Abstract
Cotton (Gossypium spp.) is an important economic crop and there is obvious heterosis in cotton, fertility has played an important role in this heterosis. However, the genes that exhibit critical roles in anther development and fertility are not well understood. Here, we report an acyl-CoA N-acyltransferase (EC2.3; GhACNAT) that plays a key role in anther development and fertility. Suppression of GhACNAT by virus-induced gene silencing in transgenic cotton (G. hirsutum L. cv. C312) resulted in indehiscent anthers that were full of pollen, diminished filaments and stamens and plant sterility. We found GhACNAT was involved in lipid metabolism and jasmonic acid (JA) biosynthesis. The genes differentially expressed in GhACNAT-silenced plants and C312 were mainly involved in catalytic activity and transcription regulator activity in lipid metabolism. In GhACNAT-silenced plants, the expression levels of genes involved in lipid metabolism and jasmonic acid biosynthesis were significantly changed, the amount of JA in leaves and reproductive organs was significantly decreased compared with the amounts in C312. Treatments with exogenous methyl jasmonate rescued anther dehiscence and pollen release in GhACNAT-silenced plants and caused self-fertility. The GhACNAT gene may play an important role in controlling cotton fertility by regulating the pathways of lipid synthesis and JA biogenesis.
Similar content being viewed by others
Introduction
Cotton (Gossypium hirsutum L.) has long been the world’s most important source of natural textile fiber and is one of the first genetically modified crops. The modifications include transgene-mediated herbicide tolerance and pest-resistance1. Biotechnology has now accelerated cotton breeding programs, especially heterosis development. G. hirsutum is allotetraploid with a large and complex genome (AADD, 2n = 4x = 52); thus, it is difficult to study gene function in cotton with traditional methods of RNA interference. This approach is hindered by the inefficient production of stable transformed plants because of the long periods of plant tissue culture and the very low efficiency of transformation, which is genotype limited1. The genome of the wild cotton species G. raimondii was sequenced and showed many genes involved in cotton fiber initiation and elongation in the DD genome2. Another study analyzed the polyploidization of Gossypium genomes and the evolution of spinnable cotton fibers3. The genome sequence of G. arboreum has been completed recently, providing information about the AA genome4. In public databases, an increasing number of expressed sequence tags (ESTs) and deep sequencing data for cotton have become available. A rapid and cost-effective tool has been needed to connect the gene sequence information with gene function in cotton. As a transient, efficient expression tool, virus-induced gene silencing (VIGS) has been used in many species and there are different vector systems available for the different species5.
Transient transformation with a specific virus has become a fast and efficient method to analyze gene function at the genome-wide level. VIGS mediated by Agrobacterium is an efficient, transient posttranscriptional gene silencing method for fast, low-cost and large-scale gene function analysis in plants. VIGS exploits the innate plant defense system of posttranscriptional gene silencing (PTGS) against intracellular viral proliferation and extracellular viral movement6. Both RNA and DNA plant viruses can contribute to VIGS5, including the viral vectors derived from positive-strand RNA, Potato virus X (PVX), Tobacco mosaic virus (TMV) and Tobacco rattle virus (TRV)7. Different sources of VIGS vectors are the bipartite single-strand DNA viruses and the mono-partite DNA viruses that may require a helper virus or satellite DNA for inducing disease symptoms, including the Tomato yellow leaf curl China virus (TYLCCV)5,7,8. Currently, both types of VIGS viruses, including TRV9,10 and Cotton leaf crumple virus (CLCrV) based silencing vectors11, have been used in the analysis of cotton gene function. The CLCrV is a member of the genus Begomovirus, family Geminiviridae and it consists of two circular single-stranded DNAs called CLCrV DNA-A and CLCrV DNA-B with different, indispensable functions12,13,14. The new, modified CLCrV-based vector, pCLCrV, was established specifically for cotton infection15. In this study, we utilized the pCLCrV-VIGS system to analyze the function of the GhACNAT gene. The GhACNAT gene was acquired from the differentially expressed genes of G. hirsutum L. cv. C312 and its male sterile mutant ms1. The ms1 mutant has the same phenotype and genetic background as C312 except for male sterility. Using cDNA-AFLP, we screened for genes that showed differential expression between C312 and ms1 to obtain the genes related to male sterility. The GhACNAT gene was significantly differentially expressed with an unknown function in male sterility. The GhACNAT gene is homologous to the Arabidopsis thaliana (A. thaliana) gene At2g23390, which is a member of the superfamily that encodes acyl-CoA N-acyltransferase (EC2.3).
N-Acyltransferase is one of the important enzymes that catalyze the transfer of an acyl group to a substrate; N-acyltransferase is implicated in a variety of functions and it is conserved from bacterial antibiotic resistance to circadian rhythms in mammals. Members of the N-acyltransferase superfamily have a similar catalytic mechanism but vary in the types of acyl groups they transfer, including those of the three main nutrient substances, saccharides, lipids and proteins. These substances participate in a common metabolic pathway mediated by acetyl-CoA in the tricarboxylic acid cycle and oxidative phosphorylation reactions, which involve a variety of enzymes.
Acyl lipids have various functions in plants and the structures and properties of the acyl lipids vary greatly even though they are all derived from the same fatty acid and glycerolipid biosynthesis pathway16. Some of them, including jasmonic acid, participate in signaling pathways17.
Acyl-CoA and acyl-CoA N-acyltransferase are involved in these metabolic pathways, including pyruvate dehydrogenase and pyruvate formate lyase reactions and they are involved in the metabolism of sugars in the citric acid cycle and fatty acids and fat metabolism required for the synthesis of flavonoids and related polyketides for the elongation of fatty acids involved in sesquiterpenes, brassinosteroids and membrane sterols.
The AtGPAT1 (glycerol-3-phosphate acyltransferase) gene has been identified as a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, correlated with several fatty acids produced in flower tissues and seeds, including linoleic acid (18:2). Jasmonic acid is derived from linolenic acid (LA)18 and its biosynthesis is catalyzed by several enzymes, including lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC) and 12-oxo-phytodienoic acid reductase (OPR) followed by three cycles of β-oxidation19,20,21. The mutants of opr3, dad1 (defective in anther dehiscence1), ms35 (male sterile35) and the genes AOS, AOC, AIF (ANTHER INDEHISCENCE FACTOR) in A. thaliana and other species (tomato, tobacco, brassica, rice and maize) regulate the male or female sterility involved in JA biosynthesis22,23,24,25,26,27,28. The AOC gene is highly expressed in distinct flower organs and vascular bundles and in jasmonic acid influenced female fertility in the tomato27. The OPR7 and OPR8 genes regulate the diverse functions of JA in maize29. In G. hirsutum, GhSERK1 (somatic embryogenesis receptor-like kinase) has been shown to regulate pollen development30.
Lipid metabolism plays a very important role in the development of the tapetum and pollen grains31. We constructed the pCLCrV-GhACNAT vector to silence the target gene GhACNAT in G. hirsutum cv. C312. In the transgenic GhACNAT-silenced plants, the phenotype of the reproductive organs was investigated. Genes that were expressed at different levels in C312 and GhACNAT-silenced plants were screened and assayed by qRT-PCR to study the function of GhACNAT and related genes. The results indicated that GhACNAT played an important role in regulating the development of reproductive organs and fertility by influencing lipid and JA biosynthesis.
Results
GhACNAT is differentially expressed in the male sterile ms1 mutant and C312
Among the genes differentially expressed in G. hirsutum cv. C312 and the male sterile ms1 mutant of C312, the acyl-CoA N-acyltransferase (EC2.3) gene was significantly down-regulated with unknown function and designated GhACNAT. GhACNAT is homologous to genes in different species, including with At2g23390 (Fig. 1a), which is a member of the acyl-CoA N-acyltransferase superfamily in A. thaliana with a conserved domain that catalyzes N-acetyltransferase reactions in lipid synthesis and metabolism (Fig. 1b). We amplified the full-length 1410 bp cDNA of GhACNAT from C312. One highly homologous gene was found in the DD genome of G. raimondii.
The expression pattern of the GhACNAT gene analyzed by qRT-PCR
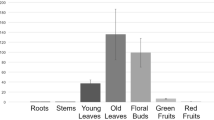
The GhACNAT gene was expressed at different levels in the tested tissues and organs, including roots, stems, leaves, bracts, petals, stamens stigmas and fibers of different development stages (0-, 5-, 10-, 15-, 20- and 25-DPA (days post anthesis)) in C312 (Fig. 2). The expression levels of the GhACNAT gene in the reproductive organs (bracts, petals, stigmas and stamens) were significantly higher than in the vegetative organs (roots, stems and leaves) and fibers of different developmental stages. The GhACNAT gene maintained significantly high-level expression in the flower organs, including the sepals, petals, stamens and stigmas.
The construction of the pCLCrV-GhACNAT vector for VIGS
The pCLCrV-based virus-induced gene-silencing system produced an efficient and persistent gene silencing in cotton. To determine whether the pCLCrV-based silencing vector could silence endogenous gene expression in cotton, a 327 bp fragment of the phytoene desaturase gene (PDS), commonly used as a positive marker for VIGS, was cloned into the pCLCrVA vector to construct pCLCrV-PDS. Agrobacteria tumefaciens strain GV3101 containing different pCLCrVA and pCLCrVB vector was infiltrated by syringe into the cotyledons of 2-week-old seedlings of C312. By 20 days postinfiltration, cotton plants infiltrated with Agrobacteria harboring both pCLCrVA-PDS and pCLCrVB showed the typical PDS gene-silencing phenotype of photobleaching in newly emerging leaves.
The photobleaching phenotype in plants infiltrated with pCLCrV-PDS was observed on the newly emerging leaves by 3 weeks postinfiltration, first appearing in the nearby veins and gradually extending to the whole leaves, which became mosaic with green spots (Fig. S1). The efficiency of the photobleaching in the plants infiltrated with pCLCrV-PDS and pCLCrVB was greater than 75%. The transgenic pCLCrV-PDS plants were confirmed by PCR and showed the photobleaching mosaic with green spots in the leaves. Transgenic plants of the pCLCrVA-empty with pCLCrVB and pCLCrVA-GhACNAT with pCLCrVB showed a slight shrinking of the new leaves (Fig. S1 a, b, c). The transgenic plants with pCLCrVA-empty and pCLCrVA-GhACNAT were identified by PCR with specific primers (Table S3). The photobleaching in the transgenic pCLCrV-PDS plants appeared in different organs, including the stem, petiole, bracts and bolls during the growth cycle (Fig. S1), indicating that the silencing caused by this vector persisted in the cotton plants. Pink stripes appeared on the stems in the transgenic pCLCrV-PDS plants (Fig. S1 e). To investigate the silencing efficiency, the levels of PDS mRNA in both pCLCrV-PDS-silenced and wild-type C312 plants were measured by quantitative real time PCR (qRT-PCR). In the transgenic pCLCrV-PDS plants with strong photobleaching of the new leaves, the expression level of the endogenous PDS gene was significantly reduced, 2–40-fold compared with wild-type C312 plants (Fig S1i). The control plants of C312 and pCLCrV-empty did not show the photobleaching phenotype at any time during the studies (Fig S1c).
A 200 bp fragment of the GhACNAT gene was amplified to construct the pCLCrV-GhACNAT vector to silence the endogenous GhACNAT gene in C312 plants.
Silencing the GhACNAT gene influenced the development of the reproductive organs of transgenic plants
There were no obvious differences in the phenotypes of the control plants, the transgenic pCLCrV-PDS plants and the transgenic GhACNAT-silenced plants during vegetative growth except for the mosaic photobleaching spots in the transgenic pCLCrV-PDS plants. In the reproductive stage of the transgenic GhACNAT-silenced plants, the distinct phenotypic changes appeared in the reproductive organs (Fig. 3). The stigmas were forked and warped compared with the stigmas of C312 (Fig. 3a) and the transgenic control plants. The stamens of the transgenic GhACNAT-silenced plants appeared indehiscent but full of viable pollen (Fig. 3b,c,d and 4a,d,g). The filaments became shorter and the number of stamens decreased significantly (Table S1). No pollen was released from the closed anthers in the transgenic GhACNAT-silenced plants. In the male sterile ms1 mutant, the anthers were closed and shrunken and no pollen formed (Fig. 4a,f,h). The anthers of the C312 plants were dehiscent and full of active pollen grains, whereas the stamens of the transgenic GhACNAT-silenced plants were indehiscent but as full of viable pollen as the wild-type C312 (Fig. 4d,g). However, the pollen could not be released from the closed stamens. In the transgenic GhACNAT-silenced plants, mature bolls were obtained successfully by pollination with normal C312 pollen. The bolls had two to four loculi containing seeds covered with fibers (Fig. 3e–i).
In the transgenic pCLCrV-GhACNAT-silenced plants, the endogenous GhACNAT gene was significantly down-regulated (2–12-fold) compared with C312 (Fig. 3j). The lines designated 36-8, 36-12 and 36-16 were selected for further analysis of the phenotypes and expression changes of related genes.
The analysis of genes differentially expressed in transgenic GhACNAT-silenced plants and control plants
To study differentially expressed genes related to the alteration of reproductive organs in the transgenic GhACNAT-silenced plants, 3 individual plants of wild type C312 and the transgenic pCLCrV-GhACNAT-silenced lines were selected for differential expression analysis by cDNA-AFLP. The stigmas and stamens of the flowers (on the day of anthesis) were picked and collected for total RNA isolation. The differentially expressed genes were selected and 450 fragments (100–200 bp) were cloned and sequenced. The function annotations of the differentially expressed genes in the reproductive organs of the control plants and the transgenic GhACNAT-silenced plants were divided into several categories (Fig. 5). The main differentially expressed genes were focused on catalytic activity, translation regulator activity and metabolic processes involved in lipid metabolism. Acyl-CoA and acyl-CoA N-acyltransferase were involved in the lipid metabolic pathways.
The expression changes of genes related to the development of reproductive organs in the transgenic GhACNAT-silenced plants
Many mutants with anther indehiscence, including those with mutations in opr322 and dad123, showed that jasmonic acid participates in the regulation of anther development, filament elongation and pollen viability32. AtAIF was identified as a repressor that controls anther dehiscence by regulating the jasmonic acid biosynthesis pathway in A. thaliana24. Linolenic acid (18:3) was released from membrane phospholipid by a lipolytic enzyme (DAD1) and the DAD1 gene participated in the regulation of anther development23.
JA plays a role in anther development. The expression levels of genes encoding key enzymes involved in the JA biosynthesis pathway were examined in transgenic GhACNAT-silenced plants and C312 plants. The genes (GhAIF, GhDAD1, GhLOX1 and GhOPR3) with high homology to AtAIF, AtDAD1, AtLOX1 and AtOPR3 in A. thaliana, which are genes related to reproductive organ development, were detected by qRT-PCR in stigmas and stamens of the C312 plants and the transgenic GhACNAT-silenced plants on the blossom day (Fig. 6). GhAIF was significantly up-regulated (29-fold) in the transgenic GhACNAT-silenced plants compared with the control plants. The GhLOX1 gene was up-regulated 3.5-fold compared with the gene in control plants. The corresponding enzyme catalyzes the conversion of linolenic acid to 13-hydroperoxylinoinic acid, which is the precursor for JA and cis-3 hexenal+traumatin using two different and competing pathways. The expression of GhAOC1 was not significantly different in C312 and transgenic GhACNAT-silenced plants. The expression of the GhDAD1 gene was significantly reduced in GhACNAT-silenced plants and the expression of the GhOPR3 gene for JA biosynthesis was reduced approximately 4.5-fold in transgenic GhACNAT-silenced plants. This showed that JA biogenesis was influenced by changes in expression of these genes, which could cause anther indehiscence and short filaments. The GhSERK gene, which was shown to regulate anther dehiscence and pollen viability in cotton30, was identified from a differentially expressed fragment by cDNA-AFLP. The expression of GhSERK was significantly reduced in the stamens of the transgenic GhACNAT-silenced plants.
The GhACNAT silencing influenced fatty acid biosynthesis in the transgenic plants
GhACNAT belongs to the gene superfamily encoding acyl-CoA N-tranferase and has high homology with the At2g23390 gene. The acyl-CoA N-tranferase superfamily primarily participates in lipid metabolism to provide acyl-CoA. To investigate the function of GhACNAT in fatty acid biosynthesis, several key genes in the fatty acid biosynthetic pathway were measured by qRT-PCR. The single transcription factor WRI1, which has been shown to directly control the transcriptional activation of the fatty acid biosynthetic pathway33, was significantly decreased in the transgenic GhACNAT-silenced plants. The glycolytic and late fatty acid biosynthetic genes, including GhPDH-E1a (homologous to At1g01090), GhBCCP2 (homologous to At5g15530), GhMAT (homologous to At2g30200), GhKASI (homologous to At5g46290), GhENR (homologous to At2g05990) and GhFATA (homologous to At3g25110), were measured in the stigmas and stamens of the control plants and the transgenic GhACNAT-silenced plants (Fig. 7a). In the transgenic GhACNAT-silenced plants, the expression levels of GhPDH-E1a, GhBCCP2 and GhMAT were reduced 2-fold compared with the control plants and the expression levels of GhKASI and GhENR were significantly down-regulated. The relative mRNA levels of GhCYP86A2 (homologous to At4g00360), GhCYP86A8 (homologous to At2g45970) and GhCYP77A6 (homologous to At3g10570), which are involved in cuticle formation, were almost the same in the transgenic GhACNAT-silenced plants and the control plants. In the GhACNAT-silenced plants, the expression of the genes involved in lipid synthesis was reduced significantly and the tendency was consistent. The changes in gene expression influenced lipid synthesis and reduced JA biosynthesis in the stigmas and stamens of the GhACNAT-silenced plants.
The AtGPAT1 gene is associated with several fatty acid composition changes in flower tissues and seeds, including JA biosynthesis, catalyzed by lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC) and 12-oxo-phytodienoic acid reductase (OPR). The genes GhGPAT1, GhGPAT3 and GhGPAT4 were detected by qRT-PCR and showed obvious differences in expression in C312 and the transgenic GhACNAT-silenced plants. The expression levels of GhGPAT1 and GhGPAT3 were significantly decreased, whereas GhGPAT4 expression increased in the transgenic GhACNAT-silenced plants. GhGPAT, GhLPAAT and GhFAD3 participate in the synthesis of phospholipids. In the transgenic GhACNAT-silenced plants, the expression of GhLPAAT did not change, but GhFAD3 expression decreased (Fig. 7b). In the JA biosynthesis pathway from the acyl-CoA pool to phospholipid to linolenic acid, the expression of genes encoding key enzymes significantly changed to influence JA biosynthesis.
Measurement of JA and rescue of transgenic GhACNAT-silenced plants treated with exogenous MeJA
Endogenous GhACNAT expression was significantly decreased by VIGS in the reproductive organs of GhACNAT-silenced plants (Fig. 8a) and it significantly influenced the expression of genes related to JA biosynthesis (Fig. 6) and caused abnormal formation of reproductive organs. In transgenic GhACNAT-silenced plants, the genes for repressing JA biosynthesis were up-regulated, whereas the expression levels of genes involved in JA biosynthesis were down-regulated and decreased the JA levels in leaves and reproductive organs (Fig. 8b,c). The amounts of JA in fresh leaves of three transgenic GhACNAT-silenced lines were significantly lower than in C312 and in the reproductive organs, the amounts of JA were all higher than those in leaves. In the flowers of three GhACNAT-silenced lines, the amounts of JA were extremely low or even undetectable by GC/MS (Fig S3) and significantly lower than in C312. However, the transcripts of genes for JA biosynthesis were apparently attenuated in transgenic GhACNAT-silenced lines compared with C312 plants and the genes for repressing JA biosynthesis were up-regulated (Fig. 6). Consistent with the kinetics of gene expression in the JA pathway, JA levels in the leaves and reproductive organs in transgenic GhACNAT-silenced plants were dramatically decreased compared with C312 plants (Fig. 8b,c).
For the rescue experiment in transgenic GhACNAT-silenced plants to determine whether the defect in jasmonic acid synthesis caused the anther dehiscence, three concentrations (100 μM, 500 μM and 1000 μM) of MeJA were used to treat the buds of transgenic GhACNAT-silenced plants before they opened. In the opening flowers of transgenic GhACNAT-silenced plants treated with MeJA, the morphologic changes in the reproductive organs were reversed and the anthers were thoroughly or partially dehiscent, similar to the normal anthers in C312 plants (Fig. 9, Table S2). With 100 μM MeJA, the anthers partially dehisced and some anthers were still occlusive (Fig. 9b; Table S2); with 500 μM and 1000 μM MeJA, all the anthers dehisced normally (Fig. 9c,d; Table S2). The bolls of transgenic GhACNAT-silenced plants were obtained by self-pollination after treatment with MeJA. The results showed that exogenous MeJA could reverse the anther phenotype and rescue the occlusive anthers caused by decreasing endogenous JA.
Discussion
VIGS provided an efficient method to analyze gene function in cotton during the entire growth cycle
VIGS has demonstrated to be an efficient method to analyze gene function in cotton10,11,15,34. By 3 weeks after the pCLCrV-Agrobacteria injection, transgenic pCLCrV-PDS plants showed obvious photobleaching in the new leaves. The efficiency of transformation reached 75% and the effects were retained in leaves, stems, flowers and even in bolls throughout the growth cycle. In the vegetative growth stage, there was no obvious phenotype except for slight wrinkling in the leaves of the transgenic GhACNAT-silenced plants. At the stage of reproductive growth, the phenotypes of the reproductive organs changed significantly, showing stigma wraped, anther indehiscence and no pollen release, which led to sterility in the transgenic GhACNAT-silenced plants. The sterility was mainly caused by the anther dehiscence and failure to release pollen. The changes of phenotypes in transgenic GhACNAT-silenced plants demonstrated that the effect of VIGS in cotton can be used for gene function analysis in vegetative growth, in reproductive growth and even in developing fibers.
The anther development was influenced by JA biosynthesis regulated by the GhACNAT gene
The GhACNAT gene belongs to the acyl-CoA N-acyltransferase gene superfamily and was selected from the genes differentially expressed in C312 and its male sterile mutant, ms1. The GhACNAT gene is homologous with At2g23390 in A. thaliana, which encodes acyl-CoA N-acyltransferase, an enzyme involved in the isopentenyl diphosphate biosynthetic pathway, maltose metabolism, regulation of proton transport and starch biosynthesis and was found to be expressed in vegetative organs and in reproductive organs35,36,37,38.
Acyl-lipid metabolism in A. thaliana is a complex process and is organized into 12 sections based on the types of lipids produced and their subcellular localization39. Acyl lipids in plants have many diverse functions, involving more than 10 membrane lipid classes in the endoplasmic reticulum (ER), including the phospholipids, glycolipids and sphingolipids39,40,41,42,43. The first committed step in fatty acid synthesis is the formation of malonyl-CoA from acetyl-CoA and bicarbonate by acetyl-CoA carboxylase (ACC)16,44. Before entering the fatty acid synthesis pathway, the malonyl-CoA group is transferred from CoA to ACP catalyzed by malonyl-CoA: acyl carrier protein malonyltransferase (MCMT). Malonyl-ACP provides two-carbon units at each step of elongation. The long-chain acyl groups hydrolyzed by acyl-ACP thioesterases release fatty acids. These fatty acids are ultimately activated to CoA esters by a long-chain acyl-CoA synthetase (LACS) and exported to the endoplasmic reticulum or possibly the PC at the plastid envelope by the action of lysophosphatidylcholine acyltransferase (LPCAT)45,46,47. Fatty acids are derived from plastid lipids desaturated in the ER and are precursors for potent signaling molecules, including jasmonates, leaf aldehydes or divinyl ethers48,49. The ER is the major site for phospholipid biosynthesis. Phosphatidic acid (PA) is the common precursor of phospholipids catalyzed by acyl-CoA: glycerol-3-phosphate acyltransferase (GPAT) and acyl-CoA: lysophosphatidic acid acyltransferase (LPAAT). PAs originating from the ER pathway exclusively contain C18 fatty acids in the sn-2 position in eukaryotic species, whereas PAs synthesized in plastids exclusively contain C16 fatty acids in the sn-2 position in prokaryotic species. The GPAT and LPAAT reactions of phospholipid synthesis or triacylglycerol synthesis utilize a mixed pool of acyl-CoA substrates that in many tissues is produced mostly from a PC acyl editing cycle. An AtGPAT1 deficiency had pleiotropic effects on glycerolipid biosynthesis and resulted in several fatty acid composition changes in flower tissues and seeds. The molar proportions of palmitic acid (16:0), stearic acid (18:0), linolenic acid (18:3) and the very-long-chain eicosenoic acid (20:1) decreased to varying degrees, whereas the proportions of oleic acid (18:1) and linoleic acid (18:2) increased31. Linolenic acid (18:3)-derived oxylipins are the biosynthetic C18 precursors of JA17. Acyl groups esterified to PC are the sites of extraplastidic FA desaturation50. The FAD251 and FAD352 enzymes convert PC-bound oleate to linoleate and then linolenate, respectively, which is the precursor of JA (Fig. 10).
The DAD1 gene encodes a lipase that hydrolyzes both TAGs (triacylglycerols) and glycerolipids23 and releases linolenic acid for the biosynthesis of jasmonic acid. The precursor of JA originates from an 18-carbon fatty acid of linolenic acid (LA) released by the phospholipid in membranes, which is regulated by DAD1. The biosynthesis of JA is catalyzed by the key enzymes LOX, AOS, AOC and OPR3 in A. thaliana. The key genes of TAG synthesis were significantly down-regulated in the stigmas and stamens of transgenic GhACNAT-silenced plants. The gene expression of GhLOX and GhOPR3 displayed down-regulation in transgenic GhACNAT-silenced plants compared with the control plants. The GhAIF gene was identified as an inhibiting factor for JA biosynthesis24 and the expression level of GhAIF was significantly up-regulated in transgenic GhACNAT-silence plants.
The JA pathway was influenced because GhACNAT-silencing changed the expression levels of some key genes related to lipid and JA biological processes. The change in JA biosynthesis in transgenic GhACNAT-silenced plants resulted in the amount of JA decreasing and the reduced JA caused anther indehiscence and other reproductive organ abnormalities. The expression changes of GhACNAT silenced by VIGS influenced many genes involved in biological processes of lipid and jasmonate acid. The GhACNAT gene and related genes involved in biological processes of lipid and jasmonate acid, regulated the development of reproductive organs and fertility by regulating lipid metabolism and JA biosynthesis.
Future work should be focused on the application of stable knockdown lines of GhACNAT, including RNA interference with indihescent anthers full of active pollen, such as the transgenic GhACNAT-silenced plants here. The stable GhACNAT-silenced plant could be used to develop two-line hybrid cotton breeding using an anther indehiscent full of pollen as a sterile line and an anther releasing pollen through MeJA treatment as a maintainer line.
Material and Methods
Plant materials
The G. hirsutum L. cv. Coker 312 (C312) was used in these experiments. Seeds were germinated and grown in a greenhouse at 28 °C with a 14 h light and 10 h dark cycle. Seedlings with a 2nd true leaf emergence were used for agroinfiltration. Infiltrated plants were grown in a growth chamber at 23–25 °C with a 14/10 h light/dark photoperiod.
Gene cloning and construction of VIGS vectors
The candidate gene was obtained from the differentially expressed genes in C312 and its male sterility mutant (ms1). The mutant ms1 had the same background as C312 except for the mutation. The full-length 1410 bp cDNA of the GhACNAT gene was amplified from the cDNA library by PCR using the forward primer 5’-CTATGGCGGCAGCACTCA-3’ and the reverse primer 5’-TGCACGATGGCTTTTCAA-3’. The PCR was incubated at 95 °C for 5 min and for 32 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 100 s and 72 °C for 10 min. The reaction was then kept at 4 °C. The PCR product was cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA) and the product was transformed into E.coli DH5α and sequenced.
The new CLCrV-based vector was modified from the CLCrV DNA-A and DNA-B components individually, which were inserted into the pCambia1300 vector to generate pCLCrVA and pCLCrVB, respectively15. The pCLCrVB is a geminivirus member and essential for the silence system in cooperation with pCLCrVA. The fragments of candidate genes were inserted into pCLCrVA to produce pCLCrVA-GhACNAT for VIGS in cotton plants as described by Gu et al., (2014)15.
A 200 bp fragment of the candidate gene GhACNAT was amplified by PCR (95 °C for 5 min and 30 cycles at 95 °C for 30 s, 59 °C for 30 s, 72 °C for 40 s and 72 °C for 10 min. The reaction was then kept at 4 °C). The phytoene desaturase gene (PDS) causes loss of chlorophyll and carotenoids53,54 and was used as a positive marker to visualize the timing and extent of endogenous gene silencing. A 327 bp fragment of the PDS gene isolated from C312 by PCR (95 °C for 5 min, 30 cycles at 95 °C for 30 s, 59 °C for 30 s, 72 °C for 40 s and 72 °C for 10 min, then 4 °C). The fragments were inserted into the pGEM-T Easy Vector (Promega, Madison, WI, USA) and the construct was transformed into E.coli DH5α and sequenced. The recombinant plasmids were digested with SpeI and AscI (New England Biolabs, Beijing, China) and the products were individually inserted into the pCLCrVA vector to generate pCLCrV-GhACNAT and pCLCrV-GhPDS.
The four vectors with or without foreign genes were transformed individually into Agrobacterium tumefaciens strain GV3101 by electroporation using a Gene Pulser Apparatus (Bio-Rad, Hercules, CA, USA) as described by Huang et al., (2009)55. Three combinations, pCLCrVA-empty and pCLCrVB (for a negative control), pCLCrV-PDS and pCLCrVB (for a positive control), pCLCrV-GhACNAT and pCLCrVB (for target gene silencing) were used. Plants were transformed with pCLCrV-PDS and the pCLCrVA-empty vectors as controls. The gene sequences and primers for cloning and detection are listed in Table S1 of the supplementary file.
Plant growth and agroinfiltration
Cotton seedlings were grown in a growth chamber at 28 °C with a 14 h light and 10 h dark cycle. Healthy 2 week old seedlings were infiltrated with different Agrobacteria carrying pCLCrVA or one of its derivatives and pCLCrVB. Agrobacteria harboring pCLCrVA or one of its derivatives was mixed with an equal volume of Agrobacteria harboring pCLCrVB. The mixed Agrobacteria solutions were infiltrated into the abaxial side of the cotyledons of the 2-week-old cotton seedlings using syringes without needles. To facilitate infiltration, a few holes were gently punched into the underside of the cotyledons using a syring needle. The Agrobacteria preparation and the process of agroinfiltration used has been described by Gu et al., (2014)15. The growth chambers for the infected plants were kept at 23 °C with a 14/10 h light/dark cycle. The agroinfiltration was repeated at least three times with approximately 30 plants for each vector in the VIGS system.
Plants infiltrated with the pCLCrVA-empty vector and the wild-type C312 were used as controls and did not show any photobleaching during the experiment. Plants infiltrated with the pCLCrV-PDS vector showed the typical photobleaching phenotype in newly developed leaves. However, the efficiency of gene silencing was judged by the intensity of photobleaching, which varied in the different organs of the transgenic plants.
Anther observation and pollen staining
In our experiments, the transgenic GhACNAT-silenced plants and the control plants were compared to observe the change in phenotypes. Pollen activity was detected by staining with 13 mM of I2-KI. The solution was prepared by adding 12 mM KI to 5 ml ddH2O and then dissolving 4 mM of I2 in the solution and adding ddH2O to 300 ml. The solution was stored in a brown glass bottle. Pollen grains were soaked in the I2-KI solution on glass slides for approximately 1 min and then the shape and color of the pollen grains were observed with a fluorescence stereomicroscope (ZEISS SteREO LuMar.V12, Germany).
DNA extraction and PCR detection
Total DNA was extracted from the leaves of pCLCrV-inoculated cotton plants using an extraction buffer containing 100 mM Tris-HCl, pH 8.0, 25 mM Na2EDTA, 2 M NaCl, 2% (w/v) CTAB, 2% (w/v) PVP and 2% (w/v) β-mercaptoethanol as described56. The presence of pCLCrV DNA in infected tissue was detected by PCR using primers specific for either pCLCrV DNA-A (CLCrVA F and CLCrVA R) or pCLCrV DNA-B (CLCrVB F and CLCrVB R). (Primers used in the experiment are listed in Table S3.) PCR was performed at 94 °C for 4 min, followed by 30 cycles of 94 °C for 30 s, 59 °C for 30 s and 72 °C for 1 min. The PCR products were visualized by electrophoresis on 1% (w/v) agarose gels.
The analysis by cDNA-AFLP of differentially expressed genes influenced by GhACNAT-silencing
The transgenic GhACNAT-silenced plants with significantly changed phenotypes were confirmed by PCR and expression analysis of the endogenous GhACNAT gene and then were selected for further analysis. The stigmas and stamens of C312 plants and transgenic GhACNAT-silenced plants were collected for RNA extraction using TRIzol® Plus RNA Purification Kits (Invitrogen, Carlsbad, CA, USA). Double-stranded cDNA was synthesized from 200 ng RNA using an iScriptTM cDNA Synthesis Kit (Quanta BioSciences, Suite 1 Gaithersburg, MD, USA) according to a standard double-stranded cDNA synthesis protocol. The analysis of differentially expressed genes by cDNA-AFLP was performed to screen for the genes affected by GhACNAT-silencing. The cDNA-AFLP analysis was carried out as described by Hawkins et al., (2005)57. The cDNA was digested with the restriction enzymes MseI and EcoR I (New England Biolabs, Beijing, China). The digested products were then ligated to adapters with the following sequences. The MseI adapters were 5’-GACGATGAGTCCTGAG-3’ and 3’-TACTCAGGACTCAT-5’. The EcoR I adapters were 5’- CTCGTATACTGCGTACC-3’ and 3’-AATTGGTACGCAGTA-5’. Adapter-ligated cDNA was the template for pre-amplification, with PCR parameters of 30 cycles of 30 s at 94 °C, 60 s at 56 °C and 60 s at 72 °C. The diluted (30-fold) amplified products were used as the templates for selective amplification. The differentially expressed bands between 100 and 200 bp were cloned into the pGEM-T Easy Vector (Promega, Madison, MI, USA), transformed into E.coli DH5α and sequenced.
The categorization of the transcript fragments on the basis of their presence or absence (qualitative variation) or differences in the amount (quantitative variation) in the control lines (wild-type C312 and transgenic pCLCrVA-empty plants) and transgenic GhACNAT-silenced lines showed that most of the variation was quantitative. The transcript fragments represented genes controlling different biological processes. The expression levels of GhACNAT and differentially expressed genes in the same biological pathway were measured in the control plants and the transgenic GhACNAT-silenced plants.
RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from fresh leaves or other tissues and organs (roots, stems, bracts, petals, stamens and stigmas and fibers of different development stages) using a TRIzol® Plus RNA Purification Kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA) and treated extensively with RNase-free DNase I. Double-stranded cDNA was synthesized from 100 ng RNA using an iScriptTM cDNA Synthesis Kit (Quanta BioSciences, Suite 1 Gaithersburg, MD, USA) according to a standard double-stranded cDNA synthesis protocol and then diluted 100-fold for qRT-PCR. The qRT-PCR included 5 μl cDNA template (diluted 1/100), 5 μl primers (2.4 μM) and 10 μl SYBR green mixture (GoTaq qPCR Master Mix, Promega, Madison, WI, USA) and was performed in an Eppendorf real-time PCR instrument (Mastercycler ep realplex, Hamburg, Germany). The incubation conditions were 95 °C for 1 min, followed by 40 cycles at 95 °C for 15 s, 60 °C for 30 s, 95 °C for 15 s and 60 °C for 15 s. The specificity of the amplified PCR product was determined by melting curve analysis. The primers for target genes were designed using Premier5 software (Premier Biosoft, Palo Alto, CA, USA) and are listed in Table S3 of the supplementary file.
The genes for the male sterility involved in JA biosynthesis, including AtOPR3, AtDAD1, AtMS35, AtGPAT1, AtAOS and AtAIF22,23,24,25,26 as well as genes for JA biosynthesis catalyzed by several enzymes, including lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC) and 12-oxo-phytodienoic acid reductase (OPR) in A. thaliana19,20,21 were used as references for the selection of homologous genes in cotton (Table S4). We obtained homologous genes in cotton by BLAST in NCBI (http://www.ncbi.nlm.nih.gov/) and CGP (Cotton Genome Project, Institute of Cotton Research of CAAS, http://cgp.genomics.org.cn/page/species/index.jsp). The expression levels of the homologous genes in cotton were analyzed in transgenic GhACNAT-silenced plants and in C312 plants.
The cotton Ubiquitin7 gene (GhUBQ7, Gen Bank accession number DQ116441) was used as an internal control for the assays. The expression levels of endogenous genes in cotton were obtained and standardized to the constitutive GhUBQ7 gene expression level. In each study, three independent experiments were conducted with three independent samples with the same phenotype per experiment.
JA Measurement
The new leaves and the whole opening flowers of C312 and transgenic GhACNAT-silenced plants were collected and immediately frozen in liquid nitrogen, then stored at −80 °C. Based on the methods of JA extraction58,59,60, samples (approximately 150 mg) were ground and extracted with 1 ml pure cold methanol and stirred overnight at 4 °C in the dark. The suspension was centrifuged at 13.8 g for 15 min at 4 °C and 900 μl of the supernatant was carefully transferred to a new 1.5-ml tube. The pellet was mixed with 1 ml pure cold methanol, shaken for 4 h at 4 °C and centrifuged. The two supernatants were combined and dried by a flow of nitrogen gas at room temperature. The residue was dissolved in 200 μl pure methanol. Before GC-MS, derivatization was necessary for JA. The formation of MeJA using trimethylsilyldiazomethane was performed according to the methods in Schmelz et al., (2004)58. The GC-MS settings have been described58,61,62,63 and were used with little modification.
An Agilent 7890 A gas chromatograph (Santa Clara, CA, USA) equipped with a split/splitless injector (splitless mode, 250 °C, injection volume 1 μl) was interfaced to an Agilent 5973C mass spectrometer (Santa Clara, CA, USA) operated in chemical ionization (CI) mode with isobutene as the ionization gas. Compounds were separated on an HP-5 MS (30 m × 0.25 mm × 0.25 μm) column held at 50 °C for 1 min after injection and then the temperature was programmed at 15 °C (1 min) to 250 °C (10 min), with helium as the carrier gas (1.2 ml /min) (Fig S3).
The transgenic GhACNAT-silenced plant treated by exogenous MeJA
The anther indehiscence phenotype of the GhACNAT-silenced plants was rescued by exogenous MeJA. The buds of 3 to 5 days-before-opening in transgenic GhACNAT-silenced plants were sprayed with methyl jasmonate (MeJA solution). Different gradient concentrations with 100 μM, 500 μM, 1000 μM dissolved in 0.05% (v/v) Tween-20 were sprayed on buds and leaves according to the description22,23,24. On the opening day of the flower, the productive organs were investigated and analyzed.
Additional Information
How to cite this article: Fu, W. et al. Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci. Rep. 5, 11790; doi: 10.1038/srep11790 (2015).
References
Wilkins, T. A., Mishra, R. & Trolinder, N. L. Agrobacterium-mediated transformation and regeneration of cotton. J. Food Agric. Environ. 2, 179–197 (2004).
Wang, K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103 (2012).
Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427 (2012).
Li, F. et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572 (2014).
Lange, M., Aravinda, L., Yellina, Svetlana, O. & Annette, B. Virus-Induced Gene Silencing (VIGS) in plants: an overview of target apecies and the virus-derived vector systems. Methods in Mol. Biol. 975, 1–14 (2013).
Baulcombe, D. Viruses and gene silencing in plants. Arch. Virol. Supp l15, 189–201 (1999).
Urkayastha, A. & Dasgupta, I. Virus induced gene silencing: a versatile tool for discovery of gene functions in plants. Plant Physiol. Biochem. 47, 967–976 (2009).
Tao, X. & Zhou, X. A modified viral satellite DNA that suppresses gene expression in plants. Plant J. 38, 850–60 (2004).
Burch-Smith, T. M., Anderson, J. C., Martin, G. B. & Dinesh-Kumar, S. P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746 (2004).
Pang, J. H. et al. Development of Agrobacterium-mediated virus-induced gene silencing and performance evaluation of four marker genes in Gossypium barbadense. PLoS ONE 8, e73211 (2013).
Tuttle, J. R., Idris, A. M., Brown, J. K., Haigler, C. H. & Robertson, D. Geminivirus-mediated gene silencing from Cotton leaf crumple virus is enhanced by low temperature in cotton. Plant Physiol. 148, 41–50 (2008).
Fauquet, C. M. et al. Geminivirus strain demarcation and nomenclature. Arch.Virol. 153, 783–821 (2008).
Jeske, H. Geminiviruses. Curr. Top. Microbiol. Immunol. 331, 185–226 (2009).
Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 51, 357–81 (2013).
Gu, Z. H., Huang, C. G., Li, F. F. & Zhou, X. P. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649 (2014).
Harwood, J. L. Recent advances in the biosynthesis of plant fatty acids. Biochim. Biophys. Acta. 1301, 7–56 (1996).
Alexandra, T. et al. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis Acyl-lipid metabolism. Plant Cell 24, 5007–5023 (2012).
Li, B. Y. et al. Acyl-lipid metabolism. Arabidopsis Book 8, e0133 (2013).
Baud, S. & Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 49, 235–249 (2010).
Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. (Lond.) 100, 681–697 (2007).
Kunst, L. & Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727 (2009).
Creelman, R. A. & Mullet, J. E. Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol. 48, 355–381 (1997).
Mueller, M. J. Enzymes involved in jasmonic acid biosynthesis. Phys. Plantarum. 100, 653–663 (1997).
Schaller, F. Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J. Exp. Bot. 52, 11–23 (2001).
Stintzi, A. & Browse, J. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U S A. 97, 10625–10630 (2000).
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I. & Okada, K. The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence and flower opening in Arabidopsis. Plant Cell 13, 2191–2209 (2001).
Shih, C. F., Hsu, W. H., Peng, Y. J. & Yang, C. H. The NAC-like gene ANTHER INDEHISCENCE FACTOR acts as a repressor that controls anther dehiscence by regulating genes in the jasmonate biosynthesis pathway in Arabidopsis. J. Exp. Bot. 65, 621–639 (2014).
Wilson, Z. A., Song, J., Taylor, B. & Yang, C. Y. The final split: the regulation of anther dehiscence. J. Exp. Bot. 62, 1633–1649 (2011).
Dawson, J. et al. Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 144, 213–222 (1999).
Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56, 393–434 (2005).
Mariani, C., Beuckeleer, M. D., Truettner, J., Leemans, J. & Goldberg, R. B. Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347, 737–741 (1990).
Mariani, C. et al. A chimaeric ribonuclease inhibitor gene restores fertility to male sterile plants. Nature 357, 384–387 (1992).
Shi, Y.l., Guo, S. D., Zhang, R., Meng, Z. G. & Ren, M. Z. The role of somatic embryogenesis receptor-like kinase 1 in controlling pollen production of the Gossypium anther. Mol. Biol. Rep. 41, 411–422 (2014).
Zheng, Z. F. et al. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell. 15, 1872–1887 (2003).
Scott, R. J., Spielman, M. & Dickinson, H. G. Stamen structure and function. Plant Cell Suppl 16, S46–S60 (2004).
Schmid, M. et al. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–6 (2005).
Obulareddy, N., Panchal, S. & Melotto, M. Guard cell purification and RNA isolation suitable for high-throughput transcriptional analysis of cell-type responses to biotic stresses. Mol. Plant-Microbe In. 26, 844–9 (2013).
Borges, F. et al. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148, 1168–81 (2008).
Heyndrickx, K. S. & Vandepoele, K. Systematic identification of functional plant modules through the integration of complementary data sources. Plant Physiol. 159, 884–901 (2012).
Gao, X. Q. et al. Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305 (2011).
Roughan, P. G. & Slack, C. R. Cellular-Organization of Glycerolipid Metabolism. Annu. Rev. Plant Phys. 33, 97–132 (1982).
Post-Beittenmiller, D., Roughan, G. & Ohlrogge, J. B. Regulation of plant fatty acid biosynthesis: analysis of acyl-coenzyme A and acyl-acyl carrier protein substrate pools in spinach and pea chloroplasts. Plant Physiol. 100, 923–930 (1992).
Ohlrogge, J. & Browse, J. Lipid biosynthesis. Plant Cell 7, 957–970 (1995).
Johnston, M. L., Luethy, M. H., Miernyk, J. A. & Randall, D. D. Cloning and molecular analyses of the Arabidopsis thaliana plastid pyruvate dehydrogenase subunits. Biochim. Biophys. Acta, Bioenerg. 1321, 200–206 (1997).
Konishi, T., Shinohara, K., Yamada, K. & Sasaki Y. Acetyl-CoA carboxylase in higher plants: Most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol. 37, 117–122 (1996).
Mou, Z. L., He, Y. K., Dai, Y., Liu, X. F. & Li, J. Y. Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–417 (2000).
Kjellberg, J. M., Trimborn, M., Andersson, M. & Sandelius, A. S. Acyl-CoA dependent acylation of phospholipids in the chloroplast envelope. Biochim. Biophys. Acta. 1485, 100–110 (2000).
Bates, P. D., Ohlrogge, J. B. & Pollard, M. Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J. Biol. Chem. 282, 31206–31216 (2007).
Browse, J., Warwick, N., Somerville, C. R. & Slack, C. R. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the “16:3” plant Arabidopsis thaliana. Biochem. J. 235, 25–31 (1986b).
Feussner, I. & Wasternack, C. The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297 (2002).
Sperling, P. & Heinz, E. Isomeric sn-1-octadecenyl and sn-2-octadecenyl analogs of lysophosphatidylcholine as substrates for acylation and desaturation by plant microsomal-membranes. Eur. J. Biochem. 213, 965–971 (1993).
Okuley, J. et al. Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6, 147–158 (1994).
Browse, J., McConn, M., James, D. J. & Miquel, M. Mutants of Arabidopsis deficient in the synthesis of alpha-linolenate: Biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Bio. Chem. 268, 16345–16351 (1993).
Rotenberg, D., Thompson, T. S., German, T. L. & Willis, D. K. Methods for effective real-time RT-PCR analysis of virus-induced gene silencing. J. Virol. Methods. 138, 49–59 (2006).
Kumar, S. M. et al. A systematic study to determine the extent of gene silencing in Nicotiana benthamiana and other Solanaceae species when heterologous gene sequences are used for virus-induced gene silencing. New Phytol. 176, 782–791 (2007).
Huang, C. J., Xie, Y. & Zhou, X. P. Efficient virus-induced gene silencing in plants using a modified geminivirus DNA1 component. Plant Biotechnol. J. 7, 254–265 (2009).
Permingeat, H. R., Romagnoli, M. V. & Vallejos, R. H. A simple method for isolating high yield and quality DNA from cotton (Gossypium hirsutum L.) leaves. Plant Mol. Biol. Rep. 16, 1–6 (1998).
Hawkins, J. S., Pleasants, J. & Wendel, J. F. Identification of AFLP markers that discriminate between cultivated cotton and the Hawaiian island endemic, Gossypium tomentosum Nuttall ex Seeman. Genet. Resour. Crop Evol. 52, 1069–1078 (2005).
Schmelzl, E. A., Engelberth, J., James, H. T., Block, A. & Alborn, H. T. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 39, 790–808 (2004).
Sun, L. Q. et al. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5, 5372 (2014).
Liu, H. B., Lim, X. H., Xiao, J. H. & Wang, S. P. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods 8, 2 (2012).
Engelberth, J. et al. Simultaneous quantification of jasmonic acid and salicylic acid in plants by vapor phase extraction and gas chromatography-chemical ionization-mass spectrometry. Anal. Biochem. 312, 242–250 (2003).
Li, C. Y. et al. Role of β-Oxidation in Jasmonate biosynthesis and systemic wound signaling in tomato. Plant Cell 17, 971–986 (2005).
Acknowledgements
The authors would like to thank Professor Zhenming Pei of Duke University for numerous helpful discussions and comments. This work was supported by Zhejiang Provincial Natural Science Foundation of China under Grant No. LR14C130001, practically funded by the National Natural Science Foundation of China under Grant No. 31200210, the State Key Laboratory of Cotton Biology Open Fund (CB2015A02) and Transgenic Major Projects- New Cotton Varieties of High Yield by Transgenic Breeding (2014ZX08005-001).
Author information
Authors and Affiliations
Contributions
Y.S. and C.X. conceived and designed the experiments. W.F., Y.S., J.H., Y.C., K.H., X.C. and B.L. performed the research and prepared the figures 1, 2, 3, 4, 5, 6, 7 and figure 9. J.W. and L.K. participated in data analysis and prepared figure 10. J.S. and M.F. performed GC-MS analysis and prepared the figure 8. Y.S., C.X. and X.Z. wrote and corrected the article. All authors reviewed the manuscript.
Phylogenetic analysis of the ACNAT family.
(a) GhACNAT had high homology with ACNAT in different species. (b) The phylogenetic cladogram of the ACNAT superfamily in different species. Pp, Prunus persica (XP_007222936.1); Tc1, Theobroma cacao1 (XP_007022847.1); Tc2, Theobroma cacao2 (XP_007022848.1); Tc3, Theobroma cacao3 (XP_007022849.1); Pm, Prunus mume (XP_008218653.1); Vv, Vitis vinifera (XP_002268159.2); Pt, Populus trichocarpa (XP_006385079.1); Es, Eutrema salsugineum (XP_006402222.1); Cr, Capsella rubella (XP_006294555.1); Br, Brassica rapa (XP_009134028.1); Nt, Nicotiana tomentosiformis (XP_009618447.1); Pv, Phaseolus vulgaris (XP_007153914.1); Os, Oryza sativa Japonica Group (NP_001052881.1); Hv, Hordeum vulgare subsp. vulgare (BAJ94372.1); Rc, Ricinus communis (XP_002527856.1); Al, Arabidopsis lyrata subsp. Lyrata (XP_002880496.1); At, Arabidopsis thaliana (At2g23390.1); Gh, G. hirsutum L.
The GhACNAT gene expression pattern in the cotton variety C312.
R: root; S: stem; L: leaves, B: bract, P: petals, St: stigmas and stamens and fibers of different development stages (0-, 5-, 10-, 15-, 20, 25-DPA) in C312. (DPA: days post anthesis). The values are the means±s.ds. for three biological replicates. The asterisks indicate statistically significant differences between the transgenic and WT plants (*P < 0.05, **P < 0.01, Student’s t-test).
The morphological changes and GhACNAT expression analysis in transgenic GhACNAT-silenced plants and C312 plants.
(a-d) the stamens and stigmas of C312 (a) and transgenic GhACNAT-silenced plants (b, c, d); (e-i) bolls of C312 (e) and transgenic GhACNAT-silenced plants pollinated with C312 pollen (f, h, i); (g) fibers and seeds of C312 and transgenic GhACNAT-silenced plants from left to right. (j) Relative expression levels of GhACNAT measured by qRT-PCR in leaves of GhACNAT-silenced plants (lines 36-2, 36-5, 36-8, 36-10, 36-11, 36-12, 36-16, 36-21 and 36-23) and in C312 plants. The Ubiquitin7 gene (GhUBQ7) was used as an internal control. The values are the means for three replicates (samples of each line collected on days 20, 35 and 50 post-infection). The asterisks indicate statistically significant differences between the transgenic GhACNAT-silenced and WT C312 plants (Significant *P < 0.05, highly significant **P < 0.01, Student’s t-test).
Phenotypic analysis of anther and pollen in WT C312 plants, transgenic GhACNAT-silenced plants and male sterile ms1 plants.
(a) comparison of anther and pollen in C312, transgenic GhACNAT-silenced plants and ms1 plants (from left to right); (b) pollen releasing from the anther in C312; (c) anther of C312 plant dehiscent and full of pollen grains; (d) indehiscent anther of the sterile transgenic GhACNAT-silenced plant, indehiscent and full of pollen grains; (e) 99% of the active pollen of WT C312 was stained with KI-I2; (f, h) the pollen of ms1 plants with no activity stained with KI-I2; (g) 95% of the active pollen of the transgenic GhACNAT-silenced plants were stained with KI-I2.
The relative expression of genes related to anther development and JA biosynthesis by qRT-PCR in transgenic GhACNAT-silenced plants and in C312 as a control (CK).
The Ubiquitin7 gene (GhUBQ7) was used as an internal control. The values are the means for three replicates (one flower for each sample of each line). The asterisks indicate statistically significant differences between the transgenic GhACNAT-silenced and the WT plants (*P < 0.05, **P < 0.01, Student’s t-test).
The relative expression of genes participating in the lipid biosynthesis pathway.
(a) The relative expression of the main genes for fatty acid synthesis; (b) the relative expression of genes for glycerolipid biosynthesis. The values are the means for three replicates (one flower for each sample of each line). The asterisks indicate statistically significant differences between the transgenic GhACNAT-silenced and the WT plants (*P < 0.05, **P < 0.01, Student’s t-test).
The morphologies of stamens and stigmas in C312 and transgenic GhACNAT-silenced plants treated with different concentrations of MeJA.
(a) normal stamens and stigma in C312 plants; (b) stamens and stigma in transgenic GhACNAT-silenced plants treated with 100 μM MeJA; (c) stamens and stigma in transgenic GhACNAT-silenced plants treated with 500 μM MeJA; (d) stamens and stigma in transgenic GhACNAT-silenced plants treated with 1000 μM MeJA.
A simple schematic diagram of fatty acid and jasmonic acid biosynthesis.
PEP, phosphoenolpyruvate; PKp, plastidial pyruvate kinase; PDH-E1a, pyruvate dehydrogenase; AcCoA, acetyl-CoA; ACP, acyl carrier protein; BCCP2, biotin carboxyl carrier protein2; MAT, malonyl-CoA: ACP transacylase; KAS, 3-ketoacyl-ACP synthase; KAR, 3-ketoacyl-ACP reducase; HD, 3-hydroxyacyl-ACP dehydratase; ENR, enoyl-ACP reductase; LCFA, long-chain fatty acid; LACS, long-chain acyl-CoA synthetase; FAT, fatty acyl-ACP thioesterase; GPAT, glycerol-3-phosphate acyltransferase; LPAAT, lysophosphaditic acid acyltransferase; LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase.
The relative expression of the GhACNAT gene in reproductive organs (a) and JA levels in C312 and transgenic GhACNAT-silenced plants in leaves (b) and in the whole flowers (c). The values are the means±s.ds for three replicates (three leaves for b and three flowers of each line for a and c). The asterisks indicate statistically significant differences between the transgenic GhACNAT-silenced and the WT plants (*P < 0.05, **P < 0.01, Student’s t-test).
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Fu, W., Shen, Y., Hao, J. et al. Acyl-CoA N-acyltransferase influences fertility by regulating lipid metabolism and jasmonic acid biogenesis in cotton. Sci Rep 5, 11790 (2015). https://doi.org/10.1038/srep11790
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep11790
This article is cited by
-
Heat-responsive microRNAs participate in regulating the pollen fertility stability of CMS-D2 restorer line under high-temperature stress
Biological Research (2023)
-
Cloning and functional analysis of GhDFR1, a key gene of flavonoid synthesis pathway in naturally colored cotton
Molecular Biology Reports (2023)
-
Sef1, rapid-cycling Brassica napus for large-scale functional genome research in a controlled environment
Theoretical and Applied Genetics (2023)
-
Dose effects of restorer gene modulate pollen fertility in cotton CMS-D2 restorer lines via auxin signaling and flavonoid biosynthesis
Plant Cell Reports (2023)
-
The cotton pectin methyl esterase gene GhPME21 functions in microspore development and fertility in Gossypium hirsutum L.
Plant Molecular Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.