Abstract
The three-dimensional organization of tightly condensed chromatin within metaphase chromosomes has been one of the most challenging problems in structural biology since the discovery of the nucleosome. This study shows that chromosome images obtained from typical banded karyotypes and from different multicolour cytogenetic analyses can be used to gain information about the internal structure of chromosomes. Chromatin bands and the connection surfaces in sister chromatid exchanges and in cancer translocations are planar and orthogonal to the chromosome axis. Chromosome stretching produces band splitting and even the thinnest bands are orthogonal and well defined, indicating that short stretches of DNA can occupy completely the chromosome cross-section. These observations impose strong physical constraints on models that attempt to explain chromatin folding in chromosomes. The thin-plate model, which consists of many stacked layers of planar chromatin perpendicular to the chromosome axis, is compatible with the observed orientation of bands, with the existence of thin bands and with band splitting; it is also compatible with the orthogonal orientation and planar geometry of the connection surfaces in chromosome rearrangements. The results obtained provide a consistent interpretation of the chromosome structural properties that are used in clinical cytogenetics for the diagnosis of hereditary diseases and cancers.
Similar content being viewed by others
Introduction
In the cell nucleus, genomic DNA molecules are associated with histone proteins and form long chromatin filaments containing many nucleosomes. The core particle of each nucleosome is a short cylinder (5.7 nm height and 11 nm diameter), which consist of a segment of 146 bp of DNA wrapped around a histone octamer1. Chromatin filaments are dynamic structures that can have different compaction degrees, ranging from extended fibres up to compact interdigitated solenoids (~30 nm diameter)2,3,4,5,6. During mitosis, each sister chromatid of a condensed chromosome contains a single DNA molecule that is densely packaged7. Metaphase chromatids of different plant and animal species show a great variety of sizes which are dependent on the amount of DNA that they contain, but all of them are elongated cylinders having relatively similar shape proportions8. Each chromatid of the largest human metaphase chromosome contains ~280 Mb of DNA9; this corresponds to a giant DNA molecule of ~9.5 cm that is confined in a cylinder of ~0.6 μm diameter and ~7 μm length8.
On the basis of results obtained using different experimental approaches, several models for the folding of the chromatin filaments within mitotic chromosomes have been proposed. From early electron microscopy images of histone-depleted chromosomes, it was suggested that chromatin fibres form radial loops attached to a protein scaffold10,11. Results obtained in vivo with chromosomes containing engineered regions with altered sequence composition12 and with chromosomes in different condensation stages13, suggested a model based on hierarchical folding of fibres of increasing diameter (from 30 to 250 nm); in this model the chromatids are segmented longitudinally into layers (~250 nm thickness) formed by the thicker fibre. Models considering that the chromatin filament is irregularly folded were proposed from stretching experiments showing that chromosomes do not have a mechanically continuous protein scaffold14 and from the analysis of cryo-sections of mitotic chromosomes15. More recently, results obtained using chromosome conformation capture methods were interpreted with polymer simulations and it was suggested that compact chromatids are formed by arrays of stochastically positioned chromatin loops that are longitudinally compressed16. All these models are based on chromatin fibres that are folded and form a three-dimensional network that fills the chromatid. However, unexpectedly, it was observed that incubation of chromosomes at 37 °C (under metaphase ionic conditions) on electron microscopy grids caused the emanation multilayered plates instead of fibres17. Further studies using polarizing microscopy, electron tomography, AFM imaging in aqueous media and AFM-based nanotribology and force spectroscopy18,19,20 showed that in each layer the chromatin filament forms a flexible two-dimensional network in which nucleosomes are irregularly oriented, allowing the compaction of the structure by interdigitation of adjacent layers (each layer has an apparent thickness of ~6 nm). This led to the proposal of the thin-plate model in which it is considered that chromatids are filled by many stacked layers of chromatin oriented perpendicular to the chromatid axis18,19. Furthermore, it was demonstrated that chromatin plates can be self-assembled from chromatin fragments obtained by micrococcal nuclease digestion of metaphase chromosomes21. The nano- and micro-mechanical data obtained from chromatin and chromosome stretching experiments22,23,24,25 and modelling studies26,27 of other laboratories, were used to estimate the nucleosome-nucleosome interaction energies between the stacked chromatin layers; it was demonstrated that the different energy components of this structure can explain the elongated cylindrical shape and the mechanical properties of metaphase chromosomes8.
On the other hand, although cytogenetics is mainly concerned with the study of chromosome structure for clinical diagnostics28, there is a wealth of cytogenetic data that may give insights about fundamental aspects of chromosome structure. Mitotic chromosomes show longitudinal heterogeneities that allowed the development of diverse cytogenetic staining procedures that produce banding patters characteristic of each human chromosome29,30. Each staining method gives rise to a continuous pattern of dark and light bands and sometimes the patterns are complementary: the Giemsa positive bands (dark G-bands) are located in the zones in which the bands obtained with reverse staining methods show a low intensity (light R-bands). Therefore, from a structural point of view bands occupy the whole chromosome. The sequence of the human genome indicated a correlation between regions of low GC content and dark G-bands and between regions high GC content and light G-bands9. The number of bands depends on the compaction degree of the chromosomes; human karyotypes corresponding to mid-prophase chromosomes yield up to 2000 bands31, but current analyses are carried out with karyotypes of 400- to 800-band levels30. Furthermore, bands can be split into several sub-bands when chromosomes are mechanically stretched32. Fluorescence in situ hybridization (FISH) allowed the positioning of many genes with respect to the chromosome bands28 and it was found that the correspondence between the cytogenetic map and the genome sequence is excellent9,33. Many structural chromosome rearrangements such as sister chromatid exchange (SCE) and chromosome fusions and translocations have been observed; most of them are related with human diseases34. Chromosome painting techniques, in particular multicolour spectral karyotyping (SKY) and multiplex (M)-FISH35,36,37,38, permitted the simultaneous discrimination of all human chromosomes and greatly facilitated the study of structural rearrangements. In this work, typical results obtained in different banding experiments and in the analyses of SCE and other rearrangements have been used to obtain information about the internal structure of chromosomes and to test the models considered in the preceding paragraph. It is demonstrated that the model based on stacked layers of chromatin can explain the main geometric characteristics of chromosome bands and translocations.
Results and Discussion
Chromosome bands are perpendicular to the chromosome axis
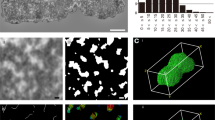
In the G-banded chromosomes shown in the example presented in Fig. 1a (reproduced from ref. 39), it seems that bands have a transverse orientation. The results of the quantitative measurement (see Methods) of the orientation angle α (defined with respect to the chromosome axis) of many bands are presented in Fig. 1d. These results correspond to G-banding patterns at ~700-band level. Equivalent results were obtained with G-banded chromosomes at a lower resolution (~400-band level) and with R-banded chromosomes (see Table 1). The conclusion of this quantitative study is that chromosome bands are orthogonal to the chromosome axis. The ideograms of chromosome banding patters (ref. 30; see Fig. 1b) are schematic drawings that are not based on quantitative measurements of the orientation angle but, in agreement with this conclusion, they represent all the bands exactly perpendicular to the longitudinal axis of the chromosome. Interestingly, even the thinnest bands in the typical karyotypes (Fig. 1a) and in the corresponding ideograms30 show this orientation and occupy the entire cross-section of the two sister chromatids. Since the analysis of the human genome showed that the thinnest bands correspond to sequences of less than 1 Mb9, this result indicates that within metaphase chromosomes chromatin containing relatively short stretches of DNA fills completely the cross-section of each chromatid. Furthermore, as top and bottom boundaries of thin bands are usually well defined, in three dimensions bands can be considered disc-like structures. All these results suggest that the chromatin filament must be able to fold forming very short cylinders having the same diameter as the chromatids and the same height as the thin bands. It will be shown below that many proposed models for chromosome structure do not have the adequate properties to satisfy these geometrical requirements.
Orientation of chromosome bands with respect to the chromosome axis.
The orientation angles (α1, α2, α3…) were measured as schematized in (a,b). (a) Example showing chromosome G-banding patterns at ~700-band level. (b) Chromosome G-bands and the corresponding ideograms before (~400-band level) and after stretching (~1400-band level). (c) Example of replication-banding patterns in which dark-stained bands indicate late-replicating regions; red arrow heads point to thin bands. (d) Histogram of the orientation angles of G-bands (~700-band level). (e) Orientation angles of G-bands of stretched chromosomes (shown in (b)). Figures in (a–c) have been reproduced, respectively, from refs 39,32,40 with permission.
Maintenance of the transverse orientation of bands after their mechanical splitting
The images from ref. 32 reproduced in Fig. 1b show that chromosome stretching causes band splitting. It can be seen that many of the resulting split bands are well defined and relatively thin. The measurements of the orientation angle α of these bands are presented in Fig. 1e. The mean of these measurements is about 90° (Table 1), indicating that splitting does not change the orthogonal orientation of bands with respect to the chromosome axis. In three dimensions such a transformation leading to defined sub-bands can only be explained considering that chromatin structure within bands is formed by planar elements that can be relatively easily pulled apart by stretching forces. It will be discussed below that many structural models of metaphase chromosomes are not compatible with these observations.
Perpendicular orientation of thin replication bands with respect to the chromosome axis
Replication bands were observed in many different types of eukaryotes29. The example shown in Fig. 1c (reproduced from ref. 40) corresponds to replication bands of plant chromosomes. In this experiment rood cells were treated with 5-bromodeoxyuridine and the resulting chromosome spreads were stained with Giemsa; dark bands are produced by late-replicating regions. The measurements of the orientation angle α of replication bands of two different species (see Table 1) indicate that they are orthogonal to the chromosome axis. The well-defined thin bands observed in these preparations (some of them are indicated with red head arrows in Fig. 1c) are particularly interesting because each one contains a short segment of DNA corresponding to different late-replicating sequences. This observation indicates again that chromatin containing a relatively short stretch of DNA can form a disk-like structure that occupies the whole cross-section of the chromatid. These results reinforce the conclusions obtained above from the analysis of thin bands in typical human karyotypes.
In the reciprocal exchanges between sister chromatids the resulting connection surfaces are flat and perpendicular to the chromatid axis
The images of SCEs shown in the example presented in Fig. 2a (reproduced from ref. 41) were obtained using different proportions of 5-bromodeoxyuridine to deoxythymidine in three successive cell division cycles, followed with Giemsa staining of the resulting metaphase spreads. Using this procedure chromatids acquire different staining degrees that allow the visualization of the interchanged chromatid segments. Note that the boundaries between the interchanged segments and the original chromatids are sharp and well defined, indicating that the connection of the two chromatids is produced in a very narrow zone. The results obtained in the measurements of the orientation angle α (with respect to the chromatid axis) of these boundaries are shown in Fig. 2f; the mean value of angle α is 90° (Table 1). Taken together, these results indicate that in three dimensions the connection of the interchanged and original chromatids is produced in a thin planar zone oriented perpendicular to the chromatid axis. To be acceptable, a structural model of the metaphase chromosome must be consistent with these observations (see below).
The connection surfaces of the multiple translocations occurring in cancer cells are planar and perpendicular to the chromosome axis
Several examples of chromosome rearrangements in different malignancies are shown in Fig. 2b–e. These images were obtained using SKY and M-FISH techniques in different laboratories (see references in the figure legend); each chromosome was painted with a different colour to clearly identify fusions leading to dicentric chromosomes (b) and translocations (c-e). It seems that both fusion and translocation junctions have a transverse orientation. The same orientation is observed in translocations associated to different non-oncological diseases35, but the many rearrangements observed in cancer cell karyotypes facilitate the study of the morphology of translocated chromatids. The results obtained in the quantitative measurement of the orientation angle α with respect to the chromosome axis of the translocation junctions of four human ovarian carcinoma cell lines (the SKY karyotype of one of them (OVCAR-8) is shown in Fig. 2e) are presented in Fig. 2g. The quantitative analysis of many translocations corresponding to different cancer cells (pancreatic, skin and cervical carcinomas and leukaemia and other haematological malignancies) led to equivalent results (see Table 1). In most of the images obtained using chromosome painting methods, the sister chromatids of each metaphase chromosome are closely joined together (Fig. 2), but in some cases the two sister chromatids are visible. In particular, M-FISH karyotypes of cervical carcinoma HeLa cells (ref. 42; reproduced in part in Fig. 2d) allowed the measurement of the orientation angle of the translocation connection surfaces of chromosomes with sister chromatids longitudinally joined and with single chromatids visible. The results obtained in both cases are equivalent (Table 1). The conclusion from all these measurements is that the angle α of translocation junctions is approximately 90°. Furthermore, many junctions are sharp and well defined (see examples in Fig. 2b–e), indicating that in three dimensions the connection between the original chromatid and the translocated part is a thin planar region orthogonal to the chromatid axis. Even when the translocations are very small (see examples in Fig. 2c–e indicated with yellow asterisks), the top and bottom boundaries between the translocated segment and the original chromatid are usually very well defined. This indicates that the relatively short sequences of genomic DNA corresponding to the small translocated chromatid segments are confined within thin discoidal structures. All these observations reinforce the conclusions obtained in the preceding sections and impose geometric constraints that have to be considered for the validation of models for chromatin folding in metaphase chromosomes.
The geometric characteristics of chromosome bands and rearrangements provide a test for the different models proposed for chromatin folding within mitotic chromosomes
The measurements of the orientation angle α of cytogenetic bands, replication bands and of the connection surfaces in SCEs and in translocations indicate that in all cases they are orthogonal to the chromatid axis. It will be shown below that this is consistent with the basic geometric features of the thin-plate model (Fig. 3f; refs 8,18, 19, 20, 21). However, these measurements show a variability (standard deviation 5.0–8.9o; Table 1) that at least in part could be attributed to the soft structure of chromosomes; chromosomes are hydrogels with good mechanical properties but they are easily deformable8,25. This reduces the capability of these measurements for direct discrimination between different chromosome models, but the perpendicular orientation of bands and connection surfaces, combined with other cytogenetic results, can be used to test different proposals for metaphase chromatin folding.
Representation of the expected shape of chromatid connections in SCEs and translocations for different chromosome structural models.
The amount of DNA required to cover completely a chromatid cross-section is different for each chromosome model (a–f) and this determines the shape of the chromatid connections in rearrangements (g). For the three categories schematized in (g), the amount of DNA necessary to cover the chromatid cross-section and the corresponding thickness of the connection zone (Δz) were calculated for human chromatids having a DNA density of 166 Mb/μm3 and a diameter of 0.6 μm8,62. The models schematized in figures (a, left), (a, right), (e) and (f) have been reproduced, respectively, from refs 44,11,16,8 with permission. Model in figure (b) has been obtained from Fig. 7B(a) of ref. 13, ©2004 Kireeva et al. The Journal of Cell Biology, 166: 775-785, doi:10.1083/jcb.200406049 (http://jcb.rupress.org/content/166/6/775.short). Figures (c,d) have been reproduced, respectively, from refs 14,15, Copyright (2002) and (2008) National Academy of Sciences, USA.
As indicated above, genomic results have shown that the thinnest bands contain less than 1 Mb of DNA9 and that there is an excellent correspondence between band location on a given mitotic chromosome and its position on the DNA sequence corresponding to this chromosome33.Therefore, the DNA molecule within each chromatid is organized in such a way that it can fill consecutively (without overlapping) the successive bands from one telomere to the other. According to the results presented in the preceding sections, the well-defined discoidal shape and orthogonal orientation of the thinnest bands indicate that DNA stretches of less than 1 Mb can fill short cylinders having the diameter of the chromatid. In agreement with this, FISH results showing that sequences separated by 0.5–1 Mb frequently produce hybridization spots on either side of the chromatid suggested many years ago that short stretches of DNA can cross the width of the chromatid43. The thin discoidal shape and orthogonal orientation of thin replication bands (Fig. 1c) and of small translocated chromatid segments (Fig. 2c–e) are also in agreement with these observations. Chromosome structural models consistent of radial loops helically folded (Fig. 3a; refs 10,11,44) and the hierarchical folding model (Fig. 3b; refs 12,13) are not compatible with these results because they need a large amount of DNA (~12 Mb) to span the cross-section of a chromatid (Fig. 3g, left scheme); this corresponds to a chromatid region having a thickness (Δz) of ~250 nm. The amount of DNA required to cover the cross-section is reduced to ~3 Mb if it is hypothesized45 that the radial loops of the 30-nm chromatin fibre are organized forming minibands. In the thin-plate model (Fig. 3f; refs 8,18, 19, 20, 21), only a monolayer of nucleosomes is needed to cover the cross-section of a chromatid. This corresponds to ~0.5 Mb of DNA (Fig. 3g, right scheme); furthermore, layers are interdigitated and have a small thickness18,19 (Δz ≈ 6 nm). In the network (Fig. 3c; ref. 14), polymer-melt (Fig. 3d; ref. 15) and compressed loop (Fig. 3d; ref. 16) models the path of the chromatin filament is irregular and the length of DNA required to cover completely a chromatid cross-section and Δz are larger than the values corresponding to the thin-plate model; the models in Fig. 3c–e cannot justify the existence of thin bands containing less than 1 Mb of DNA9.
The thin-plate model is consistent with the experimental observations considered in the preceding paragraph and it can justify the results obtained in FISH experiments with metaphase chromosomes showing that the resolution of probe ordering is 1–3 Mb46. In contrast, in the models schematized in Fig. 3a–e, the path of the chromatin filament with respect to the z axis (Fig. 3g, left and central scheme) constantly goes up and down, making difficult the correlation between the order of probes in the metaphase chromosome and the DNA sequence. For the same reasons, with these models it is difficult to explain the band splitting observed in chromosome stretching experiments, but in the thin-plate model the relative weakness of the interactions between the stacked chromatin layers8,18,19 can justify easily the observed band splitting and the maintenance of the orthogonal orientation of the resulting split bands (Fig. 1b,e and Table 1).
In SCEs and in translocations the connection between the original chromatid and the exchanged segment is planar and perpendicular to the chromatid axis. This geometry is not at all compatible with the very uneven shape of chromatid connections (Fig. 3g, left scheme) expected for the models of radial loops and of hierarchical folding (Fig. 3a,b; refs 10, 11, 12, 13,44). In the network (Fig. 3c; ref. 14) and polymer-melt (Fig. 3d; ref. 15) models the chromatin filament is irregularly folded and consequently well-defined flat connections are not expected (Fig. 3g, central scheme). Chromatin folding in these models is isotropic, making difficult the explanation of the orthogonal orientation of the junction surfaces with respect to the chromatid axis. The model consisting on a longitudinally compressed array of consecutive chromatin loops16 has a certain degree of longitudinal order but, as can be seen in Fig. 3e, the irregular folding of the chromatin fibre considered in this model cannot justify the well-defined planar connection surfaces of translocations and other rearrangements. In contrast, the thin-plate model is anisotropic8,18,19; the stacked layers perpendicular to the chromatid axis (Fig. 3f) can explain the planar geometry and orthogonal orientation (Fig. 3g, right scheme) of the connection surfaces observed in SCEs and translocations.
The simplified model in Fig. 4a illustrates the three-dimensional compatibility of the multilayer organization of chromatin with the geometric characteristics of translocations. It can be seen that the connection surface between the original chromatid and the translocated part is well defined and perpendicular to the chromatid axis. The well-defined top and bottom surfaces often observed in chromosome bands can also be explained with this model. Different experimental results indicated that the metaphase chromatids are helical structures11,47,48,49 and it was suggested that chromatin layers are connected forming a continuous helicoid8,19 with the successive turns in close contact (helicoidal pitch of ~6 nm). The detailed path of DNA in this continuous structure is unknown but, as schematized in Fig. 4b, the junction between the two chromatid fragments should be produced abruptly at the point in which the DNA of the translocated part is covalently bound to the DNA of the original chromatid. Therefore, since the thickness of chromatin sheet in the helicoidal turns is very small, this model justifies that the transition between the translocated and original chromatid is produced in a very narrow zone of the z axis, in agreement with the experimental observations. This continuous helicoidal folding permits a simple topological organization of DNA without entanglements. The absence of topological entanglement between adjacent layers is fully compatible with the band splitting observed in chromosome stretching experiments. For the maintenance of this organization, it has to be taken into account that during mitosis there is a high topoisomerase II activity50, which allows DNA segments in the chromatin filament to pass through each other51. Although this activity is mainly related with the disentanglement of DNA between sister chromatids52, if entanglements are produced within a chromatid (see drawing in Fig. 4c), it can be speculated that topoisomerase II could also be involved in the disentanglement of chromatin turns in condensed chromosomes. It seems difficult to explain any enzymatic activity inside compact chromatids. However, metaphase chromosomes are fluid and dynamic structures that can be considered as hydrogels with a lamellar liquid crystal organization8; they are highly hydrated53 and it has been found54,55 that in living cells topoisomerase II can rapidly diffuse within metaphase chromatin.
Simplified solid model representing part of a chromatid with many stacked layers.
(a) Representation of a translocation as seen in SKY and M-FISH experiments: the original chromatid (blue) is joined to a translocated chromatid (green). (b) This less compact representation illustrates that the connection with the translocated chromatid occurs in a defined region (pointed by a red arrow). (c) If some entanglement of DNA is produced between two consecutive turns (i and i + 1), topoisomerase II could regenerate the disentangled uniform structure.
Biomedical implications
In addition to give an explanation to all the geometric characteristics of chromosome bands and rearrangements, it was shown previously8 that the thin-plate model can explain the morphology and dimensions of metaphase chromatids. The energy components of this multilayer organization can also justify the outstanding mechanical properties of metaphase chromosomes8,20,25.
The multilayer organization of chromatin in metaphase chromosomes was proposed from experimental studies in which native chromosomes and plates were prepared in aqueous solutions containing metaphase cation concentrations17,18,19,20,21. In particular, in cryo-electron microscopy and AFM experiments18,20, water solutions where present even during the imaging of plates. In contrast, in the typical metaphase cell spreads used in cytogenetic studies chromosomes are fixed with methanol-acetic acid38,39,46. Early results showed that these organic solvents remove part of the histones56. However this does not imply that the multilayered organization of native chromosomes disappears when chromosomes are fixed with methanol-acetic acid. In fact, the observation of orthogonal and well-defined thin bands and orthogonal and smooth surface connections in SCEs and translocations indicates that the backbone of this multilayered structure is maintained after fixation. Thus, although methanol-acetic acid fixation may extract histones, this treatment preserves the global structure of the chromosome and its multilayered organization. This could explain the well-defined images of chromosome bands and structural rearrangements obtained in many laboratories using different cytogenetic procedures.
This study has tried to build a bridge between results obtained with chromosomes in aqueous media and results obtained with chromosomes fixed with menthol-acetic acid. It has been shown that a coherent synthesis is possible. The conclusions of this research can be useful for a better understanding of the structure of native chromosomes and of the structural basis of the cytogenetic methods considered in this work. The impact of clinical cytogenetic analyses on the diagnosis of hereditary diseases and cancers is impressive28,57. This work provides for the first time a consistent explanation of the chromosome structural elements on which these analyses are based.
Methods
The measurements of the orientation angle α of well-defined thin G-bands with respect to the chromosome axis were obtained from human karyotypes at ~400-band level (Fig. 6 of Shaffer et al.30) and ~700-band level (Fig. 2 of Venter and coworkers39; reproduced in part in Fig. 1a). Angle α is defined as indicated in Fig. 1a: the chromosome axis is equivalent to the median line (dotted red line); the dotted blue line was drawn according to the observed orientation of each analyzed band. The same procedure was used for the measurement of angle α of R-bands of human chromosomes (Fig. 3 from ref. 30). Images obtained after stretching of human chromosome 6 (Figs 1 and 3 of Hliscs et al.32; partially reproduced in Fig. 1b) were used to measure the orientation of split G-bands; angle α of these bands in homogeneously stretched regions was measured as indicated in Fig. 1b. Replication banding patters of Allium cepa and A. nigrum (Figs 1 and 4 of Cortés and Escalza40; partially reproduced in Fig. 1c) were used to measure the orientation angle α of thin bands corresponding to late-replicating regions. In the rearrangements produced by exchanges between sister chromatids, the orientation angle α of the connection surfaces with respect to the chromatid axis was measured (as indicated in Fig. 2a) from images obtained in SCE experiments with A. cepa (Figs 9.3 and 9.5 of Friebe and Cortés41; reproduced in part in Fig. 2a). Angle α of the connection surfaces between a translocated chromatid and the original chromatid was measured as shown in Fig. 2b,c. These measurements were obtained from SKY images of (i) ovarian carcinomas (OVCAR-3, 4, 5 and 8 cell lines58; OVCAR-8 is reproduced in Fig. 2e), (ii) pancreatic carcinomas (AsPc-1, Capan-1 and 2, Colo-357, FA6, MDA Panc-3, PaTu II, SUIT-2 and T3M-4 cell lines59) and (iii) haematological malignancies (Figs 1, 2, 3 of Ried and coworkers37). Angle α of translocations seen in M-FISH karyotypes was obtained from images corresponding to (i) skin carcinomas (Fig. 2 of Popp et al.60) and (ii) HeLa cells (Fig. 3 of Landry et al.42; partially reproduced in Fig. 2d). A fraction (~7%) of the analyzed translocations appeared very distorted and were not used in these measurements. In all cases angle α was measured with the angular dimension tool of CorelDRAW X3 software.
Additional Information
How to cite this article: Daban, J.-R. Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Sci. Rep. 5, 14891; doi: 10.1038/srep14891 (2015).
References
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997).
Daban, J. R. Electron microscopy and atomic force microscopy studies of chromatin and metaphase chromosome structure. Micron 42, 733–750 (2011).
Luger, K., Dechassa, M. L. & Tremethick, D. J. New insight into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell. Biol. 13, 436–447 (2012).
Rippe, K. The folding of nucleosome chains in Genome organization and function in the cell nucleus (ed. Rippe, K. ) 139–167 (Wiley-VCH, 2012).
Collepardo-Guevara, R. & Schlick, T. Chromatin fiber polymorphism triggered by variations of DNA linker length. Proc. Natl. Acad. Sci. USA 111, 8061–8066 (2014).
Boulé, J. B., Mozziconacci, J. & Lavelle, C. The polymorphisms of the chromatin fiber. J. Phys. Condens. Matter 27, 033101 (2015).
Daban, J. R. High concentration of DNA in condensed chromatin. Biochem. Cell Biol. 81, 91–99 (2003).
Daban, J. R. The energy components of stacked chromatin layers explain the morphology, dimensions and mechanical properties of metaphase chromosomes. J. R. Soc. Interface 11, 20131043 (2014).
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature409, 860-921 (2001).
Paulson, J. R. & Laemmli, U. K. The structure of histone-depleted metaphase chromosomes. Cell 12, 817–828 (1977).
Rattner, J. B. & Lin, C. C. Radial loops and helical coils coexist in metaphase chromosomes. Cell 42, 291–296 (1985).
Strukov, Y. G., Wang, Y. & Belmont, A. S. Engineered chromosome regions with altered sequence composition demonstrate hierarchical large-scale folding within metaphase chromosomes. J. Cell Biol. 162, 23–35 (2003).
Kireeva, N., Lakonishok, M., Kireev, I., Hirano, T. & Belmont, A. S. Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 166, 775–785 (2004).
Poirier, M. G. & Marko, J. F. Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. Proc. Natl. Acad Sci. USA 99, 15393–15397 (2002).
Eltsov, M., MacLellan, K. M., Maeshima, K., Frangakis, A. S. & Dubochet, J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. USA 105, 19732–19737 (2008).
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013)
Caravaca, J. M., Caño, S., Gállego, I. & Daban, J. R. Structural elements of bulk chromatin within metaphase chromosomes. Chromosome Res. 13, 725–743 (2005).
Gállego, I., Castro-Hartmann, P., Caravaca, J. M., Caño, S. & Daban, J. R. Dense chromatin plates in metaphase chromosomes. Eur. Biophys. J. 38, 503–522 (2009).
Castro-Hartmann, P., Milla, M. & Daban, J. R. Irregular orientation of nucleosomes in the well-defined chromatin plates of metaphase chromosomes. Biochemistry 49, 4043–4050 (2010).
Gállego, I., Oncins, G., Sisquella, X., Fernàndez-Busquets, X. & Daban, J. R. Nanotribology results show that DNA forms a mechanically resistant 2D network in metaphase chromatin plates. Biophys. J. 99, 3951–3958 (2010).
Milla, M. & Daban, J. R. Self-assembly of thin plates from micrococcal nuclease-digested chromatin of metaphase chromosomes. Biophys. J. 103, 567–575 (2012).
Cui, Y. & Bustamante, C. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA 97, 127–132 (2000).
Mihardja, S., Spakowitz, A. J., Zhang, Y. & Bustamante, C. Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. USA 103, 15871–15876 (2006).
Kruithof, M. et al. Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 16, 534–540 (2009).
Poirier, M., Eroglu, S., Chatenay, D. & Marko, J. F. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol. Biol. Cell 11, 269–276 (2000).
Kepper, N. et al. Force spectroscopy of chromatin fibers: extracting energetic and structural information from Monte Carlo simulations. Biopolymers 95, 435–447 (2011).
Collepardo-Guevara, R. & Schlick, T. Crucial role of dynamic linker histone binding and divalent ions for DNA accessibility and gene regulation revealed by mesoscale modeling of oligonucleosomes. Nucleic Acids Res. 18, 8803–8817 (2012).
Trask, B. J. Human cytogenetics: 46 chromosomes, 46 years and counting. Nature Rev. Genet. 3, 769–778 (2002).
Sumner, A. T. Chromosomes. Organization and function (Blackwell, 2003).
Shaffer, L. G., McGowan-Jordan, J. & Schmid, M. An international system for human cytogenetic nomenclature (Karger, 2013).
Yuris, J. J. Mid-prophase human chromosomes. The attainment of 2000 bands. Hum. Genet. 56, 293–298 (1981).
Hliscs, R., Mühlig, P. & Claussen, U. The nature of G-bands analyzed by chromosome stretching. Cytogenet. Cell Genet. 79, 162–166 (1997).
The BAC Resource Consortium. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature409, 953-958 (2001).
Kaiser-Rogers, K. & Rao, K. W. Structural chromosome rearrangements in The principles of clinical cytogenetics (eds Gersen, S. L. & Keagle, M. B. ) 139–174 (Springer, 2013).
Schröck, E. et al. Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996).
Speicher, M. R., Ballard, S. G. & Ward, D. C. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nature Genet. 12, 368–375 (1996).
Veldman, T., Vignon, C., Schröck, E., Rowley, J. D. & Ried, T. Hidden chromosome abnormalities in haematological malignancies detected by multicolour spectral karyotyping. Nature Genet. 15, 406–410 (1997).
Roschke, A. V. et al. Karyotypic complexity of the NCI-60 drug-screening panel. Cancer Res. 63, 8634–8647 (2003).
Levy, S. et al. The diploid genome sequence of an individual human. PLoS Biol. 5(10), e254 (2007).
Cortés, F. & Escalza, P. Analysis of different banding patterns and late replicating regions in chromosomes of Allium cepa, A. sativum and A. nigrum. Genetica 71, 39–46 (1986).
Friebe, B. & Cortés, F. Sister chromatid exchange and replication banding in Plant chromosomes: Laboratory methods (eds Fukui, K. & Nakayama, S. ) 171–186 (CRC Press, 1996).
Landry, J. J. M. et al. The genomic and transcriptomic landscape of a HeLa cell line. G3: Genes Genomes Genet. 3, 1213–1224 (2013).
Lawrence, J. B., Singer, R. H. & McNeil, J. A. Interphase and metaphase resolution of different distances within the human dystrophin gene. Science 249, 928–932 (1990).
Saitoh, Y. & Laemmli, U. K. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76, 609–622 (1994).
Nelson, W. G., Pienta, K. J., Barrack, E. R. & Coffey, D. S. The role of the nuclear matrix in the organization and function of DNA. Annu. Rev. Biophys. Biophys. Chem. 15, 457–475 (1986).
Trask, B. J. et al. Studies of metaphase and interphase chromosomes using fluorescence in situ hybridization. Cold Spring Harbor Symp. Quant. Biol. 58, 767–775 (1993).
Ohnuki, Y. Demonstration of the spiral structure of human chromosomes. Nature 208, 916–917 (1965).
Boy de la Tour, E. & Laemmli, U. K. The metaphase scaffold is helically folded: sister chromatids have predominantly opposite helical handedness. Cell 55, 937–944 (1988).
Baumgartner, M. et al. Genes occupy a fixed and symmetrical position in sister chromatids. Cell 64, 761–766 (1991).
Chen, S. H., Chan, N. L. & Hsieh, T. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 82, 139–170 (2013).
Salceda, J., Fernández, X. & Roca, J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 25, 2575–2583 (2006).
Chen, T., Sun, Y., Ji, P., Kopetz, S. & Zhang, W. Topoisomerase IIα in chromosome instability and personalized cancer therapy. Oncogene 10.1038/onc.2014.332 (2014).
Poirier, M. G., Monhait, T. & Marko, J. F. Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical-micromechanical techniques. J. Cell Biochem. 85, 422–434 (2002).
Christensen, M. O. et al. Dynamics of human DNA topoisomerases IIα and IIβ in living cells. J. Cell Biol. 157, 31–44 (2002).
Tavormina, P. A. et al. Rapid exchange of mammalian topoisomerase IIα at kinetochores and chromosome arms in mitosis. J. Cell Biol. 158, 23–29 (2002).
Comings, D. E. Mechanisms of chromosome banding and implications for chromosome structure. Annu. Rev. Genet. 12, 25–46 (1978).
Mitelman, F., Johansson, B. & Mertens, F. Mitelman database of chromosome aberrations and gene fusions in cancer (2015). Available at: http://cgap.nci.nih.gov/Chromosomes/Mitelman. Date of access: 06/08/15.
Cell Line NCI60 Drug Discovery Panel. Available at: http://home.ncifcrf.gov/CCR/60SKY/new/demo1.asp. Date of access: 06/08/15.
Edwards, P. SKY karyotypes and FISH analysis of epithelial cancer cell lines. Available at: http://www.path.cam.ac.uk/~pawefish. Date of access: 06/08/15.
Popp, S. et al. Genetic characterization of a human skin carcinoma progression model: from primary tumor to metastasis. J. Invest. Dermatol. 115, 1095–1103 (2000).
Gisselsson, D. et al. Chromosomal breakage-fusion-bridge events cause genetic intratumor heterogeneity. Proc. Natl. Acad. Sci. USA 97, 5357–5362 (2000).
Daban, J. R. Physical constraints in the condensation of eukaryotic chromosomes. Local concentration of DNA versus linear packing ratio in higher order chromatin structures. Biochemistry 39, 3861–3866 (2000).
Acknowledgements
The author thanks Francesca Vidal and Joaquim Roca for useful comments.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Daban, JR. Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Sci Rep 5, 14891 (2015). https://doi.org/10.1038/srep14891
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep14891
This article is cited by
-
Differentially accessible, single copy sequences form contiguous domains along metaphase chromosomes that are conserved among multiple tissues
Molecular Cytogenetics (2021)
-
Surface structures consisting of chromatin fibers in isolated barley (Hordeum vulgare) chromosomes revealed by helium ion microscopy
Chromosome Research (2021)
-
Genome maintenance in the context of 4D chromatin condensation
Cellular and Molecular Life Sciences (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.