Abstract
The transcription factors OCT4 and SOX2 are required for generating induced pluripotent stem cells (iPSCs) and for maintaining embryonic stem cells (ESCs). OCT4 and SOX2 associate and bind to DNA in different configurations depending on the arrangement of their individual DNA binding elements. Here we have investigated the role of the different OCT4-SOX2-DNA assemblies in regulating and inducing pluripotency. To this end, we have generated SOX2 mutants that interfere with specific OCT4-SOX2 heterodimer configurations and assessed their ability to generate iPSCs and to rescue ESC self-renewal. Our results demonstrate that the OCT4-SOX2 configuration that dimerizes on a Hoxb1-like composite, a canonical element with juxtaposed individual binding sites, plays a more critical role in the induction and maintenance of pluripotency than any other OCT4-SOX2 configuration. Overall, the results of this study provide new insight into the protein interactions required to establish a de novo pluripotent network and to maintain a true pluripotent cell fate.
Similar content being viewed by others
Introduction
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) following the overexpression of four transcription factors1, the most critical of which are Oct4 and Sox22. In recent years, much effort has been invested in uncovering the function of the reprogramming factors during the induction of pluripotency. CHIP-seq and global gene expression analyses have shown that the cooperative co-binding of exogenous OCT4, SOX2 and KLF4 activates many endogenous pluripotent regulators3. However, antagonistic competition among these factors has also been described and proven essential in the generation of iPSCs4. At the onset of reprogramming, the four overexpressed proteins are present at much higher levels than under physiological conditions. As a consequence, the reprogramming factors bind to both high- and low-affinity sites in accessible chromatin regions5. Overall, the initial phase of pluripotency induction is characterized by stochastic gene expression, increasing the complexity of the process and further complicating mechanistic studies5. In contrast, the final phase of reprogramming is determined by hierarchical events that involve the gradual activation of OCT4-SOX2 target genes, a process that will finally lead to the establishment of the pluripotent transcriptional network5.
OCT4 and SOX2 co-bind to the regulatory regions of most of their target genes6,7. OCT4 has a bi-partite POU domain that interacts with the major groove of the DNA8, while SOX2 has a high-mobility group (HMG) domain that interacts with the minor groove of DNA9. To accomplish their synergistic role, the OCT4 POU and SOX2 HMG domains need to assemble on closely spaced composites of ATGCAAAT-like and CATTGTC-like DNA binding sites. Predominantly, OCT4 and SOX2 cooperate to bind to a canonical composite in which their individual binding sites are juxtaposed7,10,11. This canonical composite motif, which is present in the enhancers of genes such as Utf1 and Hoxb1, can be found with minor variations in the nucleotide sequence9,11. In addition, composites with different number of spacer nucleotides between the Sox2 and Oct4 DNA binding sites have also been characterized, from which the three spacer nucleotide composite present in the Fgf4 enhancer has been mostly studied12. As previously reported, the number of spacer nucleotides determines the OCT4-SOX2 protein interactions9. Interestingly, SOX2 mutants that interfere with the formation of a specific OCT4-SOX2 heterodimer configuration have been described9. In this study, we have taken advantage of these SOX2 mutants to investigate the role played by the different OCT4-SOX2-DNA assemblies in the induction of iPSCs and the maintenance of embryonic stem cell (ESC) self-renewal. Our results show that the OCT4-SOX2 configuration formed on the canonical composite plays the most critical role in pluripotency.
Results
The different OCT4-SOX2 heterodimer configurations do not exhibit equivalent roles in reprogramming
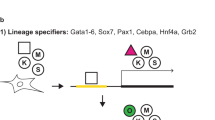
To synergistically activate their target genes, OCT4 and SOX2 proteins interact forming different protein-protein interaction interfaces according to the arrangement of their individual DNA binding motifs (Fig. 1)9,13. Indeed, several point mutations in the SOX2 HMG domain have been described to specifically abolish the OCT4-SOX2 interaction on the Utf1-like motif (HWTTSWNATGYWDGD), in which the Oct4 and Sox2 DNA binding sites are juxtaposed, or on the Fgf4-like motif (HWTTSWNNNNATGYWDWD), in which a three spacer nucleotide separates the Oct4 and Sox2 DNA binding sites (Fig. 1)9. The combination of the K95E and R98E (S95/98 mutant) or the R98E and M102E (S98/102 mutant) mutations prevents heterodimer formation on Utf1-like motifs (Fig. 1A and 2A). In contrast, the R113E mutation (S113 mutant) impedes formation of the OCT4-SOX2 surface interaction required for binding to enhancers containing Fgf4-like motifs (Figs 1B and 2A). Thus, we sought to assess the impact of these point mutations on the reprogramming of somatic cells into iPSCs. To this end, we generated SOX2 constructs containing the different sets of mutations. Retroviral defective particles coding for the different SOX2 mutants plus OCT4 and KLF4 were used to transduce Oct4-GFP mouse embryonic fibroblasts (MEFs). Our results show that the S113 mutant decreased the reprogramming ability 2-fold in comparison with the wild-type construct, as shown by the number of GFP+ colonies (Fig. 2B). In case of S98/102, only one iPSC colony was observed in 1 out of 3 independent wells transduced in parallel. In a subsequent experiment, only a single colony was also observed in 1 out of 3 independent wells, indicating that the reprogramming efficiency of the S98/102 mutant is consistently low. However, we could not detect any iPSC colonies using the S95/98 construct. Quantitative Real-Time PCR (qRT-PCR) showed that all SOX2 mutants were similarly expressed in MEFs (Fig. 2C). Two colonies of SWT, S113 and S98/102 were picked and expanded. All the stable cell lines expressed Oct4-GFP, stained positive for alkaline phosphatase (AP) (Fig. 2D) and expressed the endogenous pluripotent marker genes Oct4, Sox2, Nanog, Utf1 and Fgf4 at the same levels as ESCs (Fig. 2E). Overall, our results demonstrate that the Utf1-like OCT4-SOX2 heterodimer configuration plays a more critical role in reprogramming than the Fgf4-like configuration.
Modeling the OCT4-SOX2 interactive configurations.
The Utf1-like (A) or the Fgf4-like (B) composite is used as a DNA template and is represented in gray. The OCT4 POUS, the OCT4 POUHD and the SOX2 HMG domains are depicted in green, blue and orange, respectively. The amino acids mutated in this study are indicated in both configurations. To highlight the differences between both arrangements, the orientation of OCT4 is kept the same in both DNA templates.
The OCT4-SOX2 configuration plays an important role in reprogramming.
(A) Human SOX2 amino acid alignment indicating the position of the point mutations included in each construct. (B) iPSC reprogramming efficiency generated using each different SOX2 mutant and measured by the number of Oct4-GFP positive colonies. Error bars correspond to standard deviations between three independent biological replicates. (C) SOX2-transgene expression in MEFs measured by qRT-PCR three days after transduction. Values are normalized to the expression of C- that corresponds to non-transduced MEFs and that is considered as 1. The error bars correspond to the standard deviation between three technical replicates. (D) Images showing Oct4-GFP expression and AP staining of one independent iPSC clonal cell line generated from each SOX2 construct. (E) Expression of endogenous pluripotency markers was measured by qRT-PCR using two mESC lines and MEFs as positive and negative controls, respectively. Values are normalized to the expression of ESC line #1, which is considered as 1. The error bars correspond to standard deviations between three technical replicates.
A specific OCT4-SOX2 heterodimer configuration is essential for preventing ESC differentiation
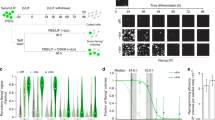
Next we sought to ascertain the impact of the different OCT4-SOX2-DNA assemblies on the maintenance of pluripotency and ESC self-renewal. Accordingly, we transfected Sox2-null ESCs, in which Sox2 is expressed from a transgene that can be repressed upon doxycycline induction14, with hygromycin-resistant PiggyBac vectors expressing the SOX2 mutants. One week after addition of hygromycin and doxycycline, the index rescue that corresponds to the number of AP-positive colonies in the presence of each SOX2 construct divided by the number of AP-positive colonies in the presence of the empty PiggyBac vector was calculated. In the presence of S113, a 4-fold reduction in the number of AP-positive colonies was observed (Fig. 3A). In contrast, the reduction was more pronounced in the presence of the S98/102 mutant and no colonies were observed in the presence of the S95/98 mutant. Moreover, we performed a western blot to assess whether the different SOX2 mutants were expressed at equivalent levels. As Sox2-null ESCs transfected with S95/98 did not proliferate, we transfected the different PiggyBac constructs in 293T cells. The western blot analysis showed that all constructs expressed similar levels of SOX2 protein (Fig. 3B). Single colonies from the Sox2-null ESCs rescued with SWT, S113 and S98/102 were picked and stable cell lines were established (Fig. 3C). SWT and S113 colonies were found to be morphologically identical. However, S98/102 colonies looked smaller, less compact and more differentiated (Fig. 3C). Furthermore, we investigated the global gene expression profile of SWT, S113 and S98/102 clonal cell lines. Pairwise scatter plots of an average of two S113 clones versus two SWT clones and of an average of three S98/102 clones versus two SWT clones showed that S113 exhibit a higher level of similarity to SWT than S98/102 clones (Fig. 3D). Indeed, S113 clones express pluripotent gene markers at levels comparable to SWT (Fig. 3E). In contrast, S98/102 clones display lower expression levels for some of these pluripotent genes (Fig. 3E), thus confirming that S98/102 Sox2-null rescued lines are prone to differentiation as suggested by their morphology (Fig. 3C). Therefore, our results demonstrate that interfering with the Utf1-like OCT4-SOX2 heterodimer configuration abolishes (S95/98) or diminishes (S98/102) the maintenance of pluripotency, whereas interfering with the Fgf4-like OCT4-SOX2 configuration also decreases the rescue of ESC self-renewal but with a lower impact on ESC morphology and global gene expression profile.
A specific OCT4-SOX2 configuration is essential for preventing ESC differentiation.
(A) Self-renewal rescue experiment. Sox2-null ESCs were transfected with hygromycin-resistant PiggyBac plasmids coding for the different SOX2 constructs, the empty vector was used as a negative control. The transfected Sox2-null ESCs were then treated with doxycycline and hygromycin for 1 week prior to AP staining. The rescue index was calculated by dividing the number of AP-positive colonies for each construct by the number of AP-positive colonies in the empty PiggyBac vector. Error bars correspond to standard deviations between three independent biological replicates. (B) Western blot analysis showing that all SOX2 constructs express equal levels of SOX2 protein after overexpression in 293T cells. α-TUBULIN is used as loading control. (C) Images of clonal cell lines established for each construct expressing the constitutive dsRed protein transgene are shown. (D) Pairwise scatter plots of global gene expression profiles comparing the average of two S113 clonal cell lines or three S98/102 clonal cell lines versus two SWT clonal cell lines. Black lines indicate a two-fold change in gene expression level between the paired populations. Color bar on the right indicates scattering density. Genes up- and downregulated are shown by red and green circles, respectively. (E) Heat map from microarray data showing the expression of a subset of pluripotent marker genes in stable Sox2-null ESC lines transfected with the different constructs. Values correspond to the average of two SWT-, two S113- and three S98/102-clonal Sox2-null ESC lines. The color bar at the top indicates gene expression in log2 scale. Blue and orange colors represent higher and lower gene expression levels, respectively.
The abundance of the bound composite motif determines the impact of each OCT4-SOX2 configuration on pluripotency
Point mutations preventing the Utf1-like heterodimer assembly have a higher impact on the induction and maintenance of a pluripotent state than the point mutation preventing the assembly on the Fgf4-like motif composites. An explanation for this observation could be that: i) OCT4-SOX2 target genes containing a Utf1-like motif are more relevant to pluripotency than the genes containing an Fgf4-like motif, or ii) pluripotent cells express more genes exhibiting a Utf1-like element than genes presenting an Fgf4-like element. Using a previous CHIP-seq study in which 3,798 loci co-bound by OCT4 and SOX2 were identified in mouse ESCs11, we quantified the number of Utf1- and Fgf4-like motifs using degenerate motif ‘words’ (Fig. 4A). In this analysis, we also included the canonical Hoxb1-like motif that differs from the Utf1-like motif in only 1 nucleotide. Importantly, OCT4 can selectively recognize this nucleotide position15. Hoxb1-like motifs (554 binding motifs) were more enriched than Utf1-like motifs (47 binding motifs) and Fgf4-like motifs (26 binding motifs). The distribution of the composite sequences in a 100-bp window centered at the peak summit is shown in Fig. 4A. As the Hoxb1-like motif is the most abundant (Fig. 4A), we sought to investigate the capacity of the mutants to heterodimerize with OCT4 on the Hoxb1 binding composite. In contrast to Remenyi et al.’s EMSA analysis, which was conducted with only the OCT4 and SOX2 DNA binding domains9, we used full-length proteins. Our EMSA results show that the S113 mutant can heterodimerize with OCT4 and assemble on the Hoxb1- and Utf1-like OCT4-SOX2 element, but, in contrast to Remenyi et al.9, S113 could also interact weakly with OCT4 and bind to the Fgf4-like OCT4-SOX2 element (Fig. 4B). The S98/102 mutation abrogates the association of both proteins on the Utf1-like motif but not on the Hoxb1- and Fgf4-like motifs (Fig. 4B). Finally, S95/98 can assemble with OCT4 on Fgf4-like motifs but not on Utf1-like motifs, as previously described9. Our results also demonstrate that S95/98 prevents the cooperative binding of SOX2 and OCT4 on Hoxb1-like motifs, as shown by lack of a supershift of the OCT4/DNA complex in the presence of S95/98 (Fig. 4B). Overall, the abundance of co-bound motifs in CHIP-seq peaks and the EMSA results correlate with the reprogramming (Fig. 2) and rescuing (Fig. 3) experiments. Indeed, the S95/98 mutant, which is unable to induce or maintain a pluripotent state, cannot cooperatively bind with OCT4 to the Hoxb1- and Utf1-like motifs that are the most abundant. In addition, the S98/102 mutant, which is severely impaired in iPSC reprogramming and ESC self-renewal rescuing, shows a reduced and an absence of the ternary OCT4/SOX2/DNA complex band on Hoxb1- and Utf1-like motifs, respectively. Furthermore, the S113 mutation has a lower impact on the pluripotent state because Fgf4-like motifs are not enriched and, additionally, binding to these motifs is not fully abolished.
The abundance of the bound composite motif determines the impact of each OCT4-SOX2 configuration on pluripotency.
(A) Distribution of the motif sequences 100 bp around the peak summit obtained from OCT4 and SOX2 co-pulling down CHIP-seq experiments is depicted in black. Gray curves show the distribution in background control sequences selected using homer (http://homer.salk.edu/homer/index.html). Logos of the OCT4-SOX2 degenerate consensus sequences generated using http://weblogo.berkeley.edu/ are shown above. (B) EMSA analysis to evaluate the heterodimerization and binding capacity of OCT4 with wild-type or mutant SOX2 constructs on Hoxb1 (left), Utf1 (center) and Fgf4 (right) motifs. OCT4/SOX2/DNA supershift band, OCT4/DNA band and free DNA are indicated as  ,
,  and
and  , respectively. Of note, full-length SOX2 binding alone cannot be observed in this assay. (C) Pairwise scatter plots exhibiting the expression level of genes co-bounded by OCT4 and SOX2 on Hoxb1-like motif (left), Utf1-like motif (center) and Fgf4-like motif (right). A red and a green line represent a 2-fold expression level increase or decrease in comparison with SWT. Genes up- and downregulated are shown by red and green dots, respectively. Expression levels below 5 were considered as background.
, respectively. Of note, full-length SOX2 binding alone cannot be observed in this assay. (C) Pairwise scatter plots exhibiting the expression level of genes co-bounded by OCT4 and SOX2 on Hoxb1-like motif (left), Utf1-like motif (center) and Fgf4-like motif (right). A red and a green line represent a 2-fold expression level increase or decrease in comparison with SWT. Genes up- and downregulated are shown by red and green dots, respectively. Expression levels below 5 were considered as background.
Finally, we investigated whether the global gene expression abnormalities observed in S98/102 and S113 Sox2-null clonal ESC lines (Fig. 3D,E) were due to impaired OCT4-SOX2 binding. To this end, we identified the genes co-bound by OCT4 and SOX2 on each of the three motifs using data from a previous CHIP-seq study11. Only genes with high-confidence OCT4 and SOX2 peaks within 50 kb of their transcription start site were considered for this analysis. Then, pairwise scatter plots were generated to compare the expression levels of the genes co-bounded by OCT4 and SOX2 on Hoxb1, Utf1 and Fgf4 binding elements (Fig. 4C). In comparison with SWT, a higher number of genes are differentially expressed in S98/102 than in S113 on the Hoxb1-like composite. These results correlate with the weaker supershift band of S98/102 in the EMSA assay (Fig. 4B) and suggest that these changes in gene expression are the result of direct binding. In contrast, the genes differentially expressed between S113 and SWT on the Hoxb1-like composite cannot be explained by direct binding since both constructs showed a similar ternary OCT4-SOX2-DNA band on this motif (Fig. 4B). The OCT4 and SOX2 interaction on the Utf1-like element is abolished by the S98/102 but not by the S113 mutant. Accordingly, the expression of genes containing the Utf1-like motif is almost identical between S113 and SWT (Fig. 4C). In contrast, a subset of genes is expressed more than 2-fold lower in S98/102 than in SWT, indicating a correlation between OCT4-SOX2 cooperative binding and transcriptional activation. In addition, no differences were observed between S98/102 and S113 on Fgf4-like motifs, suggesting that the Fgf4-like composite is not the main element driving transcription on these genes. In summary, our results show that the impact of each heterodimer conformation correlates to the total number of genes in which the OCT4-SOX2 cooperative binding is impaired.
Discussion
The developmental role of OCT4 and SOX2 on specifying and maintaining pluripotency is determined by the ability of these transcription factors to dimerize on the enhancers of pluripotent genes that contain specific composite DNA elements. Our results show that the OCT4-SOX2 configuration formed on the Utf1- and on the Hoxb1-like composite motifs plays a major role in pluripotency. Although the Utf1- and the Hoxb1-like motif sequences differ by only a single nucleotide, S98/102 mutants show a different cooperativity on these two composites. Indeed, S98/102 mutants can cooperate on the Hoxb1-binding motif but not on the Utf1-binding motif. As both Hoxb1 and Utf1 are canonical motifs, the OCT4-SOX2 interaction interface is unlikely to differ on these two sequences. Moreover, the difference between these two sequences is found in the hemisite of the second OCT4 DNA-binding subdomain, which does not interact with SOX2. Therefore, the difference between these two binding motifs might be due to the individual contribution of the amino acids located on the OCT4-SOX2 interface. Interestingly, R98 has been shown to exhibit a negative contribution to the OCT4-SOX2 cooperativity on an idealized canonical motif containing the Hoxb1-like consensus sequence. This negative contribution was higher in absolute magnitude than the positive contribution of M10210, suggesting that the double S98/S102 mutant would actually retain a positive cooperativity on this motif. Similar data for the OCT4-SOX2 cooperativity on the Utf1-like sequence is not available. However, the cooperativity on this motif may have more stringent requirements for the amino acid identities on the OCT4-SOX2 interface due to the non-consensus Utf1-like sequence, an observation that may explain the inability of the double mutant S98/S102 to bind cooperatively with OCT4 on the Utf1-like motif.
The different SOX2 mutants exhibit different iPSC reprogramming efficiencies and different rescue abilities. The inability of the S95/98 mutant to induce pluripotency is consistent with previous reprogramming experiments using an S95 mutant16. Indeed, our S95/98 double mutant completely abrogates the SOX2-OCT4 interaction on the Hoxb1- and the Utf1-binding element, a result that correlates with the decreased heterodimerization ability of the S95 single mutant on this motif16. The S98/102 mutant, which is impaired in interacting with OCT4 only in an Utf1-like protein conformation, is deficient in inducing and maintaining pluripotency. Indeed, Sox2-null ESCs rescued with the S98/102 mutant exhibit a differentiated morphology and express lineage-committed genes from all three germ layers. OCT4 and SOX2 have been described to associate with polycomb group proteins to repress the expression of developmental genes in ESCs17. Our results suggest that lineage-committed genes usually repressed by a Utf1-like OCT4-SOX2 heterodimer conformation are expressed in the S98/102 mutant, thus promoting the differentiation of ESCs.
The OCT4 protein exhibits two DNA binding domains connected by a linker. We have recently reported that specific point mutations in the OCT4 linker abolish reprogramming ability without affecting OCT4 DNA binding properties18. Indeed, we showed that the OCT4 linker might function as a protein-protein interface that recruits epigenetic regulators to OCT4 target genes. It will be interesting to investigate whether different OCT4-SOX2 heterodimer conformations are able to recruit different epigenetic remodeling proteins.
Overall, our results demonstrate that certain OCT4-SOX2 dimer conformations are more relevant than others in reprogramming somatic cells into iPSCs and in maintaining cells in the ESC state. Further studies are required to determine the key genes activated by each conformation, an approach that might eventually help to simplify the complex mechanism underlying pluripotency.
Methods
Retroviral production and mouse iPSC induction
The human SOX2 mutants were cloned into the pMX retroviral vector19. The replicative-defective retroviral particles were produced as previously described20. Briefly, 3 μg of each pMX vector1 together with 3 μg of pCL-Eco21 were transfected into 2 × 106 293T cells using Fugene® in a total volume of 10 mL. The supernatants containing the viral particles were collected after 48 hours, filtered and frozen. 500 μL of OCT4, 500 μL of KLF4 and 250 μL of one of the different SOX2 constructs were used to transduce 5 × 104 MEFs derived from transgenic mice containing an Oct4-GFP transgene in which the GFP transgene expression is driven by the Oct4 regulatory regions22. Two days after transduction, the medium was replaced with ESC medium containing LIF.
iPSC characterization
Quantitative RT-PCR and AP staining were performed as previously described20,23.
Microarray analyses
Microarray analysis was performed as previously reported24,25. The microarray data are available from the GEO (Gene Expression Omnibus) website under accession number GSE65345.
ESC self-renewal assay
Sox2-null ESCs containing a doxycycline inducible Sox2 transgene, a tet transactivator and a constitutive red-fluorescent protein transgene (DsRed) have been previously described14. The different SOX2 mutants were cloned into the hygromycin-resistant PiggyBac plasmid pPBCAG-cHA-IH, which is a modification of the previously reported pGG131 plasmid26. A total of 500 ng of each plasmid together with 500 ng of pCAG-pBase plasmid were co-transfected into Sox2-null ESCs using Lipofectamine 2000. Next, transfected cells were treated with doxycycline (1 μg/mL) and hygromycin (300 μg/mL) for 1 week prior to AP staining. Western blot analysis was performed as previously described27.
Whole-cell lysate generation and electrophoretic mobility shift assay (EMSA)
293T cells were transduced with the same retroviral defective particles used for the reprogramming experiments and whole-cell extracts were prepared as previously described28. Equal protein expression from the different constructs was analyzed by Western blot (data not shown). The protein DNA binding capacity was analyzed by EMSA using a modified version of a previous protocol29. Briefly, 2–3 μg of wild-type or mutant SOX2 protein lysates were incubated with 180 fmol of a P32-labeled double-stranded DNA probe in the presence or absence of equal amounts of OCT4 protein lysates. Binding reactions were incubated on ice for 1 hour and then loaded directly onto 6% native polyacrylamide gels and run at 10 mA (200–300 V) for 2.5 hours. After electrophoresis, the gels were dried on Whatmann paper and visualized by exposure to X-ray film at −80 C. The following probe sequences were used: HOXB1 (GGAGGAAGTGTCTTTGTCATGCTAATGATTGGGGCTCC), FGF4 (GGAGAAGAAAACTCTTTGTTTGGATGCTAATGGGATACTAAGCTCC) and UTF1 (GGAGAAGATGAGAGCCCTCATTGTTATGCTAGTGAAGTGCCAAGCTCC).
OCT4-SOX2 composite binding motif quantification and modeling
Based on a previous OCT4-SOX2 CHIP-seq dataset11, motif ‘word’ searches were performed in glbase30 using fasta-format sequence that span 100-bp centered around the CHIP-seq peak summit. Based on the IUPAC nucleotide nomenclature conventions, the following motifs were used: Fgf4-like (HWTTSWNNNNATGYWDWD), Utf1-like (HWTTSWNATGYWDGD) and Hoxb1-like (HWTTSWNATGYWDWD). Finally, the modeling of the OCT4-SOX2 cooperativity on Utf1- and Fgf4-like motifs was performed as previously described18.
Additional Information
How to cite this article: Tapia, N. et al. Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci. Rep. 5, 13533; doi: 10.1038/srep13533 (2015).
References
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Huangfu, D. et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 26, 1269–1275 (2008).
Sridharan, R. et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 (2009).
Tiemann, U. et al. Counteracting Activities of OCT4 and KLF4 during Reprogramming to Pluripotency. Stem cell Reports 2, 351–365 (2014).
Buganim, Y., Faddah, D. A. & Jaenisch, R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 14, 427–439 (2013).
Boyer, L. A. et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 (2005).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Jerabek, S., Merino, F., Scholer, H. R. & Cojocaru, V. OCT4: dynamic DNA binding pioneers stem cell pluripotency. Biochim. Biophys. Acta 1839, 138–154 (2014).
Remenyi, A. et al. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17, 2048–2059 (2003).
Merino, F. et al. Structural basis for the SOX-dependent genomic redistribution of OCT4 in stem cell differentiation. Structure 22, 1274–1286 (2014).
Aksoy, I. et al. Oct4 switches partnering from Sox2 to Sox17 to reinterpret the enhancer code and specify endoderm. EMBO J. 32, 938–953 (2013).
Ambrosetti, D. C., Scholer, H. R., Dailey, L. & Basilico, C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 275, 23387–23397 (2000).
Williams, D. C., Jr., Cai, M. & Clore, G. M. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem. 279, 1449–1457 (2004).
Masui, S. et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 9, 625–635 (2007).
Nishimoto, M. et al. The embryonic Octamer factor 3/4 displays distinct DNA binding specificity from those of other Octamer factors. Biochem. Biophys. Res. Commun. 302, 581–586 (2003).
Jauch, R. et al. Conversion of Sox17 into a pluripotency reprogramming factor by reengineering its association with Oct4 on DNA. Stem Cells 29, 940–951 (2011).
Lee, T. I. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 (2006).
Esch, D. et al. A unique Oct4 interface is crucial for reprogramming to pluripotency. Nat. Cell Biol. 15, 295–301 (2013).
Kitamura, T. et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 (2003).
Tapia, N. et al. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 3, 1279 (2012).
Naviaux, R. K., Costanzi, E., Haas, M. & Verma, I. M. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70, 5701–5705 (1996).
Yoshimizu, T. et al. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth Diff. 41, 675–684 (1999).
Fischedick, G. et al. Zfp296 is a novel, pluripotent-specific reprogramming factor. PloS One 7, e34645 (2012).
Ko, K. et al. Autologous pluripotent stem cells generated from adult mouse testicular biopsy. Stem Cell Rev. 8, 435–444 (2012).
Ko, K. et al. Brief report: evaluating the potential of putative pluripotent cells derived from human testis. Stem Cells 29, 1304–1309 (2011).
Guo, G. et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development 136, 1063–1069 (2009).
Fischedick, G. et al. Nanog induces hyperplasia without initiating tumors. Stem cell Res. 13, 300–315 (2014).
Muller-Immergluck, M. M., Schaffner, W. & Matthias, P. Transcription factor Oct-2A contains functionally redundant activating domains and works selectively from a promoter but not from a remote enhancer position in non-lymphoid (HeLa) cells. EMBO J. 9, 1625–1634 (1990).
Scholer, H. R., Balling, R., Hatzopoulos, A. K., Suzuki, N. & Gruss, P. Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 8, 2551–2557 (1989).
Hutchins, A. P., Jauch, R., Dyla, M. & Miranda-Saavedra, D. glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regen. (Lond) 3, 1 (2014).
Acknowledgements
We would like to thank Martina Sinn and Boris Burr for technical assistance. The Sox2-null ESCs and the PiggyBac constructs were kind gifts from Hitoshi Niwa and Austin Smith, respectively. This project was mainly carried out by funds of the Max Planck Society. R.J. is supported by the 2013 MOST China-EU Science and Technology Cooperation Program [2013DFE33080]. V.C. is supported by DFG (priority program SPP1356 on pluripotency and cellular reprogramming). The NRW Stem Cell Network and the Heinrich Heine University Düsseldorf are funding N.T. Junior Research Group.
Author information
Authors and Affiliations
Contributions
N.T., D.E. and H.R.S. designed the concept of the experiment. N.T., C.M., A.G.M., U.T., M.J.A.-B., R.J. and V.C. performed the experiments and assembled the data. N.T. and H.R.S. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Tapia, N., MacCarthy, C., Esch, D. et al. Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci Rep 5, 13533 (2015). https://doi.org/10.1038/srep13533
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep13533
This article is cited by
-
Dissecting OCT4 defines the role of nucleosome binding in pluripotency
Nature Cell Biology (2021)
-
Biological importance of OCT transcription factors in reprogramming and development
Experimental & Molecular Medicine (2021)
-
Key features of the POU transcription factor Oct4 from an evolutionary perspective
Cellular and Molecular Life Sciences (2021)
-
Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals
Nature Communications (2019)
-
Deconstructing the pluripotency gene regulatory network
Nature Cell Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.