Abstract
Arabidopsis VQ motif-containing proteins have recently been demonstrated to interact with several WRKY transcription factors; however, their specific biological functions and the molecular mechanisms underlying their involvement in defense responses remain largely unclear. Here, we showed that two VQ genes, VQ12 and VQ29, were highly responsive to the necrotrophic fungal pathogen Botrytis cinerea. To characterize their roles in plant defense, we generated amiR-vq12 transgenic plants by using an artificial miRNA approach to suppress the expression of VQ12 and isolated a loss-of-function mutant of VQ29. Phenotypic analysis showed that decreasing the expression of VQ12 and VQ29 simultaneously rendered the amiR-vq12 vq29 double mutant plants resistant against B. cinerea. Consistently, the B. cinerea-induced expression of defense-related PLANT DEFENSIN1.2 (PDF1.2) was increased in amiR-vq12 vq29. In contrast, constitutively-expressing VQ12 or VQ29 confered transgenic plants susceptible to B. cinerea. Further investigation revealed that VQ12 and VQ29 physically interacted with themselves and each other to form homodimers and heterodimer. Moreover, expression analysis of VQ12 and VQ29 in defense-signaling mutants suggested that they were partially involved in jasmonate (JA)-signaling pathway. Taken together, our study indicates that VQ12 and VQ29 negatively regulate plant basal resistance against B. cinerea.
Similar content being viewed by others
Introduction
In nature, plants are constantly threatened by various microbial pathogens. To protect themselves against pathogen infection, resisant plants have evolved an effective innate immune system. Upon infection by virulent pathogens, plants detect microbes or pathogen-associated molecular patterns (PAMPs) via transmembrane pattern recognition receptors (PRRs), resulting in PAMP-triggered immunity (PTI)1. However, Gram-negative bacterial pathogens can successfully interfere with PTI by secreting effector proteins into plant cells2. When pathogens overcome the PTI, plant hosts recognize those effector molecules by specific disease resistance (R) proteins and activate highly efficient immune responses, known as effector-triggered immunity (ETI)1.
R proteins-mediated resistance is often associated with activation of the salicylic acid (SA)-signaling pathway that induces a subset of PATHOGENESIS-RELATED GENE (PR) genes3. Arabidopsis mutants deficient in SA biosynthesis (e.g., sid2) or responsiveness (e.g., npr1) are compromised to establish both basal defense and systemic acquired resistance4,5. Besides SA, ethylene (ET) and JA also play crucial roles in plant defense responses to pathogen attack6,7. The ET signal transduction components ETHYLENE INSENSITIVE2 (EIN2), EIN3 and EIN3-Like1 (EIL1) function as critical positive regulators of plant disease resistance toward necrotrophic pathogens8,9,10,11. In addition, the JA receptor CORONATINE INSENSITIVE1 (COI1) also positively regulates plant tolerance against necrotrophic pathogens12,13,14.
The Arabidopsis WRKY transcription factor family comprises 74 members which are subdivided into three major structural groups15,16. Accumulating evidence has indicated that WRKY proteins act as both positive and negative regulators in modulating plant defense responses17,18,19. For example, WRKY33 positively regulates plant resistance to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola20. Disruption of the structurally related WRKY46, WRKY70 and WRKY53 compromised plant basal defense against biotrophic pathogen Pseudomonas syringae21. In contrast, several WRKY family members negatively modulate plant pathogen resistance. For instance, the evolutionarily related WRKY18, WRKY40 and WRKY60 function as negative regulators of plant resistance against P. syringae22. Mutations of WRKY11 and WRKY17 resulted in increased defense-related gene expression and enhanced basal defense to P. syringae23.
Recently, several WRKY transcription factors have been found to physically interact with a class of novel proteins, defined as VQ proteins, to regulate various physiological processes24,25,26,27,28,29. The name of VQ proteins are derived from the FxxxVQxxTG motif, a conserved amino acid region shared by all members of this family in Arabidopsis27,30. Increasing studies have demonstrated that VQ proteins play crucial roles in modulating plant defense responses. For example, VQ21 functions as one substrate of MAP kinase 4 (MPK4) and is involved in MPK4-mediated resistance24,31,32. The structurally related VQ16 and VQ23, previously identified as sigma factor binding proteins, redundantly regulate plant defense response against B. cinerea26,30,33. In addition, the JA-associated VQ protein VQ22 modulates JA-mediated defense responses34. Besides defense responses, VQ proteins are also involved in abiotic stress responses. Our previous study revealed that VQ9 interacts with WRKY8 and participates in plant salinity tolerance28. In an earlier study, VQ15 was identified as a calmodulin (CaM)-binding protein that affects osmotic stress tolerance35. Moreover, several developmental processes are also modulated by VQ proteins; for example, VQ14 was reported to play an essential role in seed development25,36. Very recently, Li et al. showed that VQ29 acts as a negative transcriptional regulator of light-mediated inhibition of hypocotyl elongation by interacting with PHYTOCHROME-INTERACTING FACTOR1 (PIF1)37.
Cheng et al. showed that a number of Arabidopsis VQ genes were responsive to plant defense signals27. However, the specific biological functions of VQ genes and the exact mechanisms underlying their involvement in defense responses remain largely unknown. To further clarify the functions of Arabidopsis VQ genes in plant defense, we chose VQ12 and VQ29 for further investigation. VQ12 and VQ29 were strongly induced by JA treatment and B. cinerea infection; and the proteins encoding by VQ12 and VQ29 were exclusively localized in the nucleus. Phenotypic analysis indicated that the resistance of vq29 mutant plants to B. cinerea was enhanced compared with that of wild type. Moreover, decreasing the expression of VQ12 and VQ29 simultaneously conferred the amiR-vq12 vq29 double mutant plants even greater resistance against B. cinerea. In contrast, the transgenic plants overexpressing VQ12 or VQ29 were much more susceptible to B. cinerea. Further investigation revealed that VQ12 and VQ29 physically interacted with themselves and each other to form homodimers and heterodimer. Taken together, our results provide evidence that VQ12 and VQ29 negatively regulate plant basal resistance against B. cinerea.
Results
VQ12 and VQ29 genes are strongly responsive to B. cinerea
Arabidopsis VQ12 (AT2G22880) and VQ29 (AT4G37710) encode two VQ motif-containing proteins with 114 and 123 amino acids, respectively27. To characterize their biological functions, we generated homozygous T3 lines of promoterVQ12:GUS and promoterVQ29:GUS transgenic plants. β-Glucuronidase (GUS) staining showed that VQ12 was mainly expressed in the root, leaf, hypocotyl and silique base (Fig. 1A), which is similar to the basic expression pattern of VQ2937. To determine the expression profiles of VQ12 and VQ29 more precisely, we further analyzed their induced expression in response to various defense-related hormones. As shown in Fig. 1B, expression of VQ12 was induced by methyl jasmonate (MeJA) and SA, but not by abscisic acid (ABA) and 1-aminocyclopropane-1-carboxylate (ACC). Similarly, the expression level of VQ29 was also upregulated by MeJA treatment (Fig. 1C). Further quantitative RT-PCR (qRT-PCR) analysis showed that the VQ12 and VQ29 transcripts accumulated high levels in B. cinerea-infected plants (Fig. 2A); and these results were confirmed by GUS staining, as high GUS activities were detected in leaves of promoterVQ12:GUS and promoterVQ29:GUS transgenic plants after B. cinerea infection (Fig. 2B). However, the expression of VQ12 and VQ29 was not responsive to PstDC3000 infection (Fig. 2C). Together, these results indicate that VQ12 and VQ29 mainly respond to MeJA and B. cinerea-infection and may be involved in disease resistance against B. cinerea.
Analysis of VQ12 and VQ29 expression.
(A) GUS staining of whole eight-day-old Arabidopsis transgenic seedlings and various tissues expressing the GUS reporter gene under the control of the VQ12 promoter. (B,C) qRT-PCR analysis of VQ12 (B) and VQ29 (C) expression in response to defense-related hormones. Total RNA was extracted from thirty-day-old wild-type plants at given times after spraying with H2O, MeJA (100 μM), SA (1 mM), ACC (2 mM) or ABA (100 μM). Error bars indicate SD from three independent RNA extracts; statistics by Student’s t test; *p < 0.05; **p < 0.01.
Pathogen-induced expression and subcellular localization of VQ12 and VQ29.
(A) qRT-PCR analysis of VQ12 and VQ29 expression in response to B. cinerea. Total RNA was extracted from thirty-day-old wild-type plants (WT) at given times after spraying with B. cinerea or Sabouraud maltose broth (SMB) buffer (CK). Error bars indicate SD from three independent RNA extracts. (B) GUS staining of WT, proVQ12-GUS and proVQ29-GUS leaves treated with B. cinerea or SMB buffer (CK) for 24 h. (C) qRT-PCR analysis of VQ12 and VQ29 expression in response to P. syringae. Total RNA was extracted from thirty-day-old wild type at given times after infiltration with a suspension of PstDC3000 or MgCl2 (CK). Error bars indicate SD from three independent RNA extracts; statistics by Student’s t test; *p < 0.05; **p < 0.01. (D) Subcellular localization of VQ12 and VQ29 proteins. VQ12-GFP, VQ29-GFP and free GFP were transformed into N. benthamiana epidermal cells. DAPI staining marks the nucleus.
To determine the properties of VQ12 and VQ29 in more detail, we next analyzed their subcellular localizations. The full-length VQ12 and VQ29 were fused to the green fluorescent protein (GFP) protein under the control of the Cauliflower mosaic virus (CaMV) 35S promoter and these constructs were transiently expressed in leaves of tobacco (Nicotiana benthamiana). As shown in Fig. 2D, the transiently expressed VQ12-GFP and VQ29-GFP fused proteins displayed fluorescence exclusively in the nucleus, as revealed by 4’,6-diamidino-2-phenylindole (DAPI) staining. In the control, free GFP was observed in both the cytoplasm and the nucleus (Fig. 2D). These observations indicate that VQ12 and VQ29 are nuclear proteins and may function in the nucleus.
Decreasing the expression of VQ12 and VQ29 simultaneously enhances plant resistance against B. cinerea
To characterize the function of VQ12 in plant defense against B. cinerea, we generated amiR-vq12 transgenic plants by using an artificial miRNA approach to repress VQ12 expression38. qRT-PCR analysis showed that the transcripts of VQ12 in amiR-vq12 transgenic lines 5 and 7 (amiR-vq12–5 and amiR-vq12–7) were reduced compared with those in wild-type plants with or without MeJA treatment (Supplementary Figure S1). We also identified a T-DNA insertion mutant for VQ29 from the Salk T-DNA population. vq29 (Salk_061438) mutant harbors a T-DNA insertion in the promoter region (−136 bp from the translation start site) of VQ29 (Supplementary Figure S2)37. Further examination indicated that the expression of VQ29 was significantly decreased in vq29 compared with that in wild type (Supplementary Figure S2)37. To clarify the possible functional cooperation between VQ12 and VQ29, we generated amiR-vq12 vq29 double mutant plants by crossing amiR-vq12–5 with vq29.
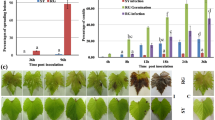
To determine the possible functions of VQ12 and VQ29 in plant defense, we analyzed the performance of amiR-vq12 lines, vq29 and amiR-vq12 vq29 in response to B. cinerea infection. Thirty-day-old plants were challenged with a B. cinerea spore suspension (5 × 105 spores/ml). As shown in Fig. 3A, no significant difference between the amiR-vq12 lines and wild type in disease symptom development was observed. However, following infection with B. cinerea, the vq29 mutant plants showed reduced disease symptom with restricted disease lesions in comparison with wild-type plants (Fig. 3A). Moreover, the amiR-vq12 vq29 double mutant plants were substantially more resistant against B. cinerea compared with vq29 and wild type (Fig. 3A). To confirm these disease symptoms, we quantified the biomass of the infecting pathogen by examining the transcripts of β-tubulin gene of B. cinerea in inoculated plants. As shown in Fig. 3B, lower levels of β-tubulin mRNA of B. cinerea was detected in vq29 and amiR-vq12 vq29 plants at 4 d after post-inoculation (dpi). Since vq29 and amiR-vq12 vq29 plants acquired more resistance against B. cinerea, the pathogen-induced expression of PDF1.2, the antifungal gene THIONIN2.1 (Thi2.1) and the pathogenesis response gene PR4, were also analyzed34,39,40. Compared to those in wild-type plants, PDF1.2, Thi2.1 and PR4 transcripts were increased in vq29 and amiR-vq12 vq29 plants after B. cinerea infection (Fig. 3C). Taken together, these results indicate that decreasing the expression of VQ12 and VQ29 simultaneously enhances plant resistance against B. cinerea infection.
Responses of vq29, amiR-vq12 and amiR-vq12 vq29 plants to B. cinerea.
(A) Leaves of various plants were drop-inoculated with B. cinerea spores (2 × 105 spores/ml). The disease symptoms were photographed at 3dpi. (B) Accumulation of B. cinerea β-tubulin. Total RNA was isolated from inoculated plants at 0 and 4 dpi and the expression levels of β-tubulin were analyzed using B. cinerea β-tubulin gene-specific primers. (C) qRT-PCR analysis of PDF1.2, Thi2.1 and PR4 expression levels. Total RNAs were extracted from B. cinerea-inoculated leaves harvested at 0, 1 and 2 dpi. In (B) and (C) values are mean SE (n = 3 experiments) and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05).
Overexpression of VQ12 or VQ29 confers plants susceptible to B. cinerea
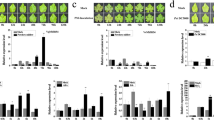
To further investigate the roles of VQ12 and VQ29 in plant defense, we generated transgenic plants overexpressing VQ12 or VQ29 under the control of the CaMV 35S promoter. qRT-PCR analysis showed that several overexpression lines constitutively expressed elevated levels of VQ12 or VQ29 transcripts even without any treatment (Supplementary Figure S3). Two lines of VQ12 (VQ12OX1 and VQ12OX4) and two lines of VQ29 (VQ29OX2 and VQ29OX3) were selected for further study (Supplementary Figure S3). The F2 progeny of those homozygous transgenic plants showed the same morphology as wild-type plants under normal growth conditions for thirty days. Then, we analyzed the performances of those overexpression plants in response to B. cinerea infection. After B. cinerea inoculation, those overexpression plants showed enhanced susceptibility to this fungal pathogen with more obvious leaf decay and greater accumulation of fungal biomass, compared to wild-type plants (Fig. 4A,B). Moreover, the plants constitutively expressing both VQ12 and VQ29 (VQ12OX4/VQ29OX3) simultaneously exhibited even more severe disease symptoms than VQ12OX4, VQ29OX3 and wild type (Fig. 4A,B). Consistent with the disease symptoms, the B. cinerea induced-expression levels of PDF1.2, Thi2.1 and PR4 genes in VQ12 or VQ29 over-expressing plants were reduced compared with those in wild type (Fig. 4C). Thus, overexpression of VQ12 and/or VQ29 increases plant susceptibility to the fungal pathogen B. cinerea.
Responses of transgenic plants overexpressing VQ12 or VQ29 to B. cinerea.
(A) Leaves of various transgenic plants were drop-inoculated with B. cinerea spores and the disease symptoms were photographed at 2 dpi. (B) Accumulation of B. cinerea β-tubulin. Total RNA was isolated from inoculated plants at 0 and 4 dpi and the expression levels of β-tubulin were analyzed using B. cinerea β-tubulin gene-specific primers. (C) qRT-PCR analysis of PDF1.2, Thi2.1 and PR4 expression levels. Total RNAs were extracted from B. cinerea-inoculated leaves harvested at 0, 1 and 2 dpi. In (B) and (C) values are mean SE (n = 3 experiments) and different letters above columns indicate significant differences based on Tukey’s test (P < 0.05).
VQ12 and VQ29 physically interact to form heterodimer and homodimers
To understand how VQ12 and VQ29 modulate plant resistance against B. cinerea, we employed the yeast two-hybrid system to identify their potentially interacting partners. As the full-length VQ12 and VQ29 proteins showed strong transcriptional activation activities, the VQ12 and VQ29 proteins with deleted activation domains (deleted amino acids 1 to 22 for VQ12 and deleted amino acids 1 to 21 for VQ29) were fused to the Gal4 DNA-binding domain of the bait vector (BD-VQ12 and BD-VQ29). After screening, more than 30 independent colonies were isolated. Among these positive clones, clones encoding VQ proteins were frequently represented. To confirm their physical interactions, the full-length coding sequences of all 34 Arabidopsis VQ proteins were cloned and introduced into the prey vector (AD-VQ). The BD-VQ12 or BD-VQ29 and AD-VQ plasmids were co-transformed into yeast. As shown in Fig. 5A, VQ12 strongly interacted with eight VQ proteins (VQ3, VQ8, VQ10, VQ12, VQ17, VQ18, VQ29 and VQ32) and slightly interacted with ten other VQ proteins (VQ1, VQ2, VQ4, VQ5, VQ11, VQ21, VQ22, VQ25 and VQ30) in yeast. Similarly, VQ29 strongly interacted with seventeen VQ proteins (VQ1, VQ3, VQ6, VQ8, VQ9, VQ10, VQ11, VQ12, VQ13, VQ17, VQ18, VQ21 VQ25, VQ26, VQ29, VQ32 and VQ34) and slightly interacted with three other VQ proteins (VQ5, VQ15 and VQ30) (Fig. 5B). To determine whether VQ12 and VQ29 interact with themselves or each other in plant cells, we used BiFC assay for further analysis. Full-length VQ12 and VQ29 proteins were fused to the N-terminal region of the yellow fluorescent protein (nYFP) and C-terminal region of YFP (cYFP). When VQ12-cYFP was co-infiltrated with VQ12-nYFP or VQ29-nYFP in tobacco leaves, strong YFP fluorescence was detected in nuclei (Fig. 5C). Similarly, tobacco leaves co-expressing VQ29-cYFP and VQ29-nYFP also showed strong YFP fluorescence (Fig. 5C). We did not detect any fluorescence in all negative controls (Supplementary Figure S4). These results indicate that VQ12 and VQ29 physically interact to form heterodimer and homodimers.
Physical interactions between VQ proteins.
(A,B) Yeast two-hybrid analysis of VQ12 (A) and VQ29 (B) interactions with all VQ proteins. Interaction was indicated by the ability of cells to grow on selective media lacking Leu/Trp/His/Ade and containing 10 mM 3-aminotriazole. WRKY33 interactions with VQ12 (A) and VQ29 (B) were used as positive controls (P). (C) BiFC assay showing the fluorescence complementation of the C-terminal part of YFP fused with VQ12 or VQ29 and the N-terminal part of YFP fused with VQ12 or VQ29. DAPI staining marks the nucleus.
C-terminal parts are required for VQ12-VQ29 physical interaction
To characterize which domain of VQ12 or VQ29 is responsible for the interactions with themselves or each other, VQ12 and VQ29 were divided into N-terminal parts containing the VQ motif and C-terminal portions (Fig. 6). Moreover, we also mutated the amino acids (LVQR) in VQ motifs of VQ12 and VQ29 to EDLE (Fig. 6). The directed yeast two-hybrid analysis showed that deleting the N-terminal residues or replacing the VQ motifs of VQ12 and VQ29 did not affect the physical interactions with themselves or each other (Fig. 6A,B). However, deletion of the C-terminal parts of VQ12 or VQ29 eliminated those interactions (Fig. 6A,B). To further characterize whether the C-terminal fragments are sufficient for their interactions, the C-terminal portions of VQ12 and VQ29 were cloned and introduced into the prey vector. The yeast two-hybrid results show that the C-terminal part of VQ12 or VQ29 interacted with themselves or each other (Fig. 6C). To further determine the interaction from yeast two-hybrid assay, we used the BiFC assay to analyze their physical interactions. The BiFc results also indicate that the C-terminal parts of VQ12 and VQ29 interacted with themselves or each other in plant cells (Fig. 6D and Supplementary Figure S5).
C-terminal parts are required for VQ proteins to form hetero- or homo-dimers.
(A) The C-terminal part of VQ12 was required for interaction with itself or VQ29. (B) The C-terminal part of VQ29 was essential for interaction with itself or VQ12. (C) The C-terminal fragments of VQ12 and VQ29 are sufficient for their interactions. Interactions were indicated by the ability of cells to grow on selective media lacking Leu/Trp/His/Ade and containing 10 mM 3-aminotriazole. The empty pGADT7 prey vector was used as a negative control. (D) BiFC analyses.
We further re-examined the properties of different domains of VQ12 and VQ29 in more detail by analyzing their sub-cellular localization. Different domains of VQ12 and VQ29 were fused to the GFP protein and these fused proteins were transiently expressed in leaves of tobacco. As shown in Supplementary Figure S6, the transiently expressed VQ12-CT-GFP, VQ29-CT-GFP, VQ12△VQ-GFP and VQ29△VQ-GFP fusion proteins were exclusively localized in the nucleus. These results were consistent with their abilities to interact with themselves or each other in nuclei. However, the VQ12-NT-GFP and VQ29-NT-GFP were localized in both the nucleus and the cytoplasm (Supplementary Figure S6). These results indicate that the C-terminal parts of VQ12 and VQ29 are critical for their nuclear localizations.
VQ12 and VQ29 are partially involved in JA-mediated signaling pathway
In Arabidopsis, several signaling pathways are involved in defense responses, such as the SA-, ET- and JA-signaling pathways. To analyze which pathway VQ12 and VQ29 are involved in, we monitored their expression in various defense-related mutants, including sid2 (SA-biosynthesis mutant), npr1 (SA-signaling mutant), coi1 (JA-signaling mutant) and ein2 (ET-signaling mutant)4,8,12,14,41. Before infection with B. cinerea, the basic expression levels of VQ12 and VQ29 were not affected in those mutants (Fig. 7A,B). When inoculated with B. cinerea, the expression levels of VQ12 and VQ29 were strongly induced in sid2, npr1 and ein2 mutants, as those in wild-type plants. However, their induced-expression levels were remarkably reduced in the coi1 mutant (Fig. 7A,B). As a negative control, their expression levels in sid2 and npr1 mutants after PstDC3000 infection were not altered compared with those in wild type (Supplementary Figure S7). Those observations indicate that the B. cinerea-induced expression of VQ12 and VQ29 may be partially dependent on the JA-signaling pathway.
Partial involvement of VQ12 and VQ29 in JA-signaling pathway.
qRT-PCR analysis of VQ12 (A) and VQ29 (B) expression in response to B. cinerea in wild type (WT) and various defense-related mutants. Total RNA was extracted from thirty-day-old wild-type or mutant plants at 0, 1 and 2 dpi. Error bars indicate SD from three independent RNA extracts; statistics by Student’s t test; *p < 0.05.
Discussion
Recently, several studies demonstrated that members of VQ family are responsive to pathogen and/or defense-related hormones. For example, Cheng et al. showed that a number of Arabidopsis VQ genes responded to SA treatment and/or the bacterial pathogen PstDC3000 infection27. Several VQ genes of Arabidopsis and tomato were also strongly induced by fungal pathogen infection and/or treatment with fungal elicitors26,42. In addition, Kim et al. reported that the expression of several rice VQ genes was up-regulated by the bacterial pathogen Xanthomonas oryzae pv. Oryzae43. These results suggested that VQ proteins may play regulatory roles in plant defense responses; however, direct evidence for their associations with defense responses remains largely limited. Investigating specific fuctions of VQ proteins and VQ-mediated signaling pathways may provide new insight on the molecular basis of plant defense responses.
In this study, we found that VQ12 and VQ29 were highly responsive to JA treatment and B. cinerea infection, but not responsive to PstDC3000 infection (Figs 1 and 2). However, in Fig. 2C, it appears that VQ12 expression is induced by infiltration with MgCl2, indicating that the expression of VQ12 may also be responsive to wounding. Interestingly, accumulating evidence has indicated that some defense-related genes are also wounding-induced. For example, the VQ family member VQ22 which modulates JA-mediated defense responses is also responsive to wounding treatments34. In addition, our earlier study showed that the defense-associated WRKY8 is also strongly induced by wounding44. These results suggest that there may be some associations between wounding and defense responses.
Further phenotypic analysis showed that VQ12 and VQ29 negatively regulate plant resistance against B. cinerea, as the amiR-vq12 vq29 double mutant plants displayed greater resistance compared with vq29 single mutant and wild type (Fig. 3). Consistent with those findings, the induced expression levels of defense-related PDF1.2, Thi2.1 and PR4 genes were increased in B. cinerea-infected amiR-vq12 vq29, compared with that in wild type (Fig. 3). In contrast, transgenic plants constitutively-expressing VQ12 or VQ29 were much more susceptible to B. cinerea (Fig. 4). Taken together, these JA-responsive VQ12 and VQ29 proteins function as negative regulators in plant basal defense against B. cinerea.
Similarly, several other reported VQ proteins, such as VQ5, VQ20, VQ21 and VQ22, also negatively mediate plant basal defense against B. cinerea27,32,34. For example, Hu et al. showed that decreasing the expression of VQ22 gene enhanced plant resistance against B. cinerea34. However, Lai et al. showed that two structurally related VQ16 and VQ23 positively regulate plant defense responses against B. cinerea26. This means that they function as positive regulators of plant defense against B. cinerea. Moreover, several VQ proteins were shown to be involved in plant resistance against herbivorous insects and/or the biotrophic pathogen P. syringae24,27,29,34. The multiple roles of VQ proteins in plant defense may suggest that the signal transduction of defense responses require tight regulation and fine-tuning. It’s possible that VQ proteins play crucial roles in maintaining proper balance of different signaling pathways, resulting in appropriate tolerance against pathogens and/or insects parasitism while minimizing detrimental effects on plant growth. However, the exact molecular mechanisms underlying their involvement in defense responses are still largely unclear. Further researches are required to identify their putative interacting proteins and illustrate the signaling pathways they are involved in.
Over the past several years, several proteins have been reported to physically interact with VQ family members, including WRKYs, MPKs and PIF124,25,27,28,29,37. For example, Cheng et al. revealed that VQ proteins interacted with group I and IIc WRKY transcription factors and their VQ motifs were essential for those interactions27. Interestingly, accumulating evidence has demonstrated that WRKY transcription factors play crucial roles in regulating plant defense responses17. Therefore, the physical interactions between WRKY factors and VQ proteins may provide an important mechanism for regulating plant defense responses. In this study, we further found that VQ12 and VQ29 physically interacted with themselves and several other VQ members to form homodimers and heterodimer (Fig. 5). Further investigation revealed that the C-terminal fragment but not the VQ motif was required for VQ–VQ protein interactions (Fig. 6). Thus, it’s possible that VQ12 and VQ29 interact with themselves or each other via their C-terminal parts, while interact with WRKY33 via the VQ motif to form a big protein complex to mediate plant defense responses against B. cinerea. Nevertheless, future experiments are needed to analyze the regulatory effect of VQ12 and/or VQ29 on WRKY33 factor in modulating defense responses.
The phytohormone JA, as a crucial defense signal, positively regulates plant resistance against necrotrophic pathogens and herbivorous insects. The JA receptor COI1 has been shown to play critical roles in plant defense responses, as its mutations result in enhanced susceptibility to necrotrophic pathogens and herbivorous insects12,13,14. In this study, we observed that, after infection with B. cinerea pathogen, the coi1 mutant plants showed decreased expression of VQ12 and VQ29, compared to wild type (Fig. 7). This observation suggested that the B. cinerea-induced expression of VQ12 and VQ29 is involved in JA-mediated signaling pathway. Similarly, Hu et al. showed that the induced expression of VQ22 was also associated with JA signaling34. Interestingly, further analysis in their study revealed that the regulation or degradation of VQ22 protein was also involved in JA signaling. Therefore, JA signaling plays dual roles to regulate VQ22 during defense responses. It’s interesting to illustrate the exact molecular mechanisms underlying the regulation of VQ members by JA signaling in future studies.
Materials and Methods
Materials and Arabidopsis growth conditions
The vq29 (Salk_061438) mutant and all transgenic plants used in this study were derived from the Arabidopsis Columbia (Col-0) ecotype. Arabidopsis plants were grown in growth chambers at 22°C with a 10-h light/14-h dark photoperiod. The mutant lines npr1, sid2, coi1 and ein2 were kindly gifted by Dr. Zhixiang Chen (Purdue University, USA). The plant hormones MeJA, SA, ABA and ACC were purchased from Sigma-Aldrich. Taq DNA polymerase was purchased from TaKaRa Biotechnology Co. Ltd. Other common chemicals were obtained from Shanghai Sangon Biotechnology Co. Ltd.
Pathogen infection and induction treatments
PstDC3000 infection was performed as described in21. Collection of B. cinerea spores and plant inoculation was performed as described previously44. For GUS staining and single-leaf drop inoculations, a single 3-μl drop of a suspension of 2 × 105 spores/ml in Sabouraud maltose broth (SMB) buffer was placed on each leaf. Induction treatments with the plant hormones MeJA, SA and ACC were performed as described in Chen et al.44.
Subcellular localization
Full-length VQ12 and VQ29 fused to GFP were cloned into the pOCA30 binary vector, downstream of the CaMV 35S promoter. The constructs were then transformed into Agrobacterium tumefaciens strain EHA105. For transient expression, N. benthamiana leaves were infiltrated with the bacterial cell suspensions (optical density at 600 nm [OD600] = 0.05, 10 mM MES, 10 mM MgCl2 and 100 mM acetosyringone). After infiltration, the plants were incubated at 24 °C for 4 h and then the infected leaves were sectioned for observation. GFP and DAPI fluorescence were observed under a confocal laser scanning microscope (Olympus, Japan).
Identification of T-DNA insertion mutants and construction of transgenic plants
The vq29 mutant contains a T-DNA insertion in the promoter of VQ29 gene. We confirmed the T-DNA insertion by PCR using a combination of a T-DNA border primer (5′-AAACGTCCGCAATGTGTTAT-3′) and a gene-specific primer (vq29m-A: 5′-GATCAAGGAATCCGATTAGATTCA-3′; vq29m-B: 5′- GAAGGTTGTTTACGTGTCGAATG-3′). The insertion mutant was further confirmed by qRT-PCR. To generate the VQ12 and VQ29 overexpression constructs, the full-length VQ12 and VQ29 gene coding sequences were PCR-amplified from Arabidopsis gDNA and cloned into the pOCA30 vector in the sense orientation behind the CaMV 35S promoter45. To suppress VQ12 expression we used an artificial miRNA approach as described in Liang et al.38. An amiR-vq12 sequence was designed using WMD3 (http://wmd3.weigelworld.org), with an AthmiR319a backbone to drive its expression38,46. Then, the amiR-vq12 precursor was cloned into the pOCA30 vector. All constructs were transformed into Arabidopsis plants using the Agrobacterium-mediated flower dip method. Homozygous transgenic amiR-vq12–5 line were crossed with vq29 to generate amiR-vq12 vq29 homozygous plants. VQ12OX4/VQ29OX3 plants were generated through genetic crosses of VQ12OX4 and VQ29OX3 homozygous transgenic lines.
RNA extraction and qRT-PCR
Total RNA was extracted using the Trizol reagent (Invitrogen). One microgram of DNase-treated RNA was reverse-transcribed in a 20 μl reaction mixture using Superscript II (Invitrogen). After the reaction, 1 μl cDNA was subjected to qRT-PCR using a SYBR Premix Ex Taq kit (Takara) on a Roche LightCycler 480 real-time PCR machine. At least three independent biological samples were used for qRT-PCR analysis for each reported result. ACTIN2 (AT3G18780) was used as an internal control. The gene-specific primers for qRT-PCR are listed in Supplementary Table S1.
GUS reporter analysis
The putative promoters of VQ12 and VQ29 were amplified from genomic DNA using gene-specific primers (Supplementary Table S1). The constructs proVQ12:GUS and proVQ29:GUS were cloned into the pOCA28 binary vector. Transgenic plants were subjected to GUS staining as described in Chen et al.44.
Yeast two-hybrid screening and confirmation
The truncated VQ12 and VQ29 CDSs were cloned into the bait vector pGBKT7 and then transformed into the yeast strain Y2HGold (Clontech). The cDNA library was obtained from Clontech (Catalog number 630487). Yeast two-hybrid screening was performed as described in Hu et al.47. To confirm protein–protein interactions, full-length CDSs of all 34 VQ proteins were cloned into the prey vector pGADT7. Primers used for amplifying these fragments for yeast two-hybrid assays are listed in Supplementary Table S1.
Bimolecular fluorescence complementation (BiFC) assay
The cDNA sequences for the N-terminal 173-amino acid eYFP (N-YFP) and C-terminal 64-amino acid (C-YFP) fragments were cloned into pFGC5941 to generate pFGC-nYFP and pFGC-cYFP, respectively28. Full-length coding sequences of VQ12 and VQ29 were cloned into pFGC-nYFP to generate N-terminal in-frame fusions with N-YFP, while VQ12 and VQ29 CDSs were inserted into pFGC-C-YFP to form C-terminal in-frame fusions with C-YFP. The plasmids were transformed into A. tumefaciens strain EHA105 and infiltration of N. benthamiana was performed as described previously48. Infected leaves were analyzed at 48 h after infiltration. YFP fluorescence and DAPI staining were observed under a confocal laser scanning microscope (Olympus, Japan).
Additional Information
Accession codes: Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: VQ12 (AT2G22880), VQ29 (AT4G37710), PDF1.2 (AT5G44420), VQ1 (AT1G17147), VQ2 (AT1G21320), VQ3 (AT1G21326), VQ4 (AT1G28280), VQ5 (AT1G32585), VQ6 (At1G32610), VQ7 (AT1G35830), VQ8 (AT1G68450), VQ9 (AT1G78310), VQ10 (AT1G78410), VQ11 (AT1G80450), VQ13 (AT2G33780), VQ14 (AT2G35230), VQ15 (AT2G41010), VQ16 (AT2G41180), VQ17 (AT2G42140), VQ18 (AT2G44340), VQ19 (AT3G15300), VQ20 (AT3G18360), VQ21 (AT3G18690), VQ22 (AT3G22160), VQ23 (AT3G56710), VQ24 (AT3G56880), VQ25 (AT3G58000), VQ26 (AT3G60090), VQ27 (AT4G15120), VQ28 (AT4G20000), VQ30 (AT4G39720), VQ31 (AT5G08480), VQ32 (AT5G46780), VQ33 (AT5G53830), VQ34 (AT5G65170).
How to cite this article: Wang, H. et al. Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci. Rep. 5, 14185; doi: 10.1038/srep14185 (2015).
References
Jones, J. D. & Dangl, J. L. The plant immune system. Nature 444, 323–329 (2006).
Chisholm, S. T., Coaker, G., Day, B. & Staskawicz, B. J. Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814 (2006).
Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227 (2005).
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S. & Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63 (1997).
Durrant, W. E. & Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209 (2004).
Penninckx, I. A., Thomma, B. P., Buchala, A., Metraux, J. P. & Broekaert, W. F. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113 (1998).
Browse, J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 60, 183–205 (2009).
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S. & Ecker, J. R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152 (1999).
Thomma, B. P., Eggermont, K., Tierens, K. F. & Broekaert, W. F. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1102 (1999).
Chen, H. et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21, 2527–2540 (2009).
Boutrot, F. et al. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. 107, 14502–14507 (2010).
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M. & Turner, J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094 (1998).
Yan, J. et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236 (2009).
Sheard, L. B. et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405 (2010).
Ulker, B. & Somssich, I. E. WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498 (2004).
Eulgem, T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78 (2005).
Pandey, S. P. & Somssich, I. E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655 (2009).
Rushton, P. J., Somssich, I. E., Ringler, P. & Shen, Q. J. WRKY transcription factors. Trends Plant Sci. 15, 247–258 (2010).
Pierre, B. & Susana, R. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46 (2014).
Zheng, Z., Abu-Qamar, S., Chen, Z. & Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605 (2006).
Hu, Y., Dong, Q. & Yu, D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci. 185–186, 288–297 (2012).
Xu, X., Chen, C., Fan, B. & Chen, Z. Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors. Plant Cell 18 1, 310–1326 (2006).
Journot-Catalino, N., Somssich, I. E., Roby, D. & Kroj, T. The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302 (2006).
Andreasson, E. et al. The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589 (2005).
Wang, A. et al. The VQ motif protein IKU1 regulates endosperm growth and seed size in Arabidopsis. Plant J. 63, 670–679 (2010).
Lai, Z. et al. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 23, 3824–3841 (2011).
Cheng, Y. et al. Structural and functional analysis of VQ motif containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol. 159, 810–825 (2012).
Hu, Y. et al. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 74, 730–745 (2013).
Pecher, P. et al. The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-containing proteins to regulate immune responses. New Phytol. 203, 592–606 (2014).
Xie, Y. D. et al. The Arabidopsis gene SIGMA FACTOR-BINDING PROTEIN 1 plays a role in the salicylate- and jasmonate-mediated defence responses. Plant, Cell & Environ. 33, 828–839 (2010).
Qiu, J. L. et al. Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221 (2008).
Petersen, K. et al. Arabidopsis MKS1 is involved in basal immunity and requires an intact N-terminal domain for proper function. PLoS One 5, e14364 (2010).
Morikawa, K., Shiina, T., Murakami, S. & Toyoshima, Y. Novel nuclear-encoded proteins interacting with a plastid sigma factor, Sig1, in Arabidopsis thaliana. FEBS Lett. 514, 300–304 (2002).
Hu, P. et al. JAV1 controls jasmonate-regulated plant defense. Mol. Cell 50, 504–515 (2013).
Perruc, E. et al. A novel calmodulin-binding protein functions as a negative regulator of osmotic stress tolerance in Arabidopsis thaliana seedlings. Plant J. 38, 410–420 (2004).
Garcia, D. et al. Arabidopsis haiku mutants reveal new controls of seed size by endosperm. Plant Physiol. 131, 1661–1670 (2003).
Li, Y., Jing, Y., Li, J., Xu, G. & Lin, R. Arabidopsis VQ MOTIF-CONTAINING PROTEIN29 represses seedling deetiolation by interacting with PHYTOCHROME-INTERACTING FACTOR1. Plant Physiol. 164, 2068–2080 (2014).
Liang, G., He, H., Li, Y. & Yu, D. A new strategy for construction of artificial miRNA vectors in Arabidopsis. Planta 235, 1421–1429 (2012).
Reymond, P. & Farmer, E. E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411 (1998).
Reymond, P., Weber, H., Damond, M. & Farmer, E. E. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720 (2000).
Wildermuth, M. C., Dewdney, J., Wu, G. & Ausubel, F. M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565 (2001).
Durrant, W. E., Rowland, O., Piedras, P., Hammond-Kosack, K. E. & Jones, J. D. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12, 963–977 (2000).
Kim, D. Y. et al. Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 529, 208–214 (2013).
Chen, L., Zhang, L. & Yu, D. Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant-Microbe Interact. 23, 558–565 (2010).
Chen, C. & Chen, Z. Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716 (2002).
Liang G., He H., Li Y., Ai Q. & Yu D. MYB82 functions in regulation of trichome development in Arabidopsis. J. Exp. Bot. 65, 3215–3223 (2014).
Hu, Y., Jiang, L., Wang, F. & Yu, D. Jasmonate Regulates the INDUCER OF CBF EXPRESSION–C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 25, 2907–2924 (2013).
Hu, Y. & Yu, D. BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26, 4394–4408 (2014).
Acknowledgements
We thank Dr. Zhixiang Chen (Purdue University, USA) for Arabidopsis sid2, npr1, coi1 and ein2 mutants and Arabidopsis Resource Center at Ohio State University for vq29 mutant. This work was supported by the Natural Science Foundation of China (31401040 to Yanru Hu), the Youth Innovation Promotion Association CAS (to Yanru Hu), the CAS “Light of West China” Program (to Yanru Hu) and Yunnan provincial government (2014HC017 to Diqiu Yu).
Author information
Authors and Affiliations
Contributions
H.P.W., Y.R.H. and D.Q.Y. designed the experiments. H.P.W., Y.R.H. and J.J.P. performed the experiments and analyzed the data. H.P.W. and Y.R.H wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, H., Hu, Y., Pan, J. et al. Arabidopsis VQ motif-containing proteins VQ12 and VQ29 negatively modulate basal defense against Botrytis cinerea. Sci Rep 5, 14185 (2015). https://doi.org/10.1038/srep14185
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep14185
This article is cited by
-
The role and pathway of VQ family in plant growth, immunity, and stress response
Planta (2024)
-
Genome-wide identification and expression analysis of VQ gene family under abiotic stress in Coix lacryma-jobi L.
BMC Plant Biology (2023)
-
Genome-wide analysis of the VQ motif-containing gene family and expression profiles during phytohormones and abiotic stresses in wheat (Triticum aestivum L.)
BMC Genomics (2022)
-
Characterization of the wheat VQ protein family and expression of candidate genes associated with seed dormancy and germination
BMC Plant Biology (2022)
-
Identification and expression analysis of phospholipase C family genes between different male fertility accessions in pepper
Protoplasma (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.