape. However, physical barriers and climate rather than geographic distance significantly affected the population genetic differentiation of M. spicatum, as revealed at the country scale in China (Wu et al. unpublished). Different results confirmed that the main factor driving genetic divergence might vary in different regions or at different scales35. A variety of studies are needed to help us obtain a deeper understanding of how abiotic factors influence the evolutionary processes of freshwater plants.
Methods
Sample collection and DNA extraction
A total of 206 individuals of M. spicatum were collected at 13 sites of the QTP and its adjacent highlands in August 2012 (Table 1). We sampled between 6 and 20 plants from each population according to the population size. The plants were collected randomly at intervals of at least 20 m to avoid collecting ramets from a single genet in the clonal Eurasian watermilfoil. Hybridization between M. spicatum and its closely related species M. sibiricum has been reported in North America36,37. In our previous study, we confirmed that hybridization between these two species also occurred in the QTP of China38. Hybrid populations identified by morphological traits and genetic data were not included in the present study.
Fresh leaves were dried in silica gel in the field and frozen at −20 °C after being transported to the laboratory. Total genomic DNA was extracted using the DNA Secure Plant Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s protocol.
Genetic diversity and population genetic structure
We previously isolated 20 microsatellite loci from M. spicatum39. Fourteen of these loci (Myrsp1–7, Myrsp9, Myrsp12–16 and Myrsp18) were used in the present study because of their successful amplification in all populations. The protocols for polymerase chain reaction (PCR) amplification and the analyses of the obtained PCR products followed Wu et al.39. Genotyping was performed using GeneMapper 4.0 software (Applied Biosystems, Foster City, California, USA).
The indices of genetic diversity, the number of alleles (NA), the number of effective alleles (NEA), the number of genotypes (clone assignment) and Nei’s40 diversity index (corrected for sample size) (D) were calculated using GenoType and GenoDive41 for each population. Only the genotypes of the genets were kept for subsequent analysis (Table 1).
Because the allelic copies of microsatellites were ambiguous in polyploid species, we could not determine the exact genotypes of heterozygotes in M. spicatum due to dosage problems. Additionally, the statistics of microsatellite markers were developed for diploid organisms and were not suitable for polyploid organisms42. Therefore, we converted the microsatellite data to a binary format as dominant data43,44 for genetic structure analysis. In this study, the Bruvo distance (DB42), based on the shared presence or/and absence of alleles, was recruited for the pairwise individual genetic relatedness. The values were calculated in R software version 3.1.1 (R Development Core Team, 2014) with the package POLYSAT45. This index has been proven to be an adequate estimator for polyploids when full genotypes are not required46. Because individuals from the same site cannot be considered as independent samples, we randomly picked one individual per sampling site to calculate pairwise Bruvo distances. We repeated the process 100 times and averaged the values of the genetic distance obtained across all of the random sub-samples of individuals for each pair of populations47.
Distinct genetic clusters of the 13 Eurasian watermilfoil populations were identified using an individually based assignment approach as implemented in STRUCTURE 2.3.4 48,49 and the current version of the software can accommodate dominant markers50. Twenty independent runs were performed for each K value (K = 1 to 10) with a burn-in period of 20,000 iterations and 100,000 MCMC iterations under the admixture model. The best-fit number of clusters was determined based on the ∆K method51. The geographical distribution of different clusters identified was mapped using ArcGIS 9.0 (Esri, Redlands, CA, USA). In order to assess potential hierarchical structure, each identified cluster was also analyzed separately.
An alternative method, Discriminant Analysis of Principal Components (DAPC52), was also used for the genetic structure. DAPC is an adequate analysis of genetic clustering for polyploidy organisms and does not require the populations to be in Hardy-Weinberg equilibrium46. The method required a priori clustering algorithms determined by k-means and the best number of clusters was assessed using the Bayesian information criterion (BIC)52. We evaluated up to k = 10 groups, but the value of BIC kept decreasing with the increase of k. We therefore set the identical k value of DAPC as the K of STRUCTURE for comparison. DAPC was implemented in R with “adegenet” version 1.4–153.
Isolation by distance
Mantel tests with 10,000 permutations were used to detect IBD pattern based on matrices of pairwise Bruvo distance and geographic distance among all populations, as implemented in GenAlEx 6.5 software54. Because two genetic clusters corresponding to the QTP and YGP were identified (see results), we also performed Mantel tests in each region separately. The geographic distance (log10 standardized) was calculated with the software PASSaGE 2 55 based on the coordinates.
Landscape genetics analysis
The climatic variables of the studied sites (to a resolution of 1 km) were extracted from the BioClim dataset with GIS information56,57 using ArcGIS. The properties of the water were also considered. When sampling, the pH, salinity and dissolved oxygen of the aquatic environment were measured using a handheld multiparameter meter (PROPLUS, YSI, USA) and ammonium nitrogen, nitrate nitrogen, the total nitrogen and phosphorus concentrations were determined with a Palintest 7500 Photometer (Palintest, UK).
We reduced the environmental variables by the components of principal component analysis (PCA) based on the 27 environmental variables (including 19 BioClim variables, 7 variables of water conditions and the elevation of the study sites) using the “prcomp” function in R. Prior to analysis, all Bioclim variables and elevation values were standardized.
The correlations between genetic differentiation and geographic/environmental factors were determined by a combination of partial Mantel tests58 and matrix regression analysis with a distance matrix. The environmental distance between populations was the Euclidean distance calculated with the values of PCA axes. Partial Mantel tests with 10,000 permutations were performed between genetic factors and one factor under the influence of the other, as implemented in R using the “ecodist” package59. Multiple matrix regression with randomization (MMRR) is a novel and robust approach for estimating the independent effects of potential factors, especially in situations of low-to-moderate gene flow60. The analysis was implemented with 10,000 permutations in R with the MMRR function script60. Because two genetic clusters corresponding to the QTP and YGP were identified (see results), partial Mantel tests and MMRR were also conducted in each region separately.
Additional Information
How to cite this article: Wu, Z. et al. Great influence of geographic isolation on the genetic differentiation of Myriophyllum spicatum under a steep environmental gradient. Sci. Rep. 5, 15618; doi: 10.1038/srep15618 (2015).
References
Sork, V. L., Nason, J., Campbell, D. R. & Fernandez, J. F. Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol. Evol. 14, 219–224 (1999).
Slatkin, M. Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–430 (1985).
Bohonak, A. J. Dispersal, gene flow and population structure. Q. Rev. Biol. 74, 21–45 (1999).
Orsini, L., Vanoverbeke, J., Swillen, I., Mergeay, J. & De Meester, L. Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol. Ecol. 22, 5983–5999 (2013).
Wang, I. J. & Bradburd, G. S. Isolation by environment. Mol. Ecol. 23, 5649–5662 (2014).
Shafer, A. B. A. & Wolf, J. B. W. Widespread evidence for incipient ecological speciation: a meta-analysis of isolation-by-ecology. Ecol. Lett. 16, 940–950 (2013).
Lee, C.-R. & Mitchell-Olds, T. Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Mol. Ecol. 20, 4631–4642 (2011).
Barrett, S. C. H., Eckert, C. G. & Husband, B. C. Evolutionary processes in aquatic plant populations. Aquat. Bot. 44, 105–145 (1993).
Santamaría, L. Why are most aquatic plants widely distributed? Dispersal, clonal growth and small-scale heterogeneity in a stressful environment. Acta Oecol. 23, 137–154 (2002).
Wright, S. Isolation by distance. Genetics 28, 114–138 (1943).
Wang, I. J. & Summers, K. Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Mol. Ecol. 19, 447–458 (2010).
De Meester, L., Gómez, A., Okamura, B. & Schwenk, K. The monopolization hypothesis and the dispersal–gene flow paradox in aquatic organisms. Acta Oecol. 23, 121–135 (2002).
Storfer, A., Murphy, M. A., Spear, S. F., Holderegger, R. & Waits, L. P. Landscape genetics: where are we now? Mol. Ecol. 19, 3496–3514 (2010).
Zhao, Y. et al. Are habitat fragmentation, local adaptation and isolation-by-distance driving population divergence in wild rice Oryza rufipogon? Mol. Ecol. 22, 5531–5547 (2013).
Couch, R. & Nelson, E. Myriophyllum spicatum in North America. Anderson, L. W. J. (Ed.), Proceedings of the first international symposium on watermilfoil (Myriophyllum spicatum) and related Haloragaceae species, Vancouver, Canada, 8–18 (1985).
Reed, C. F. History and distribution of Eurasian watermilfoil in United States and Canada. Phytologia 36, 417–436 (1977).
Aiken, S. G., Newroth, P. R. & Wile, I. The biology of Canadian weeds. 34. Myriophyllum spicatum L. Can. J. Plant Sci. 59, 201–215 (1979).
Yu, D., Wang, D., Li, Z. & Funston, A. M. Taxonomic revision of the genus Myriophyllum (Haloragaceae) in China. Rhodora 104, 396–421 (2002).
Lacoul, P. & Freedman, B. Environmental influences on aquatic plants in freshwater ecosystems. Environ. Rev. 14, 89–136 (2006).
Viana, D. S. et al. Environment and biogeography drive aquatic plant and cladoceran species richness across Europe. Freshwater Biol. 59, 2096–2106 (2014).
Suren, A. M. & Ormerod, S. J. Aquatic bryophytes in Himalayan streams: testing a distribution model in a highly heterogeneous environment. Freshwater Biol. 40, 697–716 (1998).
Lacoul, P. Aquatic macrophyte distribution in response to physical and chemical environment of the lakes along an altitudinal gradient in the Himalayas, Nepal. Ph. D. thesis, Dalhousie University (2004).
Figuerola, J. & Green, A. J. Dispersal of aquatic organisms by waterbirds: a review of past research and priorities for future studies. Freshwater Biol. 47, 483–494 (2002).
Olney, P. J. S. The food and feeding habits of teal Anas crecca. P. Zool. Socie. 140, 169–210 (1963).
Brochet, A. L., Guillemain, M., Fritz, H., Gauthier-Clerc, M. & Green, A. J. Plant dispersal by teal (Anas crecca) in the Camargue: duck guts are more important than their feet. Freshwater Biol. 55, 1262–1273 (2010).
MacKinnon, J. R. et al. A biodiversity review of China. WWF International, Hong Kong (1996).
Sibley, C. G. & Monroe, B. L. Distribution and taxonomy of birds of the world. Yale University Press, New Haven (1990).
Wang, Y. et al. Population genetic structure of an aquatic herb Batrachium bungei (Ranuculaceae) in the Hengduan Mountains of China. Aquat. Bot. 92, 221–225 (2010).
Chen, J., Du, Z., Sun, S., Gituru, R. W. & Wang, Q. Chloroplast DNA phylogeography reveals repeated range expansion in a widespread aquatic herb Hippuris vulgaris in the Qinghai-Tibetan Plateau and adjacent areas. PLoS ONE 8, e60948 (2013).
Ohsawa, T. & Ide, Y. Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Global Ecol. Biogeogr. 17, 152–163 (2008).
Shi, M., Michalski, S. G., Chen, X. & Durka, W. Isolation by elevation: genetic structure at neutral and putatively non-neutral loci in a dominant tree of subtropical forests. Castanopsis eyrei. PLoS ONE 6, e21302 (2011).
Frei, E. R., Ghazoul, J., Matter, P., Heggli, M. & Pluess, A. R. Plant population differentiation and climate change: responses of grassland species along an elevational gradient. Global Change Biol. 20, 441–455 (2014).
Wang, Z., Xia, C., Yu, D. & Wu, Z. Low-temperature induced leaf elements accumulation in aquatic macrophytes across Tibetan Plateau. Ecol. Eng. 75, 1–8 (2015).
Hirao, A. S. & Kudo, G. Landscape genetics of alpine-snowbed plants: comparisons along geographic and snowmelt gradients. Heredity 93, 290–298 (2004).
Storfer, A. et al. Putting the ‘landscape’ in landscape genetics. Heredity 98, 128–142 (2007).
Moody, M. L. & Les, D. H. Evidence of hybridity in invasive watermilfoil (Myriophyllum) populations. P. Natl. Acad. Sci. USA 99, 14867–14871 (2002).
Moody, M. L. & Les, D. H. Geographic distribution and genotypic composition of invasive hybrid watermilfoil (Myriophyllum spicatum × M. sibiricum) populations in North America. Biol. Invasions 9, 559–570 (2007).
Wu, Z., Ding, Z., Yu, D. & Xu, X. Influence of niche similarity on hybridization between Myriophyllum sibiricum and M. spicatum. J. Evol. Biol. 28, 1465–1475 (2015).
Wu, Z., Yu, D. & Xu, X. Development of microsatellite markers in the hexaploid aquatic macrophyte, Myriophyllum spicatum (Haloragaceae). Appl. Plant Sci. 1, 1200230 (2013).
Nei, M. Molecular evolutionary genetics. Columbia University Press, New York (1987).
Meirmans, P. G. & Van Tienderen, P. H. GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes 4, 792–794 (2004).
Bruvo, R., Michiels, N. K., D’Souza, T. G. & Schulenburg, H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 13, 2101–2106 (2004).
Lynch, M. The similarity index and DNA fingerprinting. Mol. Biol. Evol. 7, 478–484 (1990).
Samadi, S., Mavárez, J., Pointier, J. P., Delay, B. & Jarne, P. Microsatellite and morphological analysis of population structure in the parthenogenetic freshwater snail Melanoides tuberculata: insights into the creation of clonal variability. Mol. Ecol. 8, 1141–1153 (1999).
Clark, L. V. & Jasieniuk, M. POLYSAT: an R package for polyploid microsatellite analysis. Mol. Ecol. Resour. 11, 562–566 (2011).
Dufresne, F., Stift, M., Vergilino, R. & Mable, B. K. Recent progress and challenges in population genetics of polyploid organisms: an overview of current state-of-the-art molecular and statistical tools. Mol. Ecol. 23, 40–69 (2014).
Ortego, J., Gugger, P. F. & Sork, V. L. Climatically stable landscapes predict patterns of genetic structure and admixture in the Californian canyon live oak. J. Biogeogr. 42, 328–338 (2015).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003).
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes 7, 574–578 (2007).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 14, 2611–2620 (2005).
Jombart, T., Devillard, S. & Balloux, F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11, 94 (2010).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405 (2008).
Peakall, R. & Smouse, P. E. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28, 2537–2539 (2012).
Rosenberg, M. S. & Anderson, C. D. PASSaGE: pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol. Evol. 2, 229–232 (2011).
Busby, J. R. BIOCLIM—A bioclimate analysis and prediction system. Margules, C. R. & Austin, M. P. (Eds.), Nature conservation: cost effective biological surveys and data analysis, Melbourne, Australia, 64–68 (1991).
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G. & Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978 (2005).
Smouse, P. E., Long, J. C. & Sokal, R. R. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35, 627–632 (1986).
Goslee, S. C. & Urban, D. L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19 (2007).
Wang, I. J. Examining the full effects of landscape heterogeneity on spatial genetic variation: a multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution 67, 3403–3411 (2013).
Acknowledgements
We thank Nikos Andreakis, Sarah Goslee and Ian Wang for their recommendations and guidance in the data analysis as well as members of Dan Yu’s group for field assistance. This study was supported by grants from the National Natural Science Foundation of China to Xinwei Xu (31070190 and 31270265) and Dan Yu (30930011).
Author information
Authors and Affiliations
Contributions
X.X. and D.Y. designed the research; Z. Wu, Z. Wang and X.L. conducted the field work and laboratory experiments; and Z. Wu and X.X. analyzed the data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wu, Z., Yu, D., Wang, Z. et al. Great influence of geographic isolation on the genetic differentiation of Myriophyllum spicatum under a steep environmental gradient. Sci Rep 5, 15618 (2015). https://doi.org/10.1038/srep15618
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep15618
This article is cited by
-
Population genetic analysis illustrated a high gene diversity and genetic heterogeneity in Himalayacalamus falconeri: a socio-economically important Indian temperate woody bamboo taxon
Journal of Plant Biochemistry and Biotechnology (2023)
-
Influence of environmental factors on the genetic variation of the aquatic macrophyte Ranunculus subrigidus on the Qinghai-Tibetan Plateau
BMC Evolutionary Biology (2019)
-
Bridging population genetics and the metacommunity perspective to unravel the biogeographic processes shaping genetic differentiation of Myriophyllum alterniflorum DC.
Scientific Reports (2019)
-
Population genetic structure of the whorl-leaf watermilfoil Myriophyllum verticillatum shaped by topography and geographic distance
Hydrobiologia (2019)
-
The Tanggula Mountains enhance population divergence in Carex moorcroftii: a dominant sedge on the Qinghai-Tibetan Plateau
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.