Abstract
Castration-resistant (CR) prostate cancer (PCa) partly arises due to persistence of androgen receptor (AR) transcriptional activity in the absence of cognate ligand. An emerging mechanism underlying the CRPCa phenotype and predicting response to therapy is the expression of the constitutively-active AR-V7 splice variant generated by AR cryptic exon 3b inclusion. Here, we explore the role of the RNA-binding protein (RBP) Sam68 (encoded by KHDRBS1), which is over-expressed in clinical PCa, on AR-V7 expression and transcription function. Using a minigene reporter, we show that Sam68 controls expression of exon 3b resulting in an increase in endogenous AR-V7 mRNA and protein expression in RNA-binding-dependent manner. We identify a novel protein-protein interaction between Sam68 and AR-V7 mediated by a common domain shared with full-length AR and observe these proteins in the cell nucleoplasm. Using a luciferase reporter, we demonstrate that Sam68 co-activates ligand-independent AR-V7 transcriptional activity in an RNA-binding-independent manner and controls expression of the endogenous AR-V7-specific gene target UBE2C. Our data suggest that Sam68 has separable effects on the regulation of AR-V7 expression and transcriptional activity, through its RNA-binding capacity. Sam68 and other RBPs may control expression of AR-V7 and other splice variants as well as their downstream functions in CRPCa.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) onset and progression is driven by androgen steroid hormones binding to their cognate androgen receptor (AR) nuclear hormone receptor transcription factor. Androgen deprivation therapy (ADT) is the mainstay of treatment for locally-advanced/metastatic PCa1 and inactivates AR signaling for a period of time by suppressing gonadal androgen biosynthesis. However, PCa inevitably becomes castration-resistant (CRPCa) due to a number of molecular mechanisms2, including the presence of a ligand-independent AR transcriptional programme3.
An evolving mechanism for the persistence of AR signaling in CRPCa is the generation of constitutively-active AR variants sharing a common amine (N)-terminus domain (NTD) and DNA-binding domain (DBD) but lacking a ligand-binding domain (LBD)4. These include several variants generated through alternative pre-mRNA splicing resulting in inclusion of cryptic exons containing a premature termination codon (PTC) resulting in translation of a truncated AR protein5. Of the AR splice variants, AR-V7 mRNA (Ensembl: ENST00000504326), which is generated by cryptic exon 3b inclusion, is an important predictive biomarker of response to second-line endocrine therapy6.
Although ligand-activated AR can affect overall expression of both full-length AR and AR-V77,8, there is only one report to date studying the mechanisms of AR-V7 mRNA splicing8. Therein, the trans-acting RNA-binding proteins (RBPs) SRSF1 (encoded by SRSF1) and U2AF65 (encoded by U2AF2) were found to be involved in an ADT-mediated increase in AR-V7 expression. We have previously identified the RBP Sam68 (encoded by KHDRBS1) as an AR-interacting protein partner and co-regulator of AR-dependent transcription and splicing9. Here, we test the functions of Sam68 on AR-V7 expression and transcriptional activity.
Results
Sam68 regulates AR exon 3b, AR-V7 mRNA and protein expression
Since Sam68 interacts with AR in vitro and co-activates AR-dependent transcription and splicing9, we hypothesised that Sam68 may also have an effect on the expression of transcripts encoding AR-V7. To test this hypothesis, we firstly used qRT-PCR to monitor expression of mRNAs containing AR cryptic exon 3b as well as constitutive exons 3 and 4 under the transcriptional control of the constitutively-active cytomegalovirus (CMV) promoter in a minigene reporter (Fig. 1A)8. HEK293 cells, which do not express endogenous AR protein, were cultured in full medium and transfected with the minigene and expression constructs encoding Sam68 protein, or a RNA-binding-deficient Sam68V229F mutant10, or the Sam68-interacting RBP hnRNPA2 (encoded by HNRNPA2B1)11 as negative controls (Fig. 1B).
Sam68 regulates exon 3b expression.
(A) AR-V7 minigene contains cryptic exon 3b with flanking constitutive exons 3 and 4 cloned downstream of a constitutively-active CMV promoter. Vertical line marks position of exonic splicing enhancer (ESE) near to the cryptic exon 3b 3′ splice site (SS). Arrows show location of primers used for qRT–PCR. (A = polyadenylation site). (B,C) HEK293 cells were cultured in full medium prior to transfection with the AR-V7 (B) or AR-V7 ESE mutant (ESEm) (C) minigenes (1.5 μg) with expression vectors for HA-hnRNPA2, GFP-Sam68V229F, GFP–Sam68 or empty vector control (500 ng) as indicated. qRT-PCR was performed on cDNAs and levels of exon 3b and exon 4 expression were normalised to ACTB levels and compared with empty vector control conditions to obtain the mean normalised fold-change in expression ± SE (*p = 0.003; **p = 0.04).
Ectopic expression of Sam68 protein resulted in a ~3.5-fold increase in cryptic exon 3b expression compared with the empty vector control (p = 0.0003) (Fig. 1B). Consistent with the preferential selection of exon 3b, Sam68 also caused ~50% reduction in exon 4 expression as compared with the empty vector control (p = 0.04). Ectopic expression of the Sam68-interacting hnRNPA2 protein did not have a statistically-significant effect on exon 3b (or exon 4) expression (p > 0.05), suggesting that exon 3b expression may be directly controlled by Sam68 itself rather than by other Sam68-interacting RBP partners.
Since Sam68-regulated exon inclusion is typically RNA-binding-dependent10, we employed the RNA-binding deficient Sam68V229F mutant10 to determine whether the observed effects on cryptic exon 3b expression required the RNA-binding capacity of Sam68 protein. Compared with wild-type Sam68, the Sam68V229F mutant was much less efficient at stimulating expression of exon 3b (~2-fold increase above controls) and did not have an effect on expression of mRNAs containing exon 4 (p > 0.05) (Fig. 1B).
Expression of the cryptic exon 3b has been show to be controlled, in part, by the RBP SRSF1 binding a cis-acting exonic splicing enhancer (ESE) close to the 3′ splice site (SS)8. Since Sam68 typically stimulates exon inclusion, we hypothesised that Sam68 may also regulate exon 3b expression through binding to the same ESE. To test this hypothesis, we employed the same minigene reporter but this time containing a point mutation within an ESE (ESEm) close to the 3′ SS (Fig. 1A), which has been shown to abrogate SRSF1-binding8. Despite the point mutation, ectopic expression of wild-type Sam68 resulted in a trend towards an increase in exon 3b expression as compared with the empty vector control (p = 0.14) (Fig. 1C). We also observed a similar Sam68-regulated increase in exon 4 expression.
To determine whether the Sam68-dependent increase in exon 3b expression translated into an increase in endogenous AR-V7 mRNA and protein expression in vivo, we employed the full-length AR- and AR-V7-expressing CWR22 and VCaP PCa cell lines12, which express KHDRBS1 mRNA or Sam68 protein at similarly high levels to other common PCa cell lines (Supplementary Figure S1A and B). ADT/anti-androgen and androgen treatment have been shown to increase and decrease expression of AR-V7 protein, respectively, through possible recruitment of RBPs8,13. Hence, we used qRT-PCR to examine the effect of ectopic Sam68 protein expression and siRNA-mediated Sam68 knockdown on production of AR-V7 and full-length AR mRNA transcripts in CWR22 and VCaP cells grown in steroid-depleted medium as a model for ADT and in the presence of dihydrotestosterone (DHT) as the only available steroid (Fig. 2A, Supplementary Figure S1C and D).
Sam68 regulates AR-V7 mRNA and protein expression
(A) CWR22 cells were cultured in steroid-depleted medium with or without 100 nM DHT for 24 h prior to transfection with expression vectors for GFP-Sam68V229F or GFP–Sam68 or empty vector control (2 μg) as indicated. qRT-PCR was performed on cDNAs and levels of AR and AR-V7 transcript expression were normalised to ACTB levels and compared with empty vector control conditions in the absence of DHT to obtain the mean normalised fold-change in expression ± SE (*p = 0.02; **p = 0.002). (B) Representative Western blotting images of whole cell lysates from CWR22 cells cultured in full medium and transfected with the expression vector for GFP–Sam68 or empty vector (2 μg) as indicated (left panel). Densitometric band quantitation was performed to calculate the mean fold-change in endogenous Sam68, total (endogenous plus ectopic) Sam68 and AR-V7 protein expression ± SE compared to empty vector control conditions (*p = 0.03; **p < 0.001).
In CWR22 cells, there was no statistically-significant difference in levels of both full-length AR and AR-V7 mRNAs between steroid-depleted and androgen-treated conditions (p > 0.05) (Fig. 2A, compare conditions 1 and 4). Ectopic expression of wild-type Sam68, but not the Sam68V229F mutant, resulted in ~1.5-fold increase in AR-V7 mRNA expression both in the presence (p = 0.02) and absence of DHT (p = 0.002) (Fig. 2A, compare conditions 1 and 3 and 4 and 6). This increase in mRNA expression correlated with a ~2.4-fold Sam68-mediated increase in AR-V7 protein expression in cells cultured in full medium (p < 0.001) (Fig. 2B, right panel). However, there was no statistically-significant effect of ectopic Sam68 on full-length AR mRNA expression (p > 0.05) (Fig. 2A).
In the VCaP CRPCa cell line model, ectopic expression of wild-type Sam68 protein resulted in a ~1.5-fold increase in AR-V7 mRNA expression in the absence of DHT (p = 0.04) without an effect on full-length AR mRNA expression (p > 0.05) (Supplementary Figure S1C). However, siRNA-mediated depletion of Sam68 protein in CWR22 cells neither affected expression of AR-V7 nor full-length AR mRNAs in the presence or absence of DHT despite at least ~50% knockdown of Sam68 mRNA levels (Supplementary Figure S1D, compare lanes 1 and 2–4 and 5 and 6–8) which typically correlated with ~70% reduction in protein expression in cells cultured in full medium (Supplementary Figure S1E, compare lane 1 with 2–4).
Sam68 and AR-V7 proteins interact and are present in the nucleoplasm of PCa cells
Consistent with published findings in LNCaP cells9, we identified a protein-protein interaction between Sam68 and full-length AR by immunoprecipitation in whole cell extracts from CWR22 cells cultured in full medium (Supplementary Figure S2A, right panel, compare lanes 1 and 2). Since CWR22 cells also express AR-V7 protein, we hypothesised that Sam68 may also interact with AR-V7, which contains the AR NTD but lacks the LBD required for ligand-dependent activity (Fig. 3A).
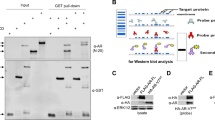
Sam68 and AR-V7 proteins interact via the AR NTD
(A) AR comprises three functional domains: An amine-terminal transactivation domain (NTD), containing the activation function (AF) subdomain 1 (AF-1); a zinc finger-type DNA-binding domain (DBD); and a carboxyl-terminal ligand binding domain (LBD) containing AF subdomain 2 (AF-2). A hinge domain, containing a bipartite nuclear localisation signal (NLS) links the LBD and DBD. (B,C) Immunoprecipitation was performed in whole cell lysates from CWR22 cells cultured in full medium using either the antibodies to Sam68 (B) or AR-V7 (C) and recovered material was subjected to Western blotting as indicated. (D) HEK293 cells were cultured in full medium prior to transfection with UASTKLuc and pCMV-β-Gal reporters and expression vectors for GAL4 DBD (pM; 100 ng) and VP16 AD (VP16; 100 ng) as indicated. After 48 hours, cells were harvested prior to analysis for luciferase and β-Galactosidase activities to provide relative luciferase activity and compared with control conditions in the absence of vectors encoding AR NTD to obtain mean fold-change in luciferase activity ± SE (*p = 0.0001). (IP = immunoprecipitation; IB: Western immunoblot).
To test this hypothesis, both Sam68 and AR-V7 proteins were separately immunopreciptated in whole cell extracts from CWR22 cells cultured in full medium using their cognate antibodies prior to Western blotting (Fig. 3B,C). The antiserum to Sam68 efficiently immunoprecipitated Sam68 protein (Fig. 3B, left panel, compare lanes 1 and 2) and also co-immunoreprecipitated AR-V7 protein (Fig. 3B, right panel, compare lanes 1 and 2). Likewise, the antibody to AR-V7 efficiently immunoprecipitated AR-V7 protein (Fig. 2C, left panel, compare lanes 1 and 2) and also co-immunoreprecipitated Sam68 protein (Fig. 2C, right panel, compare lanes 1 and 2).
To verify this novel Sam68-AR-V7 protein-protein interaction, HEK293 cells cultured in full medium were transiently transfected with expression vectors for Sam68 and AR-V7 proteins (Supplementary Figure S2B). Consistent with the interactions between endogenous proteins in PCa cells, we also observed protein-protein interactions between ectopically-expressed Sam68 and AR-V7 proteins by immunoprecipitation (Supplementary Figure 2B, right panel, compare lanes 1 and 2). In all immunoprecipitation experiments, no or weak immunoprecipitation or co-immunoprecipitation was observed using normal mouse IgG (Supplementary Figure S2, all panels, compare lanes 2 and 3) or in the absence of either antibodies (Fig. 2B,C, all panels, compare lanes 2 and 3).
The above findings identify a novel protein-protein interaction between Sam68 and AR-V7. Since both full-length AR and AR-V7 share a common NTD (Fig. 3A), we hypothesised that Sam68 protein interacts with the AR NTD. To test this hypothesis, we used the mammalian 2-hybrid system. A VP16 activation domain (AD) fusion of AR NTD was co-expressed with a GAL4 DBD fusion of full-length Sam68 in HEK293 cells grown in the full medium (Fig. 3D). Ectopic expression of the VP16 AD-AR NTD fusion protein in the absence of the GAL4 DBD-Sam68 fusion protein did not affect reporter activity. However, in the presence of the GAL4 DBD-Sam68 fusion protein, ectopic expression of the AR NTD fusion protein resulted in ~1.7-fold increase in reporter activity (p = 0.0001).
Since Sam68 interacts with both full-length AR and AR-V7 proteins in vivo, we sought to determine if these proteins also co-localise in vivo. Using indirect immunofluorescence and confocal microscopy, we compared the intracellular distributions of endogenous Sam68 protein with full-length AR in LNCaP cells (Supplementary Figure S3). As previously-described11, Sam68 exhibited a diffuse nucleoplasmic distribution and was concentrated in discrete Sam68/SLM nuclear bodies (SNBs) (Supplementary Figure S3, top left panel). AR predominantly exhibited a nucleoplasmic distribution, was excluded from the nucleolus and SC35-containing splicing speckles (Supplementary Figure S3, middle panels). Although both Sam68 and AR proteins were present in the nucleoplasm, they did not appear to co-localise to any discrete subnuclear organelle (Supplementary Figure S3, top right panel). In parallel experiments using the same combination of antibodies in PC3-M cells (which do not express endogenous AR protein) stably transfected with an expression vector for AR-V7, we observed similar localisation of Sam68 and AR-V7 proteins in the cell nucleoplasm (Supplementary Figure S3, bottom panels).
Sam68 co-activates AR-V7 transcriptional activity and regulates expression of the AR-V7-specific target gene UBE2C
In light of the above and previously-published findings of a role for Sam68 on full-length AR-dependent transcriptional activity9, we hypothesised that Sam68 may also affect AR-V7-dependent transcription. Using an (ARE)3-driven luciferase reporter assay in HEK293 cells, we investigated the effects of ectopic expression and shRNA-mediated knockdown of Sam68 protein on AR-V7-dependent transcription (Fig. 4). The (ARE)3 reporter, which responds only to AR-binding, contains androgen response elements (AREs) from the core promoter region of the KLK3 gene (see Materials and Methods). In keeping with published findings9, we observed that Sam68 protein functioned as a ligand-dependent co-activator of full-length AR-dependent transcriptional activity of the (ARE)3 reporter in an RNA-binding-independent manner (Supplementary Figure S4).
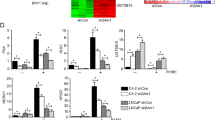
Sam68 co-activates AR-V7 transcriptional activity independent of RNA-binding capacity.
(A–C) HEK293 cells were cultured in steroid-depleted medium with or without 100 nM DHT and transfected with the p(ARE)3Luc and pRL-null reporters, c-Flag AR-V7 (100 ng), together with expression vectors for (A) GFP-Sam68 or (B) GFP-Sam68V229F or the empty vector control (50–200 ng) or (C) shRNA to Sam68 (shSam68) or the non-silencing control (shNTC) (200 ng) as indicated. After 24 hours, cells were harvested prior to analysis for luciferase and renilla activities to provide relative luciferase activity and compared with control conditions in the absence of vectors encoding Sam68 (A,B) or shRNA to Sam68 (C) and DHT (A,B) to obtain mean fold-change in luciferase activity ± SE (*p = 0.01; **p = 0.02; ***p = 0.03). Representative Western blots of whole cell lysates from HEK293 cells cultured in full medium and transfected with expression vectors for Sam68 (A,B) or shRNA to Sam68 (C) and AR-V7.
Consistent with the absence of the LBD, AR-V7-mediated reporter activity was not affected by the addition of DHT (Fig. 4A). In the presence and absence of DHT, AR-V7-mediated reporter activity was enhanced by increasing amounts of Sam68 (Fig. 4A). In the absence of DHT, we observed a maximal ~2.5-fold enhancement in reporter activity above the empty vector control (Fig. 4A) (p = 0.01). Consistent with our published data9, increasing amounts of RNA-binding-deficient mutant Sam68V229F also enhanced AR-V7-mediated reporter activity in both the presence and absence of DHT (Fig. 4B). In the absence of DHT, we observed a maximal ~2.2-fold enhancement in reporter activity above the empty vector control (p = 0.02) (Fig. 4B). Ectopic expression of Sam68 protein did not alter (ARE)3 reporter activity in the absence of the AR (data not shown).
shRNA-mediated depletion of endogenous Sam68 protein also resulted in ~0.8-fold reduction in activity as compared with AR-V7-mediated reporter activity as compared with the level in the presence of Sam68 (p = 0.04) (Fig. 4C). Western blotting demonstrated that levels of AR-V7 or endogenous Sam68 protein expression were not typically affected by Sam68 protein overexpression or knockdown in cells cultured in full medium (Fig. 4, Western blots). Similar to published findings for full-length AR9, we observed maximal change in reporter activity between low and high Sam68 expression vector conditions, despite a less significant change in ectopic Sam68 protein expression levels.
AR splice variants, including AR-V7, have been shown to regulate a transcriptional programme distinct to full-length AR in PCa cells14,15. Based on the above findings using the luciferase reporter assay, we hypothesised that Sam68 may also control expression of endogenous AR-V7 target genes in vivo. To test this hypothesis, we used qRT-PCR to examine the effect of ectopic Sam68 protein expression on canonical AR (KLK3, TMPRSS) or AR-V7-specific (UBE2C) target gene expression in CWR22 cells grown in steroid-depleted medium (Fig. 5). Ectopic expression of wild-type Sam68, but not the RNA-binding-deficient Sam68V229F mutant, resulted in a ~1.8 fold increase in KLK3 gene expression (p = 0.03) (Fig. 5A). However, neither wild-type Sam68 nor the Sam68V229F mutant had a statistically-significant effect on TMPRSS gene expression (p > 0.05) (Fig. 5A). Ectopic expression of wild-type Sam68 resulted in ~2.5-fold increase in expression of the AR-V7-specific gene target UBE2C (p = 0.02) (Fig. 5B). We also observed a trend towards an increase in UBE2C gene expression following ectopic expression of the Sam68V229F mutant (p = 0.06).
Sam68 regulates expression of the AR-V7-specific target gene UBE2C.
(A,B) CWR22 cells were cultured in steroid-depleted medium for 24 h prior to transfection with expression vectors for GFP-Sam68V229F or GFP–Sam68 or empty vector control (2 μg) as indicated. qRT-PCR was performed on cDNAs and levels of KLK3, TMPRSS (A) and UBE2C (B) were normalised to ACTB levels and compared with empty vector control conditions to obtain the mean normalised fold-change in expression ± SE (*p = 0.03; **p = 0.02).
Expression of the gene encoding Sam68 (KHDRBS1) is not altered in CRPCa
Sam68 protein is highly expressed in common PCa cell lines including the CRPCa cell line models (Supplementary Figure S1A and B). Although Sam68 protein (encoded by KHDRBS1) has been shown to be over-expressed9,16 and phosphorylated17 in primary PCa, expression of Sam68 in CRPCa is unknown. We interrogated the Grasso dataset18 (n = 122) for changes in expression of KHDRBS1 as well as SRSF1 and U2AF2, which encode the RBPs SRSF1 and U2AF65, respectively and have been implicated in AR-V7 splicing8 (Fig. 6A). There were no differences in KHDRBS1 or SRSF1 gene expression between primary localised PCa (n = 59) and CRPCa (n = 35) (p = 0.19 and p = 0.11, respectively) (Fig. 6A, left and middle panels). However, we observed a statistically-significant increase in U2AF2 gene expression in CRPCa as compared with primary PCa (p < 0.001) (Fig. 6A, right panel).
Expression of the gene encoding Sam68 (KHDRBS1) is not altered in CRPCa.
(A) Expression of genes encoding Sam68 (KHDRBS1), U2AF65 (U2AF2) and SRSF1 (SRSF1) in benign prostate (n = 28), localized PCa (n = 59) and metastatic CRPCa (n = 35) from the Grasso dataset18. Gene expression is graphically represented using a box and whisker plot with outliers are indicated as +(*p = 0.19; **p = 0.11; ***p < 0.001). (B) Scatter plots comparing expression of the major Ensembl transcripts encoding Sam68, SRSF1 and U2AF65 RBPs with AR-V7 from the TCGA dataset. A 95% confidence band is displayed for the regression lines.
To determine whether expression of genes encoding RBPs involved in AR-V7 expression (KHDRB1, SRSF1 and U2AF) correlated with expression of the AR-V7 mRNA in clinical PCa we used The Cancer Genome Atlas (TCGA) dataset (n = 417). We tested for correlations in expression of AR-V7 (ENST00000504326) and the major Ensembl transcripts encoding Sam68 (ENST00000327300), SRSF1 (ENST00000582730) and U2AF65 (ENST00000450554) (Fig. 6A). Although, there was no correlation between expression of the major transcript encoding Sam68 (n = 371; R = −0.01; FDR = 0.86) and AR-V7 (Fig. 6A, left panel), we did observe positive correlations in expression between AR-V7 and the major transcripts encoding SRSF1 (n = 371; R = 0.14; FDR = 0.01) and U2AF65 (n = 371; R = 0.23; FDR = 0.00003) (Fig. 6A, middle and right panels). Based on expression of these major Ensembl transcripts in the TCGA dataset, KHDRBS1 (encoding Sam68) appeared to be expressed at higher levels than U2AF2 and SRSF1 in primary PCa (Fig. 6A, compare left panel with middle and right panels).
Discussion
In this study, we demonstrate for the first time that the RBP Sam68 preferentially increases AR cryptic exon 3b expression and controls mRNA and protein levels of the AR splice variant AR-V7 in PCa cells largely via an RNA-binding-dependent mechanism. Consistent with our previous findings for full-length AR9, we show that Sam68 and AR-V7 interact in PCa cells mediated by an association between Sam68 and the AR NTD. We show that Sam68 protein is present with full-length AR and AR-V7 in the cell nucleoplasm, where transcription and splicing occurs. We demonstrate that Sam68 functions as a co-activator of AR-V7-dependent transcriptional activity of the (ARE)3 reporter in an RNA-independent manner. Finally, we show that Sam68 controls expression of canonical AR and AR-V7 target genes in PCa cells via both RNA-binding-dependent and –independent mechanisms.
The emergence of constitutively-active AR splice variants in CRPCa, particularly AR-V7, poses a major clinical challenge as these variants not only seem to mediate resistance to novel inhibitors of androgen biosysthesis and ligand-dependent AR activity in PCa models13,19 but also appears to accurately predict response to treatment with these drugs in CRPCa patients6. To date, little is known of the mechanisms of generation of these splice variants and their downstream transcriptional targets. Moreover, it is unclear whether expression of AR splice variants in patients should preclude treatment with currently-available second-line endocrine agents.
Controlled by trans-acting complexes of several RBPs binding cis-acting regulatory pre-mRNA elements, alternative splicing can affect the processes underlying the hallmarks of cancer20 including PCa21. A potential cis-acting effect mediating AR-V7 expression may be due to an intragenic intron 1 deletion22, although exactly how this controls exon 3b inclusion is unclear. The trans-acting RBPs SRSF1 and U2AF65 appear to mediate an increase in AR-V7 expression in response to ADT in vitro through recruitment and binding to an intronic splicing enhancer (ISE) and ESE, respectively, close to the cryptic exon 3b 3′ SS8.
Our data suggest that Sam68 also appears, at least in part, to have an effect on exon 3b expression via the SRSF1-responsive ESE (Fig. 7). This effect appears to be primarily dependent on its RNA-binding ability and not indirectly mediated by recruitment of Sam68-associated RBP complexes incorporating hnRNPA211. The ability of the Sam68V229F mutant to partially regulate exon 3b expression may be due to recruitment of endogenous wild-type protein, as Sam68 is thought to bind RNA as a homodimer23. Hence, the effect of Sam68 on exon 3b expression via the SRSF1-responsive ESE may be direct or, consistent with a role for Sam68 stabilising the SRSF1 mRNA24, possibly indirectly mediated by SRSF1. In keeping with its role in U2AF65-mediated exon definition25, recruitment of U2AF65 by Sam68 to exon 3b 3′ SS, is another possible mechanism for Sam68-mediated AR-V7 expression (Fig. 7).
Model of Sam68-regulated AR-V7 splicing and downstream gene regulation.
(A) AR cryptic exon 3b recognition is influenced by dinucleotides within splice site (SS) consensus sequences and auxiliary elements such as U2AF65 (U2AF) binding the 3′ SS; and the 5′ SS and branch point serve as binding sites for U1 and U2 snRNPs (U1 and U2), respectively. The exonic splicing enhancer (ESE) binds SRSF1 and Sam68 (S68) thereby recruit and stabilising binding of other spliceosome components such as U2AF65 to the 3′ SS. (B) Sam68 is recruited by the AR-V7 homodimer to the promoter region of target gene (e.g. UBE2C) and co-operates with other “general” transcription factors to enhance RNA polymerase (RNAPII)-mediated gene transcription from the transcription start site (TSS). Following activation (e.g. by protein phosphorylation), Sam68 is recruited to nascent pre-mRNA transcripts of target genes (e.g. AR) and in cooperation with other RNA-binding proteins (RBPs) regulates SS selection and exon inclusion.
However, the trend towards a Sam68-regulated increase in exon 3b expression from the ESEm AR-V7 minigene suggests the effect of Sam68 may not be completely mediated by the ESE. An alternative mechanism, in keeping with its ability to control transcript stability via 3′-untranslated region (UTR)-binding24, might be stabilisation of mRNAs containing exon 3b thereby increasing AR-V7 expression. Since siRNA-mediated depletion of Sam68 protein does not affect AR-V7 (or full-length AR) levels, there may be redundancy in thes above putative mechanisms of action which are substituted by SRSF1 and U2AF65 and other RBPs. These observations are in keeping with several RBPs acting both synergistically and competitively within larger protein complexes to regulate SS selection in a context-dependent manner26.
Functionally, RBP interactions with AR protein have been shown to both activate9 and inhibit27 AR-dependent transcriptional activity. Although the RNA-binding capacity of Sam68 is primarily required to regulate AR-V7 mRNA expression, our data suggest that this function is not required to co-activate AR-V7-dependent transcriptional activity. However, high total levels of Sam68 protein may be required for it to function as a co-activator of AR-V7-dependent transcriptional activity of the luciferase reporter (Fig. 4). Since both ectopic expression and shRNA-depletion of Sam68 protein affected AR-V7 dependent reporter activity, the effect of Sam68 on AR-V7-dependent transcription may be an important and non-redundant mechanism (Fig. 7). Consistent with this, we observed a trend towards an increase in expression of AR-V7-specific gene target UBE2C expression following ectopic expression of the transcriptionally-active but RNA-binding-deficient Sam68V229F mutant. Taken together, these new data are in keeping with our previous findings9, which suggest that RNA processing and regulation of gene expression by Sam68 may be separable functions.
By knowledge-based validation, we were unable to identify up-regulation of KHDRBS1 (encoding Sam68) gene expression in CRPCa and a correlation with AR-V7 expression in primary PCa, although we did reveal up-regulation of U2AF2 gene expression in CRPCa and correlations between SRSF1 (encoding SRSF1) or U2AF2 (encoding U2AF65) and AR-V7 mRNA expression in primary PCa. In the absence of any existing large datasets from CRPCa patients that allow similar transcript correlations, our observations in localised PCa suggest that absolute expression levels of U2AF2 and possibly SRSF1 may be important for regulation of AR-V7 mRNA expression in CRPCa. Since KHDRBS1 is already expressed at higher levels in primary PCa as compared with SRSF1 and U2AF2, post-translational modifications of Sam68 protein, rather than absolute expression levels, may be of greater importance for AR-V7 expression. Phosphorylation of Sam68 has been reported in primary PCa17 and is required for signal-dependent splicing regulation28.
Since AR-V7 protein appears to be able to bind to and regulate a distinct subset of genes in CRPCa cell line models14,15 and is an important predictive biomarker in clinical CRPCa6, a greater understanding of the mechanisms of AR-V7 expression and downstream functions will reveal a clearer role for AR-V7 (and other variants) in clinical CRPCa. Although cis-elements and other trans-acting RBPs (e.g. SRSF1 and U2AF65) appear to be involved, both directly and indirectly, Sam68 is the only RBP known to regulate AR-V7 expression and downstream transcription function that is over-expressed9,29 and phosphorylated17 in PCa. Hence overexpression and, moreover, activation of Sam68 protein through post-translational modification may contribute to the clinical CRPCa phenotype.
Methods
Antibodies, plasmids and oligonucleotides
The following antibodies were used: anti-Sam68 (sc333, Santa Cruz Biotechnology), anti-AR (G122-434) (BDB554225, BD Biosciences), anti-AR (N-20) (sc-816, Santa Cruz Biotechnology), anti-AR-V7 (AG10008, Precision Antibody), anti-SC35 (S4045, Sigma), anti-α-tubulin (TU-02, Santa Cruz Biotechnology), normal mouse IgG (sc-2025, Santa Cruz Biotechnology), anti-mouse IgG HRP-linked (7076, Cell Signaling), anti-rabbit IgG HRP-linked (7074, Cell Signaling), Alexa Fluor® 488 anti-Mouse IgG (A-21203, Life Technologies) and Alexa Fluor® 555 anti-Rabbit IgG (A-21429, Life Technologies). The following plasmids have been described previously or are commercially-available: pEGFP3-Sam6830; pEGFP-Sam68V229F10; pcDNA3-HA-hnRNPA2-cDNA31; pcDNA3-AR, UASTKLuc and pCMV-β-Gal32; AR-V7 minigenes8; pLKO-si-Sam6829; and control pLKO-puro Non-Target shRNA Control (SHC016, Sigma) and pRL-null (E2271, Promega). pM-Sam68 was generated by cloning of the Sam68 coding sequence from pGBTK7-Sam6833 into the empty pM vector between EcoRI and SalI. cFlag AR-V7 was generated by cloning of a PCR-amplified cDNA fragment containing the AR-V7 coding sequence from the 22Rv1 PCa cell line (CRL-2505, ATCC) into c-Flag pcDNA334 between XhoI and BamHI. p(ARE)3Luc contains 3x repeats of the 1st androgen response element (ARE) plus TATA box from KLK3 promoter region cloned into pGL3basic (E1751, Promega). The identity and integrity of newly-generated clones was confirmed by Sanger sequencing. Sequences used to generate siRNA duplexes are as previously described35 and a non- silencing control (#D-001810-01-20, Dharmacon) was also used. Sequences used to generate oligonucleotide primers for PCR were as previously described8,36 or are listed in Supplementary Table S1.
Cell Culture, DNA and RNA transfections
All cells were grown at 37 °C in 5% CO2. HEK293 (CRL-1573, ATCC), 22Rv1 (CRL-2505, ATCC), VCaP (CRL-2876, ATCC) and CWR2237 cells were maintained in RPMI-1640 medium (31870-025, Life Technologies) with 2 mM L-glutamine (25030-024, Life Technologies), supplemented with 10% foetal bovine serum (FBS) (A15-101, PAA Laboratories) or 10% Dextran Charcoal Stripped FCS (12676011, Life Technologies) to produce steroid-depleted medium as detailed in figure legends. PC3-M cells were derived as previously described38 and maintained as above. PC3-M cells stably-expressing AR-V7 were generated by transfection of parental PC-3M cells with the c-Flag AR-V7 vector. Cells were cultured in medium containing 300 μg/ml G418S (G418S, Formedium) and resistant clones were maintained under selection but removed for experiments. Transfections with plasmid DNA and siRNA duplexes were carried out as detailed in the figure legends using Lipofectamine LTX (15338-100, Life Technologies) and RNAiMax (13778-075, Life Technologies), respectively, according to manufacturer’s instructions. Where indicated, cells were treated with 100 nM dihydrotestosterone (DHT).
Immunoprecipitation, SDS-PAGE and Western blotting
Protein immunoprecipitation was performed as previously described39. Recovered material or whole cell lysates were resolved by SDS-PAGE and subjected to Western blotting as previously described36. Antibody concentrations were as follows: anti-Sam68 (1:4000), anti-AR-V7 (1:1000), anti-AR (N-20) (1:1000), anti-α-tubulin (1:1000); HRP-linked secondaries (1:2000). Immunoprecipitation data shown are representative of two independent biological experimental replicates. Where indicated, densitometric assessments of protein bands were performed using ImageJ (http://rsb.info.nih.gov/ij/) and intensities used to calculate relative normalised fold-change in protein expression. Western blotting data shown are from at least two independent biological experimental replicates.
Indirect immunofluorescence microscopy
LNCaP and PC3-M cells were grown and transfected on glass coverslips (VWR International) and stained with primary and secondary antibodies as previously described36. Antibody concentrations were as follows: anti-Sam68 (1:1000), anti-AR (G122-434;1:20) (N-20; 1:100), anti-SC35 (1:200); Alexa Fluor-linked secondaries (1:400). Images shown are representative of at least two independent biological experimental replicates.
RNA extraction, reverse transcription (RT) and quantitative PCR (PCR)
Total RNA extraction, quantitative RT-PCR (qRT-PCR) and relative gene expression analyses were performed as previously described36. Data shown are from three independent biological experimental replicates with three technical replicates.
Minigene Splicing Assays
HEK293 cells were seeded at a density of 1 × 105 cells/ml in full medium into 6-well plates (Asahi Techno Glass). Cells were transfected with DNA as detailed in figure legends. RNA was extracted using RNeasy (Qiagen) and subjected to qRT-PCR using oligonucleotides as previously described8. Data shown are from four independent biological experimental replicates with three technical replicates.
Luciferase Reporter Assays
For mammalian 2-hybrid assays, HEK293 cells were seeded at a density of 5 × 104 cells/ml in full medium into 12-well plates (Asahi Techno Glass). Cells were transfected with DNA as detailed in the figure legends. Luciferase and β–Galactosidase assays were performed to give relative activity as previously described32. Data shown are from at least two independent biological experimental replicates with four technical replicates. For transcription reporter assays, HEK293 cells were seeded at a density of 2 × 104 cells/well in steroid-depleted medium into 24-well plates (Asahi Techno Glass). Cells were transfected with DNA and supplemented with 100 nM DHT as detailed in figure legends. Firefly and Renilla luciferase assays were carried out using the Dual Luciferase Reporter Assay system (E1910, Promega) as per manufacturer’s instructions to give relative luciferase activity. Data shown are from at least three independent biological experimental replicates with three technical replicates.
Bioinformatics
Pre-processed microarray data were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO)40. For cell lines, gene expression data were extracted from the Taylor dataset41 (Accession number: GSE21034), normalized as the mean of the log2 transformed probe signal and plotted against the PCa cell lines as annotated in the sample information. For clinical samples, median-centered gene expression data were extracted from the Grasso dataset18 (Accession number: GSE35988), normalized as the mean of the array probes and plotted against clinical groups (benign prostate, localized PCa and metastatic CRPCa) as annotated in the sample information. Analysis of Variance (ANOVA) was employed to test for differences in means between groups with p < 0.05 taken to indicate statistical significance (MATLAB, MathWorks). FASTQ files containing pre-processed (Level 3) RNA-Seq data were downloaded from The Cancer Genome Atlas (TCGA) data portal (Accession number: 23700-2). Adaptors were trimmed using Skewer42 and isoform quantification performed using Sailfish43. The R software v.3.1.244 MASS (Modern Applied Statistics with S) package45 was used for robust linear modeling. Spearman correlation was performed on data from samples harboring non-zero expression in both genes of interest and q-values reported using the false discovery rate (FDR) with FDR < 0.05 taken to indicate statistical significance.
Statistical analyses
Graphical data shown represent the means ± standard error (SE) of independent experiments. The one-tailed independent sample T-Test was employed to identify differences in means between groups with p < 0.05 taken to indicate statistical significance.
Additional Information
How to cite this article: Stockley, J. et al. The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci. Rep. 5, 13426; doi: 10.1038/srep13426 (2015).
References
Mottet, N. et al. EAU guidelines on prostate cancer. Part II: Treatment of advanced, relapsing and castration-resistant prostate cancer. European urology 59, 572–583, 10.1016/j.eururo.2011.01.025 (2011).
Karantanos, T. et al. Understanding the Mechanisms of Androgen Deprivation Resistance in Prostate Cancer at the Molecular Level. European urology, 10.1016/j.eururo.2014.09.049 (2014).
Sharma, N. L. et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer cell 23, 35–47, 10.1016/j.ccr.2012.11.010 (2013).
Ware, K. E., Garcia-Blanco, M. A., Armstrong, A. J. & Dehm, S. M. Biologic and clinical significance of androgen receptor variants in castration resistant prostate cancer. Endocrine-related cancer 21, T87–T103, 10.1530/ERC-13-0470 (2014).
Sprenger, C. C. & Plymate, S. R. The Link Between Androgen Receptor Splice Variants and Castration-Resistant Prostate Cancer. Hormones & cancer, 10.1007/s12672-014-0177-y (2014).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. The New England journal of medicine 371, 1028–1038, 10.1056/NEJMoa1315815 (2014).
Quarmby, V. E., Yarbrough, W. G., Lubahn, D. B., French, F. S. & Wilson, E. M. Autologous down-regulation of androgen receptor messenger ribonucleic acid. Molecular endocrinology 4, 22–28, 10.1210/mend-4-1-22 (1990).
Liu, L. L. et al. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 33, 3140–3150, 10.1038/onc.2013.284 (2014).
Rajan, P. et al. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. The Journal of pathology 215, 67–77, 10.1002/path.2324 (2008).
Paronetto, M. P., Achsel, T., Massiello, A., Chalfant, C. E. & Sette, C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol 176, 929–939 (2007).
Rajan, P. et al. Proteomic identification of heterogeneous nuclear ribonucleoprotein L as a novel component of SLM/Sam68 Nuclear Bodies. BMC cell biology 10, 82, 10.1186/1471-2121-10-82 (2009).
Hu, R. et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer research 69, 16–22, 10.1158/0008-5472.CAN-08-2764 (2009).
Li, Y. et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer research 73, 483–489, 10.1158/0008-5472.CAN-12-3630 (2013).
Hu, R. et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer research 72, 3457–3462, 10.1158/0008-5472.CAN-11-3892 (2012).
Lu, J. et al. The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells. The Journal of urology 193, 690–698, 10.1016/j.juro.2014.08.043 (2015).
Busa, R. et al. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene 26, 4372–4382, 10.1038/sj.onc.1210224 (2007).
Paronetto, M. P. et al. Expression of a truncated form of the c-Kit tyrosine kinase receptor and activation of Src kinase in human prostatic cancer. The American journal of pathology 164, 1243–1251, 10.1016/S0002-9440(10)63212-9 (2004).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243, 10.1038/nature11125 (2012).
Mostaghel, E. A. et al. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clinical cancer research: an official journal of the American Association for Cancer Research 17, 5913–5925, 10.1158/1078-0432.CCR-11-0728 (2011).
Oltean, S. & Bates, D. O. Hallmarks of alternative splicing in cancer. Oncogene 33, 5311–5318, 10.1038/onc.2013.533 (2014).
Rajan, P., Elliott, D. J., Robson, C. N. & Leung, H. Y. Alternative splicing and biological heterogeneity in prostate cancer. Nature reviews. Urology 6, 454–460, 10.1038/nrurol.2009.125 (2009).
Li, Y. et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene 31, 4759–4767, 10.1038/onc.2011.637 (2012).
Meyer, N. H. et al. Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain. J Biol Chem 285, 28893–28901, 10.1074/jbc.M110.126185 (2010).
Valacca, C. et al. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol 191, 87–99, 10.1083/jcb.201001073 (2010).
Tisserant, A. & Konig, H. Signal-regulated Pre-mRNA occupancy by the general splicing factor U2AF. PloS one 3, e1418, 10.1371/journal.pone.0001418 (2008).
Fu, X. D. & Ares, M. Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nature reviews. Genetics 15, 689–701, 10.1038/nrg3778 (2014).
Dong, X., Sweet, J., Challis, J. R., Brown, T. & Lye, S. J. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Molecular and cellular biology 27, 4863–4875, 10.1128/MCB.02144-06 (2007).
Matter, N., Herrlich, P. & Konig, H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420, 691–695, 10.1038/nature01153 (2002).
Busa, R., Geremia, R. & Sette, C. Genotoxic stress causes the accumulation of the splicing regulator Sam68 in nuclear foci of transcriptionally active chromatin. Nucleic acids research 38, 3005–3018, 10.1093/nar/gkq004 (2010).
Ehrmann, I. et al. The tissue-specific RNA binding protein T-STAR controls regional splicing patterns of neurexin pre-mRNAs in the brain. PLoS genetics 9, e1003474, 10.1371/journal.pgen.1003474 (2013).
Ferron, L. et al. The stargazin-related protein gamma 7 interacts with the mRNA-binding protein heterogeneous nuclear ribonucleoprotein A2 and regulates the stability of specific mRNAs, including CaV2.2. The Journal of neuroscience: the official journal of the Society for Neuroscience 28, 10604–10617, 10.1523/JNEUROSCI.2709-08.2008 (2008).
Brady, M. E. et al. Tip60 is a nuclear hormone receptor coactivator. J Biol Chem 274, 17599–17604 (1999).
Thornton, J. K. et al. The tumour-suppressor protein ASPP1 is nuclear in human germ cells and can modulate ratios of CD44 exon V5 spliced isoforms in vivo. Oncogene 25, 3104–3112, 10.1038/sj.onc.1209341 (2006).
Sanjabi, S. et al. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes & development 19, 2138–2151, 10.1101/gad.1329805 (2005).
Song, L. et al. Sam68 up-regulation correlates with and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. The Journal of pathology 222, 227–237, 10.1002/path.2751 (2010).
Stockley, J. et al. The RNA-binding protein hnRNPA2 regulates beta-catenin protein expression and is overexpressed in prostate cancer. RNA biology 11, 755–765 (2014).
Wainstein, M. A. et al. CWR22: androgen-dependent xenograft model derived from a primary human prostatic carcinoma. Cancer research 54, 6049–6052 (1994).
Pettaway, C. A. et al. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clinical cancer research: an official journal of the American Association for Cancer Research 2, 1627–1636 (1996).
Logan, I. R., Sapountzi, V., Gaughan, L., Neal, D. E. & Robson, C. N. Control of human PIRH2 protein stability: involvement of TIP60 and the proteosome. J Biol Chem 279, 11696–11704 (2004).
Edgar, R., Domrachev, M. & Lash, A. E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic acids research 30, 207–210 (2002).
Taylor, B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer cell 18, 11–22, 10.1016/j.ccr.2010.05.026 (2010).
Jiang, H., Lei, R., Ding, S. W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC bioinformatics 15, 182, 10.1186/1471-2105-15-182 (2014).
Patro, R., Mount, S. M. & Kingsford, C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat Biotechnol 32, 462–464, 10.1038/nbt.2862 (2014).
Murdoch, D. & Plummer, M. The R Project for Statistical Computing, http://www.r-project.org/ (2015). Date of access: 24/07/2015.
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. 4th edn, (Springer, 2002).
Acknowledgements
The following vectors were generously gifted: pEGFP-Sam68V229F, pLKO-puro Non-Target shRNA Control and pLKO-si-Sam68 (from C. Sette, University of Rome Tor Vergata, Italy), HA-hnRNPA2 (from Y. Matsuura, Osaka University, Japan), c-Flag pcDNA3 (from S. Smale, UCLA, USA), AR-V7 minigenes (from X. Dong, University of Vancouver, Canada) and p(ARE)3Luc (from D. Gioeli, University of Virginia, USA). We thank G. Kalna (CR-UK Beatson Institute) for statistical advice. This work was funded by Cancer Research UK (C19198/A15339 to PR and C596/A17196 to HYL) and the Royal College of Surgeons of England (to PR).
Author information
Authors and Affiliations
Contributions
J.S., D.J.E., C.N.R., H.Y.L. and P.R. designed the research; J.S., E.M., D.K., J.L. and P.R. performed the research and analysed the data; Y.Z., D.J.E., C.N.R. and H.Y.L. contributed new reagents; J.S. and P.R. wrote the paper. E.M., D.K., D.J.E., Y.Z., C.N.R., J.L. and H.Y.L. edited the paper. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Stockley, J., Markert, E., Zhou, Y. et al. The RNA-binding protein Sam68 regulates expression and transcription function of the androgen receptor splice variant AR-V7. Sci Rep 5, 13426 (2015). https://doi.org/10.1038/srep13426
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep13426
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.