Abstract
Most high γ-aminobutyric acid (GABA) producers are Lactobacillus brevis of plant origin, which may be not able to ferment milk well due to its poor proteolytic nature as evidenced by the absence of genes encoding extracellular proteinases in its genome. In the present study, two glutamic acid decarboxylase (GAD) genes, gadA and gadB, were found in high GABA-producing L. brevis NPS-QW-145. Co-culturing of this organism with conventional dairy starters was carried out to manufacture GABA-rich fermented milk. It was observed that all the selected strains of Streptococcus thermophilus, but not Lactobacillus delbrueckii subsp. bulgaricus, improved the viability of L. brevis NPS-QW-145 in milk. Only certain strains of S. thermophilus improved the gadA mRNA level in L. brevis NPS-QW-145, thus enhanced GABA biosynthesis by the latter. These results suggest that certain S. thermophilus strains are highly recommended to co-culture with high GABA producer for manufacturing GABA-rich fermented milk.
Similar content being viewed by others
Introduction
γ-Aminobutyric acid (GABA), a non-protein amino acid, is widely found in plants, microorganisms and vertebrates1,2. GABA-rich foods that are naturally produced have been popular for decades and have shown anti-hypertensive effect as an important function2,3,4,5,6,7. In general, GABA content in plant and animal products is very low for delivering any functional benefit in human. Thus, there has been an increasing interest in using high GABA-producing microorganisms for manufacturing GABA-rich fermented milk products such as yogurt and cheese.
Currently, most high GABA producers belong to Lactobacillus species and Lactobacillus brevis has been identified as a key species for producing GABA8. It has been well documented that glutamic acid decarboxylase (GAD) operon comprise a transcriptional regulator (gadR), glutamate decarboxylases (gadA or/and gadB) and a glutamate/GABA antiporter (gadC) in GABA-producing microorganisms9. Moreover, high GABA-producing L. brevis of plant origin has been isolated from Korean kimchi or other fermented vegetables8. Genomic analysis indicated the absence of genes encoding extracellular or cell wall-anchored proteinases in the sequenced L. brevis ATCC 367 (a starter culture for beer, sourdough and silage) and L. brevis KB290 (an isolate from traditional Japanese fermented vegetable). This may suggest that L. brevis of plant origin may not able to survive in milk environments because of its poor proteolytic nature. It is known to us that mammalian milks contain lactose and casein as the major sugar and protein sources, but these are not ideal sources of nutrients for the growth of non-proteolytic lactic acid bacteria (LAB).
GABA-producing LAB shows great promise for manufacturing GABA-rich fermented milk. For instance, milk fermented by L. casei Shirota and Lactococcus lactis YIT 2027 contained 10 to 12 mg of GABA per 100 mL of fermented milk, this functional food has shown the functionality of lowering the blood pressure in mildly hypertensive patients5; L. helveticus ND01 yielded 165.11 mg of GABA per 1 kg of fermented milk after 20 h fermentation at 37 °C10; Lactococcus lactis DIBCA1 and L. plantarum PU11 supplemented with 20 mmol/L of glutamic acid produced 144.5 mg of GABA per 1 kg of fermented milk after 48 h fermentation at 37 °C11. It is known to us that Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus (hereafter L. bulgaricus) are important starter microorganisms required for the manufacture of fermented dairy foods such as yogurt and certain cheese varieties12,13,14. In addition, monosodium glutamate (MSG) is normally added to milk as the substrate for manufacturing GABA-rich fermented milk because of low content of free glutamate in milk8.
High GABA producer of plant origin may not be able to survive in milk, or may not even ferment milk. Although their viability in milk could be enhanced by adding particular nutrients to milk base, this practice may not be of interest for dairy industry. Although some probiotics or novel LAB strains were adopted as adjunct starters for milk fermentation, conventional dairy starters including S. thermophilus and L. bulgaricus are required to add into the milk because of the regulations in most countries. Till now, there is very little information on the synergistic effect of high GABA producers and dairy starters. In the present study, we report a new strategy of manufacturing GABA-rich fermented milk by co-culturing high GABA producer with dairy starter including S. thermophilus and L. bulgaricus in skimmed milk supplemented with MSG and provide new insights into the effects of dairy starters on the cell viability of L. brevis NPS-QW-145 (a high GABA producer; hereafter L. brevis 145) and its GABA biosynthesis ability in milk.
Results
Two GAD genes were detected in the genome of L. brevis 145
The amplification result of partial GAD gene (~408 bp) in the eight dairy starters and L. brevis 145 is shown in Fig. 1a. Normally, the full length of GAD gene is ~1400 bp. As shown in the Fig. 1a, this gene in all the selected dairy starters including S. thermophilus and L. bulgaricus was not detected, while it existed in L. brevis 145. Moreover, there was no GABA production from these dairy starters when cultured in milk and M17/MRS broth (data not shown). Thus, it was concluded that the GABA was only produced by L. brevis 145.
Amplification of GAD gene(s) and 16S rRNA gene from L. brevis 145 and eight dairy starters.
(a) detection of GAD gene using degenerate primers DP1 and DP2; (b) amplification of GAD gene(s) in L. brevis 145 using degenerate primers PGDG-2F and PGDG-4R; (c) specificity of primers s-Lbre-F and s-Lbre-R for amplifying 16S rRNA gene from L. brevis. Denotation: M, DNA ladders (Promega 1 kb DNA ladder in Fig. 1a; Invitrogen 1 Kb Plus DNA Ladder in Fig. 1b and Fig. 1c); B, amplification without DNA; Lane 1, L. brevis 145; Lane 2, S. thermophilus ASCC 1275; Lane 3, S. thermophilus ASCC 1303; Lane 4, S. thermophilus YI-B1; Lane 5, S. thermophilus YI-N1; Lane 6, S. thermophilus YI-M1; Lane 7, L. bulgaricus ASCC 756; Lane 8, L. bulgaricus ASCC 859; Lane 9, L. bulgaricus YI-B2.
The partial GAD gene from L. brevis 145 was successfully amplified and sequenced (Fig. 1b). The size of PCR product was about 1014 bp based on the alignment of amino acids sequence of GAD gene in L. brevis (Fig. 2). Interestingly, two GAD genes, gadA and gadB, were found in L. brevis 145 after analyzing the sequences of the PCR product. Excluding the length of degenerate primers PGDG-2R (35 bp) and PGDG-4R (32 bp), the length of the amplified gadA and gadB was 948 bp and 921 bp, respectively. The GenBank accession numbers for the partial gadA and gadB sequences of L. brevis 145 are KM875632.1 and KM875633.1, respectively. The nucleotides sequences of partial gadA and gadB showed a similarity of 99% with GAD gene in other L. brevis strains (KB290, 877G, CGMCC 1306, ATCC 367, BH2 and IFO 12005). The predicated amino acids sequence of amplified gadA (316 aa) only have 164 aa in common with that of amplified gadB (307 aa) after ClustalW alignment. Besides the above genetic analysis, we have confirmed high GABA production from this organism15. These sequences were further used to design qPCR primers for quantifying the expression of GAD genes in L. brevis 145 as shown in Table 1.
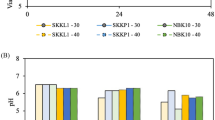
The pH of fermented milks using mixed-cultures or mono-culture
The pH of the fermented milks is shown in Fig. 3. As shown in the Figure, the pH in the milk fermented by L. brevis 145 alone was similar to that of the blank milk suggesting that L. brevis 145 was not able to ferment milk. Co-culturing of L. brevis 145 with S. thermophilus in milk after 24 h fermentation at 37 °C resulted in an average pH of ~4.50, whereas co-culturing of L. brevis 145 with L. bulgaricus showed an average pH of ~3.70. The pH of the milk fermented by three cultures of L. brevis 145, S. thermophilus YI-B1 and L. bulgaricus YI-B2 was ~3.90. It was observed that the pH of the milk fermented by co-cultures of L. brevis 145 and S. thermophilus or L. bulgaricus was not significantly (P ≥ 0.05) changed after the supplementation with MSG. This suggests that the addition of MSG did not influence the pH of the milk. Additionally, the pH (~4.50) of milk fermented by S. thermophilus and L. brevis 145 was similar to that of commercial yogurts. This implies that using S. thermophilus and L. brevis 145 could be used to produce a yogurt-like product.
Cell viabilities of S. thermophilus and L. bulgaricus in milk
Cell viabilities of eight dairy starters in fermented milks are shown in Fig. 4. As indicated in the figure, the viabilities of S. thermophilus and L. bulgaricus co-cultured with L. brevis 145 in fermented milks were not significantly (P ≥ 0.05) changed after the supplementation with MSG. This suggests that MSG supplemented (2 g/L) to milk did not have much influence on the viabilities of both S. thermophilus and L. bulgaricus cells. Also, the cell counts of both dairy starters were above 8.5 Log10 CFU/mL in milk.
Cell viability of L. brevis 145 after co-culturing with dairy S. thermophilus or/and L. bulgaricus in milk
The primers (s-Lbre-F and s-Lbre-R; Table 1) showed strong specificity for amplifying partial 16S rRNA gene in L. brevis 145 (Fig. 1c). The efficiency of this qPCR assay was 98.435%. This indicated that this pair of primers was suitable for qPCR quantitation of L. brevis 145 cells and for further gene expression experiments. The equation of standard curve is y = –4.1026x + 52.009 (R2 = 0.9851; y, Ct value; x, cell counts]. The standard curves showed a good correlation coefficient value (R2 = 0.9851), suggesting that the Ct values were linear over the range of cell count tested (3.2 × 104 ~ 3.2 × 109 CFU/mL). The analysis of the melting curves did not show the formation of non-specific fragments or primer-dimers indicating that the qPCR assay was accurate and reproducible.
The viability of L. brevis 145 in milk during co-culturing is shown in Fig. 5. Before the fermentation, the initial counts of L. brevis 145 cells in milk were ~3 × 107 CFU/mL (~7.48 Log10 CFU/mL). However, the counts of this strain decreased to ~6.50 Log10 CFU/mL after 24 h of fermentation (Fig. 5). This indicates that viability of L. brevis 145 was not maintained in milk during fermentation. In general, it was observed that the viability of L. brevis 145 decreased slightly but not significantly (P ≥ 0.05) after supplementation with MSG to milk, except the fermentation using co-cultures of L. brevis 145 and L. bulgaricus ASCC 756. Interestingly, the average cell counts of L. brevis 145 after co-culturing with S. thermophilus was ~7.90 Log10 CFU/mL, which was significantly (P < 0.01) higher than that of co-culturing with L. bulgaricus, co-culturing with both S. thermophilus and L. bulgaricus and the control fermentation with only L. brevis 145. For co-culturing with L. bulgaricus, the viability of L. brevis 145 decreased significantly (P < 0.01) as compared with the control using only L. brevis 145. Thus, it was suggested that the presence of L. bulgaricus in co-culture with L. brevis 145 had a negative effect on the viability of L. brevis 145.
Cell counts of L. brevis 145 in fermented milks.
ST, S. thermophilus; Lbu, L. bulgaricus; Lbre 145, L. brevis 145. Star (*P < 0.05) is for the comparison of data between fermentation with and without the supplementation of MSG; Capital letters (A, B, C and D) are designated to indicate the significance of the group data of L. brevis counts, the same letter among each group indicates no significance (P ≥ 0.05). The initial cell counts of L. brevis 145 after inoculation in milk was ~3 × 107 CFU/mL (7.48 Log10 CFU/mL).
GABA yield and residual MSG content in fermented milks
The content of GABA and residual MSG in milk supplemented with or without MSG is shown in Table 2. As shown in the table, GABA was not detected in the milk fermented by only L. brevis 145. This is mainly because the milk composition was not able to support the growth of this organism (Fig. 3), which was also evidenced by the decreased viability of this strain in milk grown alone (Fig. 5). However, after co-culturing with certain S. thermophilus strains (ASCC 1275, YI-B1 and YI-N1), GABA production was increased in milk supplemented with MSG after 24 h fermentation. Co-culturing of S. thermophilus YI-B1 and L. brevis 145 in milk containing 2 g/L of MSG yielded the highest level (~314 mg per 1 kg of fermented milk) of GABA after 24 h of fermentation. Till now, this may be the highest known amount of GABA content in fluid milk products that were fermented by LAB5,10,11.
MSG content at a high level in milk may not be appreciated because of its flavor. In the present study, it was found that GABA production did not correlate with the reduction in MSG level (Table 2). Hence, the level of MSG supplemented to milk could be modified. In general, the level of residual MSG in milk fermented by S. thermophilus and L. brevis 145 was lower than that by L. bulgaricus and L. brevis 145. Because there was very limited GABA production converted from MSG, the high glutamate in milk fermented by L. bulgaricus and L. brevis 145 may be due to the better extracellular proteolytic activity of L. bulgaricus than that of S. thermophilus. This also suggests that L. bulgaricus may have obtained sufficient glutamate from milk proteins after hydrolysis. In addition, MSG content in milk fermented by S. thermophilus YI-N1 and L. brevis 145 was the lowest, which indicates that S. thermophilus YI-N1 may have utilized more MSG than that by other selected dairy starters. Thus, S. thermophilus strains could be used for reducing the MSG level in fermented milk.
GAD gene expression in L. brevis 145
We wanted to find a suitable housekeeping gene in L. brevis for normalization; however, the expressed stable genes in L. brevis including tuf (elongation factor Tu)9, proC (amino acid biosynthesis) and rpoB (RNA polymerase)16 are not specific for L. brevis. These genes also exist in S. thermophilus. Clear bands were observed in agarose gel after electrophoresis when amplification was carried out for five strains of S. thermophilus using the primers reported in above studies9,16. Thus, 16S rRNA gene was used as a housekeeping gene for real-time qPCR assay using the primers exhibited in Table 1 and its efficiency was assessed as well. The efficiencies of this qPCR assay using 16S rRNA gene, gadA and gadB were 91.78%, 99.54% and 101.39%, respecitively.
The result of qPCR quantitation of gadA and gadB mRNA level in L. brevis 145 is shown in Fig. 6. Interestingly, it was observed that only the gadA mRNA level in L. brevis 145 was significantly (P < 0.01) up-regulated by certain S. thermophilus strains (ASCC 1275, YI-B1 and YI-N1) as compared with other two S. thermophilus strains (ASCC 1303 and YI-M1), whereas the gadB mRNA level in L. brevis 145 was not regulated by all selected S. thermophilus strains. The improved gadA mRNA level may suggest an enhanced GABA biosynthesis in L. brevis 145 resulting in an increased GABA production after co-culturing with S. thermophilus ASCC 1275, YI-B1 and YI-N1 in milk supplemented with 2 g/L of MSG (Table 2).
Relative gene expression of gadA and gadB in L. brevis 145 after co-cultured with S. thermophilus in milk supplemented with 2 g/L of MSG.
The levels of gadA and gadB mRNA from L. brevis 145 after co-cultured with S. thermophilus YI-M1 was used as a reference for comparision. Comparative critical threshold method (R = 2–ΔΔCт) was carried out for data analysis of three indendent exeriments, a positive value indicates up-regulation while a negative value indicates down-regulation. Lowercase letters (a & b) are designated to indicate the significance of gadB and gadA mRNA levels, the same letter above/below each bar indicates no significance (P ≥ 0.01). ST, S. thermophilus; Lbre 145, L. brevis 145.
Discussion
S. thermophilus and L. bulgaricus are two common dairy starters that are highly recommended for the manufacture of yogurt and several type of cheeses. Thus, in this study the above starters were co-cultured with L. brevis 145 for making GABA-rich fermented milk. Also, MSG was supplemented as the substrate for GABA production. However, MSG is additional sodium salt in milk and its effects on milk fermentation needs to be demonstrated. It was observed that MSG supplemented at the level of 2 g/L did not show much effect on the pH of milk (Fig. 3), cell viabilities of S. thermophilus and L. bulgaricus (Fig. 4) and the viability of L. brevis 145 (Fig. 5). These results suggest that supplementation with MSG at 2 g/L or below this level to milk base may be an option for making functional fermented milk.
Without MSG supplementation, GABA production from L. brevis 145 was very low when co-cultured with L. bulgaricus, whereas GABA was not detected when co-cultured with S. thermophilus (Table 2). However, MSG supplemented at 2 g/L in milk improved the GABA production greatly from L. brevis 145 when only co-cultured with certain S. thermophilus strains, while its production was not significantly increased during co-culturing with L. bulgaricus (Table 2). Thus, it appears that MSG supplementation was necessary for an improved GABA production from L. brevis 145. However, further documentation on the flavor of this fermented milk is necessary because of the introduction of MSG in milk. Interestingly, it was found that S. thermophilus could utilize more MSG than that of L. bulgaricus. This may be of particular interest for dairy industry.
Use of L. bulgaricus for co-culturing with L. brevis 145 in milk may not be ideal because of the generation of low counts of L. brevis 145 by this species (Fig. 5). This is possibly due to competition and because they belong to the same Lactobacillus genus17. However, use of S. thermophilus could be a promising option due to its ability to maintain the viability of L. brevis 145 in milk. A previous study revealed that formic acid, folic acid and fatty acids released from S. thermophilus supported the growth of Lactobacillus genus in milk18. Moreover, dairy S. thermophilus possesses good extracellular proteolytic property and could also supply L. brevis 145 with sufficient amino acids or peptides19,20. This may explain that S. thermophilus was able to support the growth of L. brevis 145 in milk during co-culturing. However, only certain strains of S. thermophilus (ASCC 1275, YI-B1 and YI-N1) improved the GABA yield from L. brevis 145 (Table 2), which was closely associated with an increased gadA mRNA level in L. brevis 145 when co-cultured with above strains (Fig. 6). This implies that the GABA biosynthesis in L. brevis 145 could be up-regulated by certain S. thermophilus strains.
Interestingly, gadA and gadB were found to be independently conserved in L. brevis based on their amino acids sequences and some strains only possessed gadA (NCL912 and IFO12005) or gadB (BH2, 877G and OPK-3), while some strains (KB290, ATCC367, BSO 464, AG48, EW and DmCS_003) may have both genes in their genomes. It has been demonstrated that gadA in previously studied L. brevis strains (NCL912 and IFO12005) and gadB in L. brevis strains (BH2, 877G and OPK-3) have shown their capability of producing high amount of GABA in their host. This indicates that both two glutamate decarboxylases are functional and may exhibit similar enzymatic activity because they may possess the same core conformation. Moreover, certain S. thermophilus strains (ASCC 1275, YI-B1 and YI-N1) regulated the gadA expression in L. brevis 145, while the level of gadB mRNA transcripts was not affected by the former. Other S. thermophilus strains (ASCC 1303 and YI-M1) were not able to influence the level of both gadA and gadB transcripts. This may be related with the strain-specific interactions between S. thermophilus and L. brevis 145 regarding to the metabolism of purine, amino acid and long-chain fatty acid18. It was found that the location of gadA and gadB in the genome of sequenced L. brevis strains (KB290, ATCC367, BSO 464, AG48, EW and DmCS_003) was not close to each other, but only one GAD gene (gadA or gadB) was found in the gad operon. This suggests that there may be different mechanism for the regulation of gadA and gadB gene expression in their hosts. This merits further investigation on the regulation by certain S. thermophilus strains.
Concluding remarks
In this study, two glutamate decarboxylase gene, gadA and gadB, were found in high GABA-producing L. brevis 145. However, this organism was not able to ferment milk. It was observed that all the selected dairy S. thermophilus strains, but not L. bulgaricus, improved the viability of L. brevis 145 when co-cultured in milk. Only certain S. thermophilus strains improved GABA production from L. brevis 145, which was evidenced by the increased gadA mRNA transcripts in the latter. Moreover, co-cultures of S. thermophilus and L. brevis 145 utilized more MSG than co-cultures of L. bulgaricus and L. brevis 145 suggesting the use of S. thermophilus for reducing MSG content if supplemented in milk. This study provides a new insight of using S. thermophilus for co-culturing with high GABA producer of plant origin for manufacturing GABA-rich fermented milk.
Methods
Bacterial strains and culture conditions
Non-dairy starter L. brevis NPS-QW-145, a high GABA-producing strain isolated from Korean kimchi, was used in this study as a model of high GABA producer15. Eight dairy starters (Table 1) were used for co-culturing with this organism. Lactobacillus strains were activated in DifcoTM lactobacilli MRS broth (BD Company, MD, USA), while S. thermophilus strains were cultivated in M17 broth (BD Company). Working cultures were propagated three times consecutively using 1% inoculation in the above medium (MRS or M17) at 37 °C for 18 h.
Alignment of the amino acids of glutamic acid decarboxylase (GAD) from L. brevis
In order to amplify the GAD gene from L. brevis 145, degenerated primers were designed according to the conserved regions of this enzyme from the species of L. brevis. The full-length sequences of amino acids of GAD from L. brevis strains were downloaded from the database of the National Center for Biotechnology Information (NCBI) and were aligned using BioEdit software (version 7.2.5). The conserved region [NAIDKSEYPR(K)TA] was used for designing the forward primer, whereas another conserved sequence [GWQVPA(T)YPLPKN] was for designing the reverse primer (Fig. 2). The degenerate primers, PGDG-2F and PGDG-4R, are shown in Table 1.
Amplification of GAD gene in selected dairy starters and L. brevis 145
After growing the selected bacteria in the respective medium (MRS or M17), genomic DNAs from eight dairy starters and L. brevis 145 were isolated and purified by using ChargeSwitch® gDNA Mini Bacteria Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. One pair of degenerate primers DP1 and DP2, PGDG-2F and PGDG-4R (Table 1) was applied for amplification of partial GAD gene using AmpliTaq® Gold 360 master mix (Applied Biosystems, Foster, CA, USA). Based on the manufacturer’s instruction, the PCR reaction volume (25 μL) included 12.5 μL of master mix, 0.5 μL of each primer (10 μM), 2 μL (~1 ng) of DNA template and 8.5 μL of DNase-free water. The amplification was carried out in a GeneAmp® PCR system 2700 (Applied Biosystems) with 35 cycles (94 °C for 30 s, 60 °C for 30 s and 72 °C for 90 s) for partial GAD gene. Agarose gel (1%; w/v) electrophoresis was carried out for all PCR products. The size of the PCR products was ~1014 bp.
Sequencing of GAD gene in L. brevis 145
After amplification of partial GAD gene from L. brevis 145, the PCR products from agarose gel were excised and purified according to the manufacture’s instruction of S.N.A.P.TM Gel Purification Kit (Invitrogen). The purified DNAs were ligated with pCR™4-TOPO® TA vector based on the manufacturer’s instructions of the TOPO® TA Cloning® Kit (Invitrogen) and the ligated plasmids were further transformed into One Shot® TOP10 Chemically Competent Escherichia coli (Invitrogen). After white/blue agar screening and colony-PCR amplification, the plasmids from positive colony were extracted and the amplification of inserted sequence was carried out using M13 primers. Then, PCR products were purified and sequenced in 3130xl Genetic Analyzer (Applied Biosystems) using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Sequence reads were further assembled and aligned. The sequence of partial GAD gene was used for designing qPCR primers for quantifying the expression of GAD gene in L. brevis 145.
Mixed cultures and culture conditions for skimmed milk fermentation
The cell counts of the working cultures were enumerated on MRS or M17 agar plates using plate counting method prior to inoculation in skimmed milk. The initial cell count for L. brevis 145 was ~1 × 109 CFU/mL, while the cell counts for S. thermophilus and L. bulgaricus were ~1 × 109 CFU/mL and ~1 × 108 CFU/mL, respectively.
Milk fermentation using cultures with L. brevis alone, or in co-culture with single dairy starter or in co-culture with both S. thermophilus YI-B1 and L. bulgaricus YI-B2 were performed. Monoculture fermentation of L. brevis 145 was carried out in 10% (w/v) skimmed milk supplemented with or without 2 g/L of MSG at 3% (v/v) inoculation level. For co-culturing of L. brevis 145 with one dairy starter in skimmed milk with or without MSG, the inoculation level of L. brevis 145 was 3% (v/v) and that of the dairy starter was 1% (v/v). For co-culturing of L. brevis 145 with two different dairy starters, S. thermophilus YI-B1 and L. bulgaricus YI-B2 were used as conventional starters, while L. brevis 145 was used as an adjunct culture. The inoculation level of each of the S. thermophilus YI-B1 and L. bulgaricus YI-B2 was 0.5% (v/v), while the inoculation level of L. brevis 145 was 3% (v/v). All the fermentation experiments were carried out at three occasions under static condition at 37 °C for 24 h.
Measurement of the pH of fermented milks
The pH of the fermented milk was measured using Orion Model 250A portable pH Meter (Thermo Scientific, Wilmington, DE, USA).
Selective enumeration of S. thermophilus and L. bulgaricus in milk
After milk fermentation, enumeration of S. thermophilus and L. bulgaricus was carried out using selective medium as previously described21. Briefly, the viable counts of above two species were enumerated by plating aliquots of serial dilutions on M17 agar and MRS agar (pH 5.2) plates, respectively. The M17 agar plates for S. thermophilus were incubated aerobically at 37 °C for 24 h, while MRS agar (pH 5.2) plates for enumerating L. bulgaricus were anaerobically kept at 45 °C for 48 h, followed by counting colonies.
Real-time qPCR assay for measuring the cell counts of L. brevis 145 in milk
Since there was no available selective medium for enumeration of L. brevis cells, real-time qPCR was used for assessing the cell count of L. brevis 145 after co-culturing with dairy starters in skimmed milk. Genomic DNA was extracted using the bead-beating extraction method as previously described22. Briefly, 200 μL of fermented milk, 0.40 g of glass beads (0.1 mm diameter; BioSpec Products, Bartlesville, OK, USA) and 600 μL of extraction solution [500 mM of NaCl, 50 mM of Tris, 50 mM of EDTA, 4% SDS (w/v), pH 8.0] and 200 μL of phenol/chloroform/isoamyl alcohol (25:24:1) were added into 2-mL microcentrifuge tubes, followed by disrupting the cells in a BR-2000 Vortexer (Bio-Rad, Hercules, CA, USA) at the highest speed for 5 min. Then, the mixture was separated by centrifugation (12,000× g; 15 min; 4 °C) and upper aqueous phase containing DNAs was transferred to a new tube. The aqueous phase was washed twice with 600 μL of phenol/chloroform/isoamyl alcohol (25:24:1) and the DNAs were precipitated by adding sodium acetate and isopropanol followed by centrifugation (12,000× g; 15 min; 4 °C). The DNA pellet was washed with pre-cooled 75% ethanol. Finally, the precipitated DNAs were dissolved in 30 μL of TE buffer and stored at −30 °C for further analysis.
The amplification was carried out in a StepOnePlusTM Real-Time PCR system (Applied Biosystem). For amplification, 25 μL reaction mixture contained 12.5 μL of SYBR Green master mix, 1 μL of 10 mM of each primer – s-Lbre-F and s-Lbre-R (Table 1) and 2 μL of template DNA. Real-time qPCR was performed with initial denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 10 s, primer annealing 55 °C for 30 s and extension at 72 °C for 30 s. At the end of PCR run, melting curve analysis was carried out from 60 °C to 95 °C (0.5 °C/s) for detection of primer-dimers. The efficiency of this qPCR assay using primers s-Lbre-F and s-Lbre-R was examined by 10-fold serially diluting the genomic DNA from L. brevis 145 cultures and 5 dilutions were used for qPCR assay. The standard curve generated from threshold cycle (Ct) value and viable cell counts of L. brevis ranging from 3.2 × 104 CFU/mL to 3.2 × 109 CFU/mL was prepared in milk as well. The bead-beating extraction procedure was also carried out for isolating the DNA from L. brevis 145 diluted in skimmed milk. Real-time qPCR amplification was carried out in duplicates for each sample and three independent experiments were carried out.
Reversed-phase HPLC analysis of glutamate and GABA
Carrez solutions were used to remove milk proteins before reversed phase HPLC analysis for glutamate and GABA23. Briefly, one gram of fermented milk was added into 4.0 mL of distilled water, followed by addition of 0.25 mL of Carrez I solution (0.25 M potassium ferocyanide) and 0.25 mL of Carrez solution II (0.50 M zinc acetate). Then, the mixture was thoroughly mixed and kept for 30 min at room temperature until the complex formation and precipitation of milk proteins, followed by centrifugation at 25 °C and 5,000 × g for 30 min. Supernatants were collected and filtered through 0.20 μm millipore filter. Filtrates were then freeze-dried, followed by re-dissolving in double distilled water and removing residues by centrifugation at 4 °C and 5,000 × g for 30 min. The clear supernatants with free amino acids were again filtered through 0.20 μm millipore filter. Dansyl derivatization of free amino acids including MSG and GABA was carried out, followed by HPLC analysis of dansyl amino acids as previously described15.
RNA isolation and cDNA synthesis
A modified hot SDS/hot phenol extraction method was used to obtain high quality RNA from Gram-positive bacteria24. Approximately 4 mL of fermented milk was re-suspended in 36 mL of ice-cold sterile water containing 1% (v/v) β-mercaptoethanol, followed by addition of 4 mL of ice-cold ethanol and vortexing for 1 min. Milk proteins were removed by centrifugation at 4 °C and 233 × g for 5 min and supernatants were collected and centrifuged at 4 °C and 5,000 × g for another 10 min. The harvested bacterial pellet was resuspended thoroughly in 1 mL of RNAlaterTM buffer (Qiagen, Limburg, The Netherlands) and incubated at room temperature for 5 min. Then, the bacterial suspension was centrifuged again and the cell pellet was washed with ice-cold sterile water containing 1% (v/v) β-mercaptoethanol to remove residual salts. Cell pellets were resuspended in 600 μL of lysis buffer consisting of 1% (v/v) β-mercaptoethanol and 0.5 mg/mL lysozyme (Sigma-Aldrich, St. Louis, MO) in TE buffer and 200 mg of glass beads (0.1 mm diameter; BioSpec Products) was added into the suspension. Then, the suspension was vortexed in a BR-2000 Vortexer (Bio-Rad) at the highest speed for 5 min. After that, 60 μL of 10% (w/v) SDS solution and 66 μL of 1 M sodium acetate (pH 5.2) was mixed with the lysate. Additionally, 600 μL of phenol:chloroform:isoamyl alcohol (25:24:1) was added, mixed and incubated at 64 °C for 10 min. The tubes were inverted several times every 2 min. The mixture was chilled in an ice bath for 5 min and centrifuged at 4 °C and 21,000 × g for 10 min. The aqueous layer was transferred and washed with equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) twice and centrifuged at 4 °C and 21,000 × g for 5 min. The aqueous layer was transferred to 1.5 mL Eppendorf tubes and RNA was precipitated with ethanol by adding 1/10 volume of 3 M sodium acetate (pH 5.2) and 2 volume of cold ethanol to each tube. The samples were mixed and incubated at −30 °C overnight. The RNA was pelleted by centrifuging at 4 °C and 21,000 × g for 25 min followed by washing with ice-cold 75% ethanol. The pellet was re-suspended in 20 μL of RNase-free water. RNAs isolated after DNase I (Invitrogen) treatment were further converted into cDNAs by reverse transcription according to the manufacture’s instruction of High-Capacity RNA-to-cDNATM Kit (Applied Biosystems).
Real-time qPCR quantitation of gadA and gadB mRNA transcripts in L. brevis 145
The amplification was also carried out in a StepOnePlusTM Real-Time PCR system (Applied Biosystem). The 25 μL reaction mixture contained 12.5 μL of SYBR Green master mix, 1 μL of 10 mM of each primer for 16S rRNA gene (reference gene) and GAD genes (Table 1) and 2 μL of template cDNA. Real-time qPCR was carried out with initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing and extension at 60 °C for 60 s. The efficiency of this qPCR assay using three pair of primers including s-Lbre-F and s-Lbre-R, gadA-757F and −945R, gadB-364F and −499R (Table 1) was also examined by 10-fold serially diluting the genomic DNA from L. brevis 145 cultures and 5 dilutions were used for the qPCR assay. RT-qPCR analysis was carried out for each sample in duplicate and all the experiments were replicated three times.
Statistical analysis
All presented data in the bar charts and tables correspond to means ± standard deviation. Significant difference (P < 0.05 or P < 0.01) among the groups was carried out by one-way analysis of variance (ANOVA) using IBM SPSS Statistics 20.0 version.
Additional Information
How to cite this article: Wu, Q. et al. Dairy Streptococcus thermophilus improves cell viability of Lactobacillus brevis NPS-QW-145 and its γ-aminobutyric acid biosynthesis ability in milk. Sci. Rep. 5, 12885; doi: 10.1038/srep12885 (2015).
References
Cherubini, E., Gaiarsa, J. L. & Benari, Y. GABA - an excitatory transmitter in early postnatal life. Trends Neurosci 14, 515–519 (1991).
Dianaa, M., Quílez, J. & Rafecasa, M. Gamma-aminobutyric acid as a bioactive compound in foods: a review. J Funct Foods 10, 407–420 (2014).
Chuang, C. Y., Shi, Y. C., You, H. P., Lo, Y. H. & Pan, T. M. Antidepressant effect of GABA-rich Monascus-fermented product on forced swimming rat model. J Agr Food Chem 59, 3027–3034 (2011).
Hayakawa, K. et al. Effect of a gamma-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Brit J Nutr. 92, 411–417 (2004).
Inoue, K. et al. Blood-pressure-lowering effect of a novel fermented milk containing gamma-aminobutyric acid (GABA) in mild hypertensives. Eur J Clin Nutr. 57, 490–495 (2003).
Pouliot-Mathieu, K. et al. Effect of cheese containing gamma-aminobutyric acid-producing lactic acid bacteria on blood pressure in men. PharmaNutrition 1, 141–148 (2013).
Shizuka, F. et al. Antihypertensive effect of gamma-amino butyric acid enriched soy products in spontaneously hypertensive rats. Biofactors 22, 165–167 (2004).
Li, H. X. & Cao, Y. S. Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 39, 1107–1116 (2010).
Schurr, B. C., Behr, J. & Vogel, R. F. Role of the GAD system in hop tolerance of Lactobacillus brevis. Eur Food Res Technol 237, 199–207 (2013).
Sun, T. S. et al. ACE-inhibitory activity and gamma-aminobutyric acid content of fermented skim milk by Lactobacillus helveticus isolated from Xinjiang koumiss in China. Eur Food Res Technol 228, 607–612 (2009).
Nejati, F. et al. Manufacture of a functional fermented milk enriched of Angiotensin-I Converting Enzyme (ACE)-inhibitory peptides and gamma-amino butyric acid (GABA). LWT-Food Sci Technol 51, 183–189 (2013).
Heller, K. J. Probiotic bacteria in fermented foods: product characteristics and starter organisms. Am J Clin Nutr 73, 374–379 (2001).
Lourens-Hattingh, A. & Viljoen, B. C. Yogurt as probiotic carrier food. Int Dairy J 11, 1–17 (2001).
Leroy, F. & De Vuyst, L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Tech. 15, 67–78 (2004).
Wu, Q. & Shah, N. P. Gas release-based pre-screening combined with reversed-phase HPLC quantitation for efficient selection of high γ-aminobutyric acid (GABA)-producing lactic acid bacteria. J Dairy Sci 98, 790–797 (2015).
Bergsveinson, J., Pittet, V. & Ziola, B. RT-qPCR analysis of putative beer-spoilage gene expression during growth of Lactobacillus brevis BSO 464 and Pediococcus claussenii ATCC BAA-344(T) in beer. Appl Microbiol Biot 96, 461–470 (2012).
Smid, E. J. & Lacroix, C. Microbe-microbe interactions in mixed culture food fermentations. Curr Opin Biotech 24, 148–154 (2013).
Sieuwerts, S. et al. Mixed-culture transcriptome analysis reveals the molecular basis of mixed-culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Appl Environ Microb 76, 7775–7784 (2010).
Courtin, P., Monnet, V. & Rul, F. Cell-wall proteinases PrtS and PrtB have a different role in Streptococcus thermophilus/Lactobacillus bulgaricus mixed cultures in milk. Microbiol-SGM 148, 3413–3421 (2002).
Savijoki, K., Ingmer, H. & Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl Microbiol Biot 71, 394–406 (2006).
Ashraf, R. & Shah, N. P. Selective and differential enumerations of Lactobacillus delbrueckii subsp bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium spp. in yoghurt - A review. Int J Food Microbiol 149, 194–208 (2011).
Garcia-Cayuela, T., Tabasco, R., Pelaez, C. & Requena, T. Simultaneous detection and enumeration of viable lactic acid bacteria and bifidobacteria in fermented milk by using propidium monoazide and real-time PCR. Int Dairy J 19, 405–409 (2009).
Pham, T. T. & Shah, N. P. Biotransformation of isoflavone glycosides by Bifidobacterium animalis in soymilksupplemented with skim milk powder. J Food Sci 72, M316–M324 (2007).
Jahn, C. E., Charkowski, A. O. & Willis, D. K. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Meth 75, 318–324 (2008).
Lin, Q. et al. Cloning and expression of glutamate decarboxylase gene from Streptococcus thermophilus Y2. J Gen Appl Microbiol 55, 305–310 (2009).
Matsuda, K. et al. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microb 75, 1961–1969 (2009).
Acknowledgements
We would like to thank Mr. Jing CHEN and Ms. Charis CHAN in the School of Biological Sciences at The University of Hong Kong for providing assistance in real-time qPCR design and analysis.
Author information
Authors and Affiliations
Contributions
N.P.S. and Q.W. designed this project; Y.S.L. provided technical suggestions for this project; Q.W. performed the experiments, analyzed data, prepared figures and tables and wrote the draft manuscript; N.P.S. reviewed, revised and edited the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Wu, Q., Law, YS. & Shah, N. Dairy Streptococcus thermophilus improves cell viability of Lactobacillus brevis NPS-QW-145 and its γ-aminobutyric acid biosynthesis ability in milk. Sci Rep 5, 12885 (2015). https://doi.org/10.1038/srep12885
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep12885
This article is cited by
-
Identification of Type II Toxin-Antitoxin Loci in Levilactobacillus brevis
Interdisciplinary Sciences: Computational Life Sciences (2022)
-
Gamma-aminobutyric acid fermentation in MRS-based medium by the fructophilic Lactiplantibacillus plantarum Y7
Food Science and Biotechnology (2022)
-
GABA potentiate the immunoregulatory effects of Lactobacillus brevis BGZLS10-17 via ATG5-dependent autophagy in vitro
Scientific Reports (2020)
-
Unraveling microbial fermentation features in kimchi: from classical to meta-omics approaches
Applied Microbiology and Biotechnology (2020)
-
Genomic insights into a robust gamma-aminobutyric acid-producer Lactobacillus brevis CD0817
AMB Express (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.