Abstract
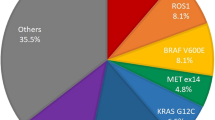
Identification of multi-gene variations has led to the development of new targeted therapies in lung adenocarcinoma patients and identification of an appropriate patient population with a reliable screening method is the key to the overall success of tumor targeted therapies. In this study, we used the Ion Torrent next-generation sequencing (NGS) technique to screen for mutations in 89 cases of lung adenocarcinoma metastatic lymph node specimens obtained by fine-needle aspiration cytology (FNAC). Of the 89 specimens, 30 (34%) were found to harbor epidermal growth factor receptor (EGFR) kinase domain mutations. Seven (8%) samples harbored KRAS mutations and three (3%) samples had BRAF mutations involving exon 11 (G469A) and exon 15 (V600E). Eight (9%) samples harbored PIK3CA mutations. One (1%) sample had a HRAS G12C mutation. Thirty-two (36%) samples (36%) harbored TP53 mutations. Other genes including APC, ATM, MET, PTPN11, GNAS, HRAS, RB1, SMAD4 and STK11 were found each in one case. Our study has demonstrated that NGS using the Ion Torrent technology is a useful tool for gene mutation screening in lung adenocarcinoma metastatic lymph node specimens obtained by FNAC and may promote the development of new targeted therapies in lung adenocarcinoma patients.
Similar content being viewed by others
Introduction
Lung cancer is the main cancer in the world today, whether considered in terms of numbers of cases or deaths, because of the high case fatality1,2,3. Non-small cell lung cancer (NSCLC) is the most common type of lung cancer and is often diagnosed at advanced stages of the disease. Lung adenocarcinoma is one of the most common subtypes in NSCLC. Conventional chemotherapeutic regimens have not significantly increased overall survival in the treatment of advanced NSCLC4. A growing body of research has focused on the association between specific genomic alterations and response of advanced-stage lung cancer to targeted therapies5. Small molecule inhibitors that target oncogenic tyrosine kinases have become an attractive treatment strategy in the therapy of several cancers6. Identification of epidermal growth factor receptor (EGFR) kinase domain activating mutations in NSCLC patients led to several retrospective studies to confirm the relationships between the EGFR mutational status and treatment response to kinase inhibitors such as erlotinib and gefitinib7,8,9,10. Recently, the identification of multiple genetic variations has led to the development of new targeted therapies in a subset of NSCLC patients11,12. Identification of an appropriate patient population with a reliable screening method is the key to the overall success of tumor targeted therapies.
Most patients with NSCLC are diagnosed at an advanced stage, when many of them have lost the opportunity of surgical treatment and have to be diagnosed by cytological specimens13. Currently, Sanger sequencing and real-time PCR have become the clinical methods used to detect gene mutations in diagnostic laboratories. However, they are expensive and time-consuming for a large number of gene detections, thus limiting their clinical applications. The recent commercial availability of the next-generation sequencing (NGS) technique has changed the way we think about scientific approaches in basic, applied and clinical research14,15,16. In this study, we used the Ion Torrent NGS technique to screen for gene mutations in lung adenocarcinoma metastatic lymph node specimens obtained by fine-needle aspiration cytology (FNAC).
Results
Performance of next-generation sequencing
The Ion Torrent platform was used to screen for gene mutations in 89 cases of lung adenocarcinoma metastatic lymph node specimens obtained by FNAC. Figure 1 depicts the procedure of mutation analysis assay using the Ion Torrent platform. Data obtained from the PGM runs were initially processed using Ion Torrent platform specific software and then the “variant caller” program, which is a plug-in of Torrent Suite Software, was performed for the initial variant calling of the Ion AmpliSeq sequencing data. To obtain results more accurately, the final data were filtered through several steps. In the first step, samples with a total target coverage of 100 x < 85% were removed. In the second step, false positive sites were removed under the following conditions: a mean total coverage depth >100, each variant coverage >20, a variant frequency of each sample >5 and P-value <0.01, not in intron and not strand-biased variants, by using the Integrative Genomics Viewer (IGV) software. Then, these variants were searched in normal database to distinguish between SNPs and mutations. Finally, mutations were searched in COSMIC database. The Sequence read distribution across 207 amplicons was normalized to 357,000 reads per sample and the mean number of reads of each amplicon was 1062 times.
EGFR mutation analysis in FNAC specimens with lung adenocarcinoma
Of the 89 samples, 30 (34%) were found to harbor EGFR kinase domain mutations and 3 (3%) were found to have multiple complex mutations (Table 1). Among them, 1 was G719A (c.2156G > C) missense point mutation in exon 18, 11 were deletions in exon 19, 4 were missense point mutation in exon 20 and 17 were L858R (c. 2573T > G) missense point mutation in exon 21. Of the 11 patients with the exon 19 deletion, 8 (73%) had the commonest 15-bp deletions (delE746_A750) and 3 (27%) had other deletions. Of the 4 patients with exon 20 mutations, 2 (50%) had T790M (c.2369C > T) mutation, 1 (25%) had S768I (c.2303G > T) mutation and 1 had A767V (c.2300C > T) mutation (Fig. 2).
Mutation spectra in KRAS, NRAS, BRAF and PIK3CA
Of the 89 samples, 7 (8%) harbored KRAS mutations at codon 12, including 3 cases of G12C (c.34G > T) mutation, 3 cases of G12A (c.35G > C) mutation and 1 case of G12D (c.35G > A) mutation (Fig. 3). Three (3%) samples had BRAF mutations involving exon 11 (G469A) and exon 15 (V600E). Eight (9%) samples harbored PIK3CA mutations, of which 5 occurred in exon 9 and 3 in exon 20. One (1%) sample had a HRAS G12C mutation.
Other mutations
Thirty-two (36%) samples harbored TP53 mutations, including 12 in exon 5, 3 in exon 6, 8 in exon 7, 5 in exon 8 and 5 in exon 10. Of the 32 TP53 mutation samples, 1 had multiple complex mutations. Other genes including APC, ATM, MET, PTPN11, GNAS, HRAS, RB1, SMAD4 and STK11 were found each in one case.
Discussion
In this study, we performed NGS to detect gene mutations in lung adenocarcinoma by using the Ion Torrent platform, demonstrating that the NGS technique had a promising potential use for multi-gene identification. The major advantage of NGS is its ability to produce an enormous volume of data less costly14.
The paraffin-embedded tissue has been considered optimal sample for molecular studies. Consequently, EGFR analysis in most published studies depended on biopsy specimens. However, biopsies are not always available, because the diagnosis of lung cancer sometimes depends on metastatic lymph node specimens obtained by FNAC through minimally invasive procedures in order to avoid more invasive techniques17,18,19 Therefore, the possibility of performing molecular methods on metastatic lymph node of FNAC samples is appealing20. Methods to detect EGFR or other gene mutations in specimens with an unfavorable tumor cell content need to be very sensitive21. The development of the NGS method represents one of the more significant technical advances in molecular biology14,22.
Some driver gene mutations are promising diagnostic and prognostic markers for lung adenocarcinomas and also represent potential therapeutic targets. Therefore, an accurate and reliable screening method for gene mutations is highly desired. To the best of our knowledge, the present study is a large series analyzing gene mutations in cytological samples. The Ion Torrent platform was performed to screen for mutations in 89 cases of lung adenocarcinoma metastatic lymph node specimens obtained by FNAC. Although the Sanger sequencing technique is the standard method to detect gene mutations, it requires a high tumor content and more starting material. NGS using the Ion Torrent technology only requires 10ng DNA for analysis. Our study has demonstrated that NGS using the Ion Torrent technology is a useful tool for gene mutation screening in lung adenocarcinoma and may promote the development of new targeted therapies in lung adenocarcinoma patients.
Methods
Patients
Data in this retrospective study were analyzed anonymously. No images and private information of the patients were released. The medical ethical committee of the Cancer Hospital of the Chinese Academy of Medical Sciences (CAMS), approved the study protocol and agreed to waive the need for consent by the patients. A total of 89 Chinese patients with lung adenocarcinoma were enrolled in this study and treated at the CAMS Cancer Hospital between July 2012 and June 2013. All the lymph node specimens in these patients were obtained by fine-needle aspiration cytology (FNAC), fixed in 10% neutral buffered formalin, embedded in paraffin and finally diagnosed as metastatic lung adenocarcinoma.
DNA preparation
DNA was extracted from paraffin-embedded tumor tissues using QIAamp® DNA Mini Kit (Qiagen, Germany), according to the manufacturer’s instructions. The quality and concentration of the DNA samples were examined by NanoDrop (Thermo).
Next-generation sequencing (NGS)
NGS was performed using the Ion Torrent platform (Life Technologies), according to the manufacturer’s specifications. An Ion Torrent adapter-ligated library was constructed with the Ion AmpliSeq Library Kit 2.0 (Life Technologies) following the manufacturer’s protocol. Briefly, 10 ng of pooled amplicons were end-repaired and DNA ligase was used to ligate Ion Torrent adapters P1 and A. The adapter-ligated products were purified with AMPure beads (Beckman Coulter) and the purified products were nick-translated and PCR-amplified for a total of 5 cycles. The resulting library was again purified with AMPure beads and the size and concentration of the library were determined by Agilent 2100 Bioanalyzer and Agilent Bioanalyzer DNA High-Sensitivity LabChip (Agilent Technologies).
Additional Information
How to cite this article: Qiu, T. et al. Next-generation sequencing for molecular diagnosis of lung adenocarcinoma specimens obtained by fine needle aspiration cytology. Sci. Rep. 5, 11317; doi: 10.1038/srep11317 (2015).
References
Parkin, D. M., Pisani, P. & Ferlay, J. Global cancer statistics. CA Cancer J Clin 49, 33–64 (1999).
Parkin, D. M., Bray, F., Ferlay, J. & Pisani, P. Global cancer statistics, 2002. CA Cancer J Clin 55, 74–108 (2005).
Jemal, A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011).
Schiller, J. H. et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med 346, 92–98 (2002).
Leighl, N. B. et al. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J Clin Oncol 32, 3673–3679 (2014).
Kancha, R. K., von Bubnoff, N., Peschel, C. & Duyster, J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res 15, 460–467 (2009).
Shepherd, F. A. et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 353, 123–132 (2005).
Natale, R. B. et al. Phase III trial of vandetanib compared with erlotinib in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol 29, 1059–1066 (2011).
Maemondo, M. et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 362, 2380–2388 (2010).
Peng, L., Song, Z. G. & Jiao, S. C. Efficacy analysis of tyrosine kinase inhibitors on rare non-small cell lung cancer patients harboring complex EGFR mutations. Sci Rep 4, 6104 (2014).
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350, 2129–2139 (2004).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Fassina, A. et al. Fine needle aspiration of non-small cell lung cancer: current state and future perspective. Cytopathology 23, 213–219 (2012).
Metzker, M. L. Sequencing technologies - the next generation. Nat Rev Genet 11, 31–46 (2010).
Han, J. Y. et al. Comparison of targeted next-generation sequencing with conventional sequencing for predicting the responsiveness to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy in never-smokers with lung adenocarcinoma. Lung Cancer 85, 161–167 (2014).
Ross, J. S. et al. Next-generation sequencing reveals frequent consistent genomic alterations in small cell undifferentiated lung cancer. J Clin Pathol 67, 772–776 (2014).
Lozano, M. D. et al. Assessment of epidermal growth factor receptor and K-ras mutation status in cytological stained smears of non-small cell lung cancer patients: correlation with clinical outcomes. Oncologist 16, 877–885 (2011).
Stigt, J. A. et al. Comparison of EUS-guided fine needle aspiration and integrated PET-CT in restaging after treatment for locally advanced non-small cell lung cancer. Lung Cancer 66, 198–204 (2009).
Gu, P. et al. Endobronchial ultrasound-guided transbronchial needle aspiration for staging of lung cancer: a systematic review and meta-analysis. Eur J Cancer 45, 1389–1396 (2009).
Gazdar, A. F. Personalized medicine and inhibition of EGFR signaling in lung cancer. N Engl J Med 361, 1018–1020 (2009).
Ellison, G. et al. EGFR mutation testing in lung cancer: a review of available methods and their use for analysis of tumour tissue and cytology samples. J Clin Pathol 66, 79–89 (2013).
Kanagal-Shamanna, R. et al. Next-generation sequencing-based multi-gene mutation profiling of solid tumors using fine needle aspiration samples: promises and challenges for routine clinical diagnostics. Mod Pathol 27, 314–327 (2014).
Acknowledgements
We thank San Valley Biotechnology inc. for technical support. This work was supported by a grant from Capital Clinical Characteristic of Applied Research Program (to Zhihui Zhang) of Cancer Hospital, CAMS (Grant number Z121107001012084).
Author information
Authors and Affiliations
Contributions
Z.Z., L.W. and T.Q. designed experiments, Z.Z., L.W. and T.Q. conducted experiments and data analysis and wrote the paper. H.G. and H.Z. performed the pathological diagnosis and performed the experiments. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Qiu, T., Guo, H., Zhao, H. et al. Next-generation sequencing for molecular diagnosis of lung adenocarcinoma specimens obtained by fine needle aspiration cytology. Sci Rep 5, 11317 (2015). https://doi.org/10.1038/srep11317
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep11317
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.