Abstract
Liver fibrosis assessment by biopsy and conventional staining scores is based on histopathological criteria. Variations in sample preparation and the use of semi-quantitative histopathological methods commonly result in discrepancies between medical centers. Thus, minor changes in liver fibrosis might be overlooked in multi-center clinical trials, leading to statistically non-significant data. Here, we developed a computer-assisted, fully automated, staining-free method for hepatitis B-related liver fibrosis assessment. In total, 175 liver biopsies were divided into training (n = 105) and verification (n = 70) cohorts. Collagen was observed using second harmonic generation (SHG) microscopy without prior staining and hepatocyte morphology was recorded using two-photon excitation fluorescence (TPEF) microscopy. The training cohort was utilized to establish a quantification algorithm. Eleven of 19 computer-recognizable SHG/TPEF microscopic morphological features were significantly correlated with the ISHAK fibrosis stages (P < 0.001). A biphasic scoring method was applied, combining support vector machine and multivariate generalized linear models to assess the early and late stages of fibrosis, respectively, based on these parameters. The verification cohort was used to verify the scoring method and the area under the receiver operating characteristic curve was >0.82 for liver cirrhosis detection. Since no subjective gradings are needed, interobserver discrepancies could be avoided using this fully automated method.
Similar content being viewed by others
Introduction
Hepatitis B is a major infectious disease in Asia. Patients with chronic hepatitis B carry high liver cirrhosis and liver cancer risks1,2. Before the development of antiviral therapies, there was no efficient strategy for preventing chronic hepatitis B from progressing to end-stage liver disease3,4. According to reports from the World Health Organization, 2 billion people are estimated to have experienced a hepatitis B virus (HBV) infection during their lifetime and more than 350 million are chronically infected. In addition, approximately 600,000 people die annually from acute or chronic HBV infection sequelae.
Liver fibrosis develops in patients with chronic hepatitis B because of repeated necroinflammation, followed by hepatocyte regeneration and the accumulation of extracellular matrix (ECM) proteins, such as collagen5,6. When hepatocytes are injured, the immune system is activated and growth factors are released that activate hepatic stellate cells to produce collagen and other fibrillar components7. These substances are deposited into the ECM space. During liver injury, the collagen degradation process is impaired, which further leads to excessive ECM protein accumulation8. The end stage of liver fibrosis is liver cirrhosis, which can lead to liver cancer or liver failure.
Liver biopsy examinations are considered the “gold standard” for liver fibrosis diagnosis, by which clinical strategies for patient management are drafted. Biopsy samples provide direct information regarding the degrees of liver injury and fibrosis. Descriptive or semi-quantitative scoring systems are used for staging the disease extent based on liver histology findings. The traditional method classifies liver biopsy samples as absent, mild, moderate, or severe fibrosis. The first semi-quantitative scoring system, which was introduced in the 1980s, comprised a number range to indicate different categories of pathological features classified based on severity9. Several routinely used scoring systems were subsequently developed, including the Knodell histological activity index (HAI)10, Scheuer11, ISHAK12 and METAVIR13 systems. However, the pathological features that must be assessed using these systems do not have strict objective definitions. Thus, grading and staging scores are to some extent based on the subjective opinions of pathologists. Inter- and intra-observer variations can be as high as 35% and obtaining highly reproducible results using these systems is difficult14,15,16.
Several studies have quantified liver fibrosis by employing image analysis systems17,18,19. These computer-aided systems are used to provide objective quantitative measurements that might exclude observer discrepancies. However, the staining-dependent processes used in these systems cause difficulties due to variations in the staining procedure between biopsy samples. Moreover, most of these systems define the liver fibrosis staging index only according to the total fibrosis area in the samples20,21,22. Other pathological features also play vital roles in liver fibrosis grading and staging23, necessitating computer-based systems that can provide measurements in addition to the total fibrosis area. Matalka et al.23 developed the first quantification system based on a pattern classification method; however, the features extracted from the images did not provide histopathological definitions. The system developed by Dioguardi24 permitted more than 10 quantitative measurements; however, these measurements were incompatible with those used by pathologists and poor correlations were found between the obtained measurements and commonly used scoring systems. Goodman et al.25 summarized the common histopathologic features of liver biopsies that can be considered in staging systems (primarily morphological features). However, incorporating all of these particular morphological features into a pathologist-readable system for clinical use is challenging.
Recently, a staining-free approach that utilizes second harmonic generation (SHG) and two-photon excitation fluorescence (TPEF) microscopy has been developed, allowing for a more quantitative method to assess fibrosis26,27. By eliminating the use of staining, this method reduces the variations commonly observed in the staining process. Furthermore, the use of SHG provides a true quantitative measurement of the collagen level in the absence of sample digestion, leading to preservation of the morphological information in images26,27. Collagen quantitation using SHG/TPEF microscopy has been evaluated in animal models but not in a large collection of human tissues with various stages of fibrosis26,27. Such studies should be completed before this technique can be applied to patient samples.
Using both monophasic and biphasic models to interpret a biological system is not uncommon; for example, biphasic waveforms are commonly observed in electrocardiograms28,29. Many automated external defibrillators are designed to incorporate this biphasic nature for defibrillation with different electric waves at different times30. In this study, we used a similar biphasic approach to create differing models for the early and late stages of fibrosis development. We measured and quantified nineteen morphological features of collagen using SHG and TPEF microscopy and developed two algorithms for assessing fibrosis using these nineteen parameters for both early and later fibrosis stages. The results were validated with the histopathological ISHAK fibrosis assessment and conventional collagen staining methods, followed by transmitted light microscopy imaging. The validation results showed that the multivariate generalized linear model was better for assessing the fibrosis progression pattern at late liver fibrosis stages, whereas a non-linear (support vector machine, SVM) approach was better for the assessment of early fibrosis stages.
Results
Advanced SHG/TPEF microscopic quantification of fibrillar collagen was highly correlated with ISHAK fibrosis scores
To elucidate the SHG/TPEF microscopy imaging capacity for liver fibrosis, 175 liver biopsy samples were obtained from chronic HBV-infected patients. The clinical parameters of the recruited patients are listed in Table 1. One hundred and five randomly selected samples were assigned to the training cohort and used for developing and optimizing the computer-based algorithm for liver fibrosis quantification. The other 70 samples were used as a validation set to assess the performances of the models.
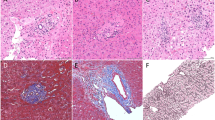
Figure 1A–D shows the images acquired using the conventional staining (Fig. 1A,B) and SHG/TPEF microscopy (Fig. 1C,D) methods. In SHG/TPEF imaging, fibrillar collagen was detected by SHG and is shown in green, whereas hepatocyte morphology was captured by TPEF microscopy and is shown in red. Visually, the SHG/TPEF technology resulted in the generation of a much more enhanced image with a better collagen distribution contrast. To minimize interference with normal-structured parenchyma and portal tract collagen, normal collagen readings were excluded. Figure 1E depicts the correlations identified by basic quantification (total collagen) or advanced quantification (Fibro-C-Index)27, excluding the normal-structured collagen signals31, with the ISHAK scores. The advanced quantification results were highly correlated with the ISHAK fibrosis scores.
Comparison between SHG/TPEF imaging and linear microscopy after Masson’s trichrome staining.
(A) and (B) represent fibrotic liver sections stained with Masson’s trichrome. Collagen fibers are stained in blue and hepatocytes are stained in red. (C) and (D) are SHG/TPEF microscopy images. Fibrillar collagen (green) was detected by an SHG signal and hepatocyte morphology (red) was observed with TPEF. (E) Correlations between the ISHAK scores and SHG/TPEF microscopy quantification results of samples from the training cohort. The total collagen levels are represented by solid squares and the advanced collagen readings are represented by solid circles.
Morphological liver fibrosis features visualized by SHG/TPEF microscopy
With the SHG/TPEF system, liver fibrosis severity could be quantitatively measured by examining the collagen fiber amount and morphology. Figure 2 shows the collagen profiles obtained from liver fibrosis qualitative imaging of the liver biopsies, including the total collagen distribution (A, green), various collagen strings (B, colored region), collagen percentage of the total area (CPA) (C) and collagen crosslinks (D, white arrow). The SHG/TPEF system employed 19 features (Table 2) to quantitatively measure liver fibrosis severity. These features were classified into three categories. The first feature category (F1-3) was collagen percentage, which included the total, aggregated and distributed collagen percentages. The second feature category (F4-13) was based on collagen string detection. Once a collagen string was detected, the contour that enclosed the collagen pixels was defined. Subsequently, the convex hull and the area and perimeter of each collagen string contour were also measured (Fig. 3A). To further describe the shapes and properties of the detected collagen strings, for each string, an ellipse was applied that enclosed all of the pixels used to calculate the global parameters (e.g., breadth and length) (Fig. 3B). Collagen connectivity (Fig. 3C) within the strings was calculated by skeletonizing them and then detecting the intersections between them.
The last set of features (F14-19) was generated according to the ratios of the different collagen string types.
Correlation between the morphological features observed with SHG/TPEF microscopy and ISHAK fibrosis stages
To identify the critical morphological features that can be used to assess liver fibrosis, the correlations between the 19 morphological features and ISHAK fibrosis scores were analyzed using univariate linear regression. Assuming that the ISHAK fibrosis scores were a linear function that increased from 1 to 6, we calculated the P values of all 19 features. The results are shown in Table 2. Note that most of the first and second category features (11 of 13 features) were highly correlated (P < 0.001), whereas only some of the third category features were correlated (P < 0.05).
Biphasic algorithm development for liver fibrosis assessment
In statistical analysis, collagen percentages (F1-3) were first employed to perform model fitting according to the ISHAK fibrosis scores (scores 1–6). Second, the collagen string properties (F4-13) were added. Finally, the collagen string ratios (F14-19) were included. A receiver operating characteristic (ROC) curve was used to assess the performance of the SHG/TPEF fibrosis score, using samples from the verification cohort (Table 3).
Overall, the sophisticated non-linear SVM model was superior to the generalized linear model. In both models, the additional morphological features significantly increased the area under the ROC curve (AUC). Additionally, some features were more important for identifying different stages, e.g., the F4-13 string properties in the SVM model were used to identify early-stage progression and the F14-19 string ratios in the generalized linear model were used to identify late-stage progression.
Most combinations possessed high AUC values (most were higher than 0.7, but some were higher than 0.8), indicating that these features were crucial indices that were highly compatible with the clinically accepted ISHAK scores (i.e., these SHG-detected morphological features might facilitate efficient liver fibrosis assessments).
Among all of the models shown in Table 3, the largest AUC was measured while distinguishing between the non-cirrhotic (ISHAK stages 1–4) and cirrhotic (ISHAK 5–6) patients with the multivariate generalized linear model, which utilized all 19 included features (AUC = 0.829). Additionally, the generalized linear model was better at distinguishing early from advanced cirrhosis patients (12345 vs. 6; AUC = 0.826). However, the SVM model was optimal for classifying the early fibrotic stages (AUCs = 0.745, 0.774 and 0.797 for 1 vs. 2–6, 1–2 vs. 3–6 and 1–3 vs. 4–6, respectively). By utilizing the strengths of both model types, a biphasic strategy was proposed, the first step of which was to classify patients as either cirrhotic or non-cirrhotic using the generalized linear model with all 19 variables. If cirrhosis was detected, the same model was used for the score calculation. Otherwise, the SVM model was used to assess patients with early stage fibrosis.
Discussion
Liver biopsy remains the standard for monitoring fibrosis progression, in which a small piece of liver tissue is removed using a biopsy needle, stained, examined under a microscope and graded based on a descriptive or semi-quantitative scoring method32. In addition to the potential complications related to the biopsy procedure, inherent drawbacks exist, including potentials for sampling errors, staining variations and inter- and intra-observer variabilities, in the interpretation of histology results32,33.
With the development of both mode-locked lasers and highly sensitive optical sensors, non-linear optical microscopy, such as multi-photon excitation fluorescence and multi-harmonic generation, has become an affordable option for tissue imaging34,35. Because TPEF microscopy has a different excitation mechanism than SHG, TPEF signals can be easily separated from SHG signals using the appropriate detectors. Combined TPEF/SHG images reveal additional fibrotic liver data, such as precise collagen distributions and undistorted liver cell morphologies. In recent studies, TPEF microscopy has been widely used for imaging organ structure components and their dynamic interactions in biological tissues. Additionally, SHG microscopy has been increasingly used for measuring highly ordered structures without central symmetry, such as type I collagen, which is the dominant collagen type in fibrotic livers36,37,38.
Compared with conventional collagen imaging, which uses transmitted light microscopy of histological tissue sections that are stained with either Masson’s trichrome or picrosirius red, SHG microscopy has several advantages, as follows: (1) this method provides superior information regarding fibrillar collagen; (2) sample staining is unnecessary; thus, the staining variations that result from different stain batches, protocols, time-dependent fading and photobleaching26,39 are eliminated; (3) fluorophores are absent from tissues; thus, signals are unaffected by dye concentrations and photobleaching; (4) infrared excitation sources can be used, resulting in less scattering in the tissue; and finally, (5) SHG microscopy enables deeper tissue penetration for imaging purposes; hence, the 3D visualization of fiber architecture can be achieved40. Additionally, TPEF microscopy can provide complementary structural and cellular information41.
Fibrillar collagen formation occurs late in fibrogenesis. A traditional fibrosis microscopy evaluation is commonly based on restricted assessments related to later fibrotic changes, whereas features of the early stages of fibrogenesis are usually neglected. However, the use of new morphological criteria in conjunction with standard histopathological fibrosis scoring could improve the assessments of relatively small amounts of fibrillar collagen within fibrous tissues. This study evaluated 19 morphological features of SHG/TPEF images that are automatically included by the Genesis program. Morphological features such as collagen string formation and connectivity may allow for the identification of several stages in the progression of fibrillar collagen formation; therefore, these morphological features may enable the evaluation of early stages of fibrogenesis. Gailhouste et al.42 have highlighted the benefits of the linear scale provided by the SHG index by comparing it with the histological METAVIR scoring system. However, they have shown that the continuous scale provides greater accuracy than the semi-quantitative scoring system because it underestimates variables that demonstrate architectural differences in collagen, resulting in the restricted SHG indexing of fibrosis stages.
In this study, a biphasic model was developed to separately assess the early and late stages of fibrosis. We compared a multivariate generalized linear model with the SVM model. The results showed that the SVM approach was superior for detecting and differentiating between fibrosis stages (ISHAK 1–4), suggesting that collagen accumulation during fibrosis progression might not be a linear process. The finding that the biphasic approach displayed better accuracy than the monophasic approach further suggests that the pace of collagen accumulation differs during the early and late stages of fibrosis development. After incorporating the SVM into the biphasic approach, we successfully achieved an ROC of >0.82 for monitoring late liver fibrosis stages (cirrhosis) and an ROC of >0.75 for monitoring early liver fibrosis stages. We also observed that the addition of many parameters with relatively high P values (P > 0.001) resulted in better performance for assessing the fibrosis degree, especially for the non-linear models. For example, F14-19, which includes the different collagen string feature ratios, was not directly correlated with the degree of fibrosis; however, the addition of these parameters to the multivariate generalized linear model resulted in significant improvements in the ROC values in most cases.
AUC analysis revealed that the inclusion of some features, such as the string properties (F4-13), into the SVM models helped to improve the distinction of early stage fibrosis stage, whereas including the string ratios (F14-19) in the generalized linear models helped to improve late fibrosis stage distinction. The first part of these results is in agreement with the results of a previous study of early stage collagen remodeling26, which revealed alterations in collagen shapes but no substantial change in the total amount of collagen. Moreover, the second part of our findings suggests that although the total amount of collagen is altered during late liver fibrosis stages, the ratio of the different collagen string features is a better method for assessing fibrosis stages.
In this study, the main reason for liver biopsy of chronic hepatitis B patients was pre-treatment evaluation. Therefore, these patients were mostly in the immune clearance stage and were not evenly distributed over all clinical stages. As a result, many patients were positive for HBeAg but were already progressing to liver cirrhosis, suggesting a failure of HBeAg seroconversion despite repeated hepatitis flares. However, even during the immune clearance phase, patients who were positive for HBeAg still had a borderline lower fibrosis score (mean ± SD, 3.42 ± 1.41 vs. 3.06 ± 1.36; HBeAg negative vs. positive, P = 0.156).
In conclusion, this study developed a fully automated and quantitative assessment system employing SHG/TREF microscopy to specifically assess HBV-related fibrosis. We systematically optimized the parameters for quantitative SHG/TPEF liver tissue imaging and developed fully automated image analysis algorithms. Compared with the traditional staining systems, the present system enables enhanced collagen fiber detection and quantification for fibrosis research and clinical diagnosis. SHG/TPEF microscopy is a standard analytical tool and its use in combination with a computer-based detection and calculation system represents a powerful technique that can be applied in multi-center comparisons of liver fibrosis.
Methods
Tissue Preparation
The study cohort consisted of 175 liver biopsy samples from chronic HBV-infected patients who underwent liver biopsies at LinKo Chang Gung Memorial Hospital between 2000 and 2012. Clinical and pathological characteristics were obtained from the patients’ medical records. As described below, liver fibrosis was staged by pathologists according to the ISHAK scoring system and was then automatically evaluated with computational algorithms according to 19 morphological variables. Of the 175 samples, 105 (training cohort) were used for developing a computer-based algorithm with a close correlation to the ISHAK fibrosis score. The other 70 samples were used to validate the scoring method derived from the training system.
This study was approved by the Ethics Committee of Chang Gung Memorial Hospital. Written informed consent was obtained from each patient and the study was carried out in accordance with the approved guidelines.
Masson’s Trichrome Staining
Masson’s trichrome staining was performed at the Chang Gung Memorial Hospital Department of Anatomic Pathology as follows. First, 5-micron FFPE sections were deparaffinized and hydrated in distilled water. Then, Bouin’s fixative was used as a mordant for 1 h at 56 °C. The FFPE sections were cooled and washed in running water until the yellow coloring disappeared. The samples were stained in Weigert’s hematoxylin stain for 10 min, thoroughly washed in tap water for 10 min, stained again in an acid fuchsin solution for 15 min and then rinsed in distilled water for 3 min. After rinsing, the slides were treated with phosphomolybdic acid solution for 10 min and then rinsed in distilled water for 10 min. Finally, the slides were stained with a light-green solution for 2 min and rinsed in distilled water. After thorough dehydration using alcohol, the slides were mounted and coverslips were placed onto them.
Image Acquisition System
Images were acquired using a GenesisTM (HistoIndex Pte. Ltd, Singapore) system, in which SHG microscopy was used to visualize collagen and the other cell structures were visualized with TPEF microscopy. Details of the optical settings used can be obtained from Sun et al.26 and Tai et al.27.
The samples were laser-excited at 780 nm, SHG signals were recorded at 390 nm and TPEF signals were recorded at 550 nm. Images were acquired at 20× magnification with 512 × 512 pixel resolution and each image had a dimension of 200 × 200 μm. Multiple adjacent images were captured to encompass large areas. Nine images (3 × 3 images, 600 × 600 μm dimension) were acquired per site. Overall, five 3 × 3 images from five sites were obtained for each sample.
SHG Quantification
Total collagen percentages and previously described collagen features, including specific collagen strings and collagen connectivity-related measurements, were used to assess correlations with the ISHAK fibrosis scores. Nineteen morphological features were used in this study. These variables were divided into three groups, as follows:
Group 1
CPA (collagen percentage of the total area), Agg (the aggregated collagen percentage of the total area) and Dis (the distributed collagen percentage of the total area). The total collagen level was detected by SHG and segmented from the raw images. The aggregated and distributed collagen were differentiated with a low-pass filter26.
Group 2
NoStr (collagen string number), NoShortStr (short collagen string number), NoLongStr (long collagen string number), NoThinStr (thin collagen string number), NoThickStr (thick collagen string number), StrArea (total string area), StrLength (total string length), StrWidth (total string width), StrPerimeter (total string perimeter) and NoXlink (total crosslink number). The collagen strings were detected using a connected-component algorithm of binary images from a previous image segmentation. Each connected component was fitted with an ellipse and then the axes of the ellipses were used to divide the strings into long and short as well as thin and thick sub-groups. The total lengths, widths, areas and perimeters of the collagen strings were also measured. We used an image skeleton algorithm to detect branch points for determining crosslink numbers.
Group 3
NoShortStr/NoStr (ratio between the short string and total string numbers), NoLongStr/NoStr (ratio between the long string and total string numbers), NoShortStr/NoLongStr (ratio between the short string and long string numbers), NoThinStr/NoStr (ratio between the thin string and total string numbers), NoThickStr/NoStr (ratio between the thick string and total string numbers) and NoThinStr/NoThickStr (ratio between the thin string and thick string numbers). We found that these relative ratios improved the system only when absolute (normalized) values were used for Group 2.
Statistical Analysis and Model Construction
Univariate analyses were performed to assess correlations between individual image features and the ISHAK scores using linear regression. Two modeling method types were used for the patient classifications, which were based on multiple image features. In the first method, the dependent variable, y, logit, which was a fibrosis stage indicator, was a linear combination of n feature variables X1 … Xn, as shown in the following equation:

The coefficients β0, β1, …βn were estimated with the maximum likelihood method.
The second method used was the SVM model. A hyperplane was constructed for optimally classifying the subjects into fibrosis stages based on the feature values in the m-dimensional space (m ≥ n). The radial basis function (RBF) kernel was used as the distance measure between two subject vectors x and x’, i.e.,

where σ controls the width of the kernel.
Additional Information
How to cite this article: Wang, T.-H. et al. Automated biphasic morphological assessment of hepatitis B-related liver fibrosis using second harmonic generation microscopy. Sci. Rep. 5, 12962; doi: 10.1038/srep12962 (2015).
References
Beasley, R. P. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer 61, 1942–1956 (1988).
Chen, C. J. et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295, 65–73 (2006).
Lok, A. S. & McMahon, B. J. Chronic hepatitis B: update 2009. Hepatology 50, 661–662 (2009).
EASL clinical practice guidelines: Management of chronic hepatitis B virus infection. J Hepatol 57, 167–185 (2012).
Bataller, R. & Brenner, D. A. Liver fibrosis. J Clin Invest 115, 209–218 (2005).
Lok, A. S. Hepatitis B: liver fibrosis and hepatocellular carcinoma. Gastroenterol Clin Biol 33, 911–915 (2009).
Friedman, S. L. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J Biol Chem 275, 2247–2250 (2000).
Benyon, R. C. & Iredale, J. P. Is liver fibrosis reversible? Gut 46, 443–446 (2000).
Standish, R. A., Cholongitas, E., Dhillon, A., Burroughs, A. K. & Dhillon, A. P. An appraisal of the histopathological assessment of liver fibrosis. Gut 55, 569–578 (2006).
Knodell, R. G. et al. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1, 431–435 (1981).
Scheuer, P. J. Classification of chronic viral hepatitis: a need for reassessment. J Hepatol 13, 372–374 (1991).
Ishak, K. et al. Histological grading and staging of chronic hepatitis. J Hepatol 22, 696–699 (1995).
Bedossa, P. & Poynard, T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology 24, 289–293 (1996).
Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. The French METAVIR Cooperative Study Group. Hepatology 20, 15–20 (1994).
Gronbaek, K. et al. Interobserver variation in interpretation of serial liver biopsies from patients with chronic hepatitis C. J Viral Hepat 9, 443–449 (2002).
Westin, J., Lagging, L. M., Wejstal, R., Norkrans, G. & Dhillon, A. P. Interobserver study of liver histopathology using the Ishak score in patients with chronic hepatitis C virus infection. Liver 19, 183–187 (1999).
Masseroli, M. et al. Automatic quantification of liver fibrosis: design and validation of a new image analysis method: comparison with semi-quantitative indexes of fibrosis. J Hepatol 32, 453–464 (2000).
O’Brien, M. J. et al. An assessment of digital image analysis to measure fibrosis in liver biopsy specimens of patients with chronic hepatitis C. Am J Clin Pathol 114, 712–718 (2000).
Wright, M., Thursz, M., Pullen, R., Thomas, H. & Goldin, R. Quantitative versus morphological assessment of liver fibrosis: semi-quantitative scores are more robust than digital image fibrosis area estimation. Liver Int. 23, 28–34 (2003).
Lazzarini, A. L., Levine, R. A., Ploutz-Snyder, R. J. & Sanderson, S. O. Advances in digital quantification technique enhance discrimination between mild and advanced liver fibrosis in chronic hepatitis C. Liver Int. 25, 1142–1149 (2005).
Friedenberg, M. A. et al. Simplified method of hepatic fibrosis quantification: design of a new morphometric analysis application. Liver Int 25, 1156–1161 (2005).
Goodman, Z. D., Becker, R. L., Jr., Pockros, P. J. & Afdhal, N. H. Progression of fibrosis in advanced chronic hepatitis C: evaluation by morphometric image analysis. Hepatology 45, 886–894 (2007).
Matalka, I. I., Al-Jarrah, O. M. & Manasrah, T. M. Quantitative assessment of liver fibrosis: a novel automated image analysis method. Liver Int 26, 1054–1064 (2006).
Dioguardi, N., Grizzi, F., Fiamengo, B. & Russo, C. Metrically measuring liver biopsy: a chronic hepatitis B and C computer-aided morphologic description. World J Gastroenterol 14, 7335–7344 (2008).
Goodman, Z. D. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol 47, 598–607 (2007).
Sun, W. et al. Nonlinear optical microscopy: use of second harmonic generation and two-photon microscopy for automated quantitative liver fibrosis studies. Journal of biomedical optics 13, 064010 (2008).
Tai, D. C. et al. Fibro-C-Index: comprehensive, morphology-based quantification of liver fibrosis using second harmonic generation and two-photon microscopy. Journal of biomedical optics 14, 044013 (2009).
Gliner, B. E., Lyster, T. E., Dillion, S. M. & Bardy, G. H. Transthoracic defibrillation of swine with monophasic and biphasic waveforms. Circulation 92, 1634–1643 (1995).
Tang, W. et al. The effects of biphasic waveform design on post-resuscitation myocardial function. Journal of the American College of Cardiology 43, 1228–1235 (2004).
Cummins, R. O. et al. Low-energy biphasic waveform defibrillation: evidence-based review applied to emergency cardiovascular care guidelines: a statement for healthcare professionals from the American Heart Association Committee on Emergency Cardiovascular Care and the Subcommittees on Basic Life Support, Advanced Cardiac Life Support and Pediatric Resuscitation. Circulation 97, 1654–1667 (1998).
Xu, S. et al. qFibrosis: A fully-quantitative innovative method incorporating histological features to facilitate accurate fibrosis scoring in animal model and chronic hepatitis B patients. J Hepatol 61, 260–269 (2014).
Bedossa, P., Dargere, D. & Paradis, V. Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology 38, 1449–1457 (2003).
Perrault, J., McGill, D. B., Ott, B. J. & Taylor, W. F. Liver biopsy: complications in 1000 inpatients and outpatients. Gastroenterology 74, 103–106 (1978).
Denk, W., Strickler, J. H. & Webb, W. W. Two-photon laser scanning fluorescence microscopy. Science 248, 73–76 (1990).
Campagnola, P. J. et al. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophys J 82, 493–508 (2002).
Han, X., Burke, R. M., Zettel, M. L., Tang, P. & Brown, E. B. Second harmonic properties of tumor collagen: determining the structural relationship between reactive stroma and healthy stroma. Opt Express 16, 1846–1859 (2008).
Lin, M. G. et al. Evaluation of dermal thermal damage by multiphoton autofluorescence and second-harmonic-generation microscopy. J Biomed Opt 11, 064006 (2006).
Theodossiou, T. A., Thrasivoulou, C., Ekwobi, C. & Becker, D. L. Second harmonic generation confocal microscopy of collagen type I from rat tendon cryosections. Biophys J 91, 4665–4677 (2006).
Strupler, M. et al. Second harmonic imaging and scoring of collagen in fibrotic tissues. Optics express 15, 4054–4065 (2007).
Cox, G. et al. 3-dimensional imaging of collagen using second harmonic generation. Journal of structural biology 141, 53–62 (2003).
Lee, H. S. et al. Optical biopsy of liver fibrosis by use of multiphoton microscopy. Opt Lett 29, 2614–2616 (2004).
Gailhouste, L. et al. Fibrillar collagen scoring by second harmonic microscopy: a new tool in the assessment of liver fibrosis. J Hepatol 52, 398–406 (2010).
Acknowledgements
The authors thank the Chang Gung Memorial Hospital Tissue Bank, LinKo, Taiwan, for their excellent tissue processing. We thank Chung-Yin Wu, Hui-Chin Chen, Chien-Chih Wang, May-Ling Tsao, Hsin-Yu Lai and Yen-Ling Chuang for the excellent technical assistance in this project. This project was funded by the Chang Gung Medical Research Program, Taiwan (CMRPG3A1193, CMRPG1B0571 and CLRPG370695).
Author information
Authors and Affiliations
Contributions
T.H.W. and C.T.Y. conceived and designed the experiments. T.H.W., T.C.C. and X.T. performed the experiments. X.T. and K.H.L. analyzed the data. T.C.C. contributed reagents/materials/analysis tools. T.H.W., X.T. and C.T.Y. wrote the manuscript. All of the authors discussed the results and commented on and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wang, TH., Chen, TC., Teng, X. et al. Automated biphasic morphological assessment of hepatitis B-related liver fibrosis using second harmonic generation microscopy. Sci Rep 5, 12962 (2015). https://doi.org/10.1038/srep12962
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep12962
This article is cited by
-
Quantitative image-based collagen structural features predict the reversibility of hepatitis C virus-induced liver fibrosis post antiviral therapies
Scientific Reports (2023)
-
Label-free multiphoton microscopy enables histopathological assessment of colorectal liver metastases and supports automated classification of neoplastic tissue
Scientific Reports (2023)
-
Quantitative assessment of liver fibrosis (qFibrosis) reveals precise outcomes in Ishak “stable” patients on anti-HBV therapy
Scientific Reports (2018)
-
Deep learning enables automated scoring of liver fibrosis stages
Scientific Reports (2018)
-
Automated evaluation of liver fibrosis in thioacetamide, carbon tetrachloride, and bile duct ligation rodent models using second-harmonic generation/two-photon excited fluorescence microscopy
Laboratory Investigation (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.